Thirty Minutes of Hypobaric Hypoxia Provokes Alterations of Immune Response, Haemostasis, and Metabolism Proteins in Human Serum
Abstract
:1. Introduction
2. Results
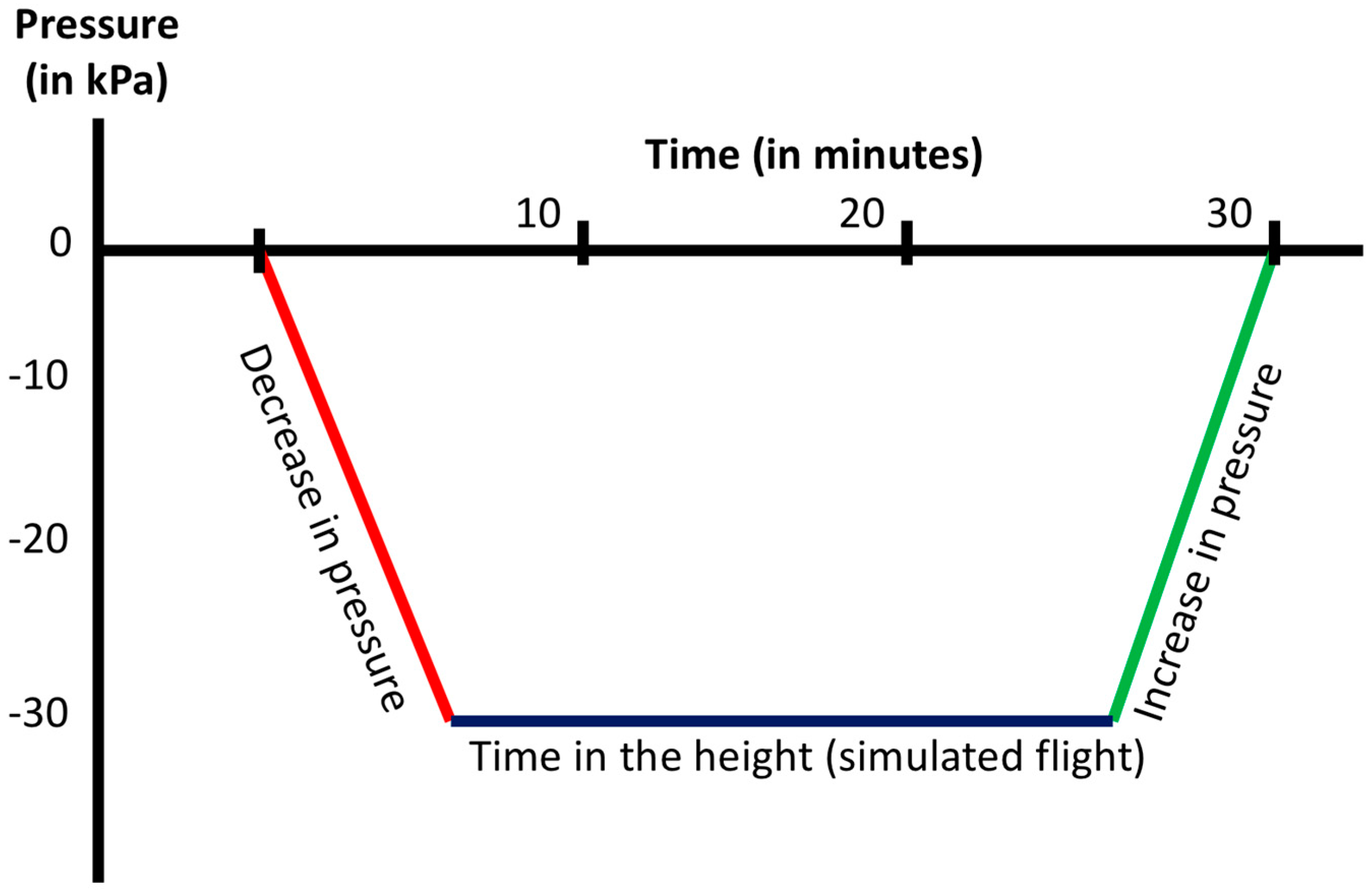
2.1. Atmospheric Profile
2.2. Participants
2.3. Proteome Profiles in 2-DIGE
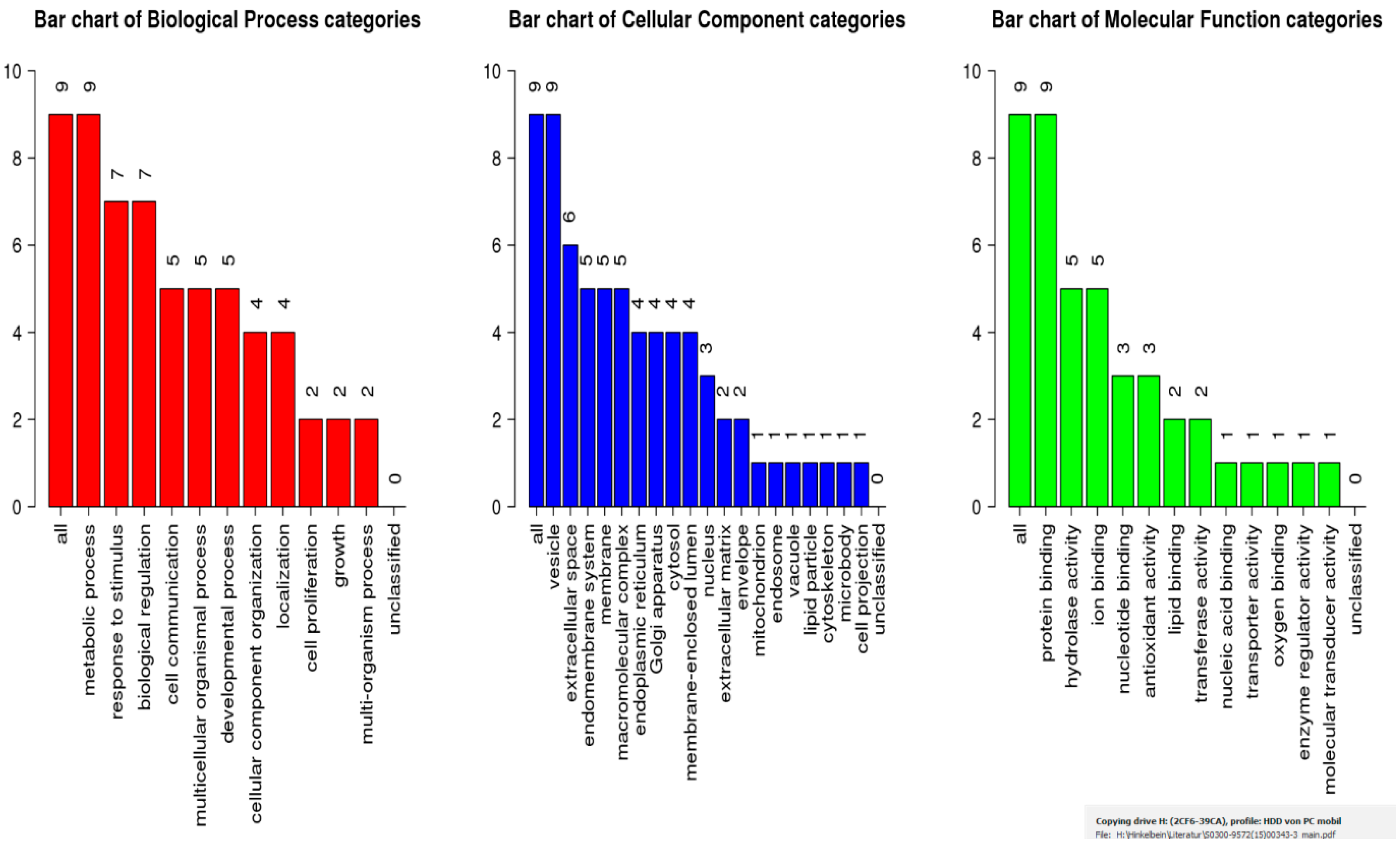
2.4. Bioinformatic Analysis
2.5. Heat Map
3. Discussion
3.1. Hypoxia Effects
3.2. Proteomics
3.3. Proteomics in High Altitude Research
3.4. Hypoxia Protein Expression in the Present Study
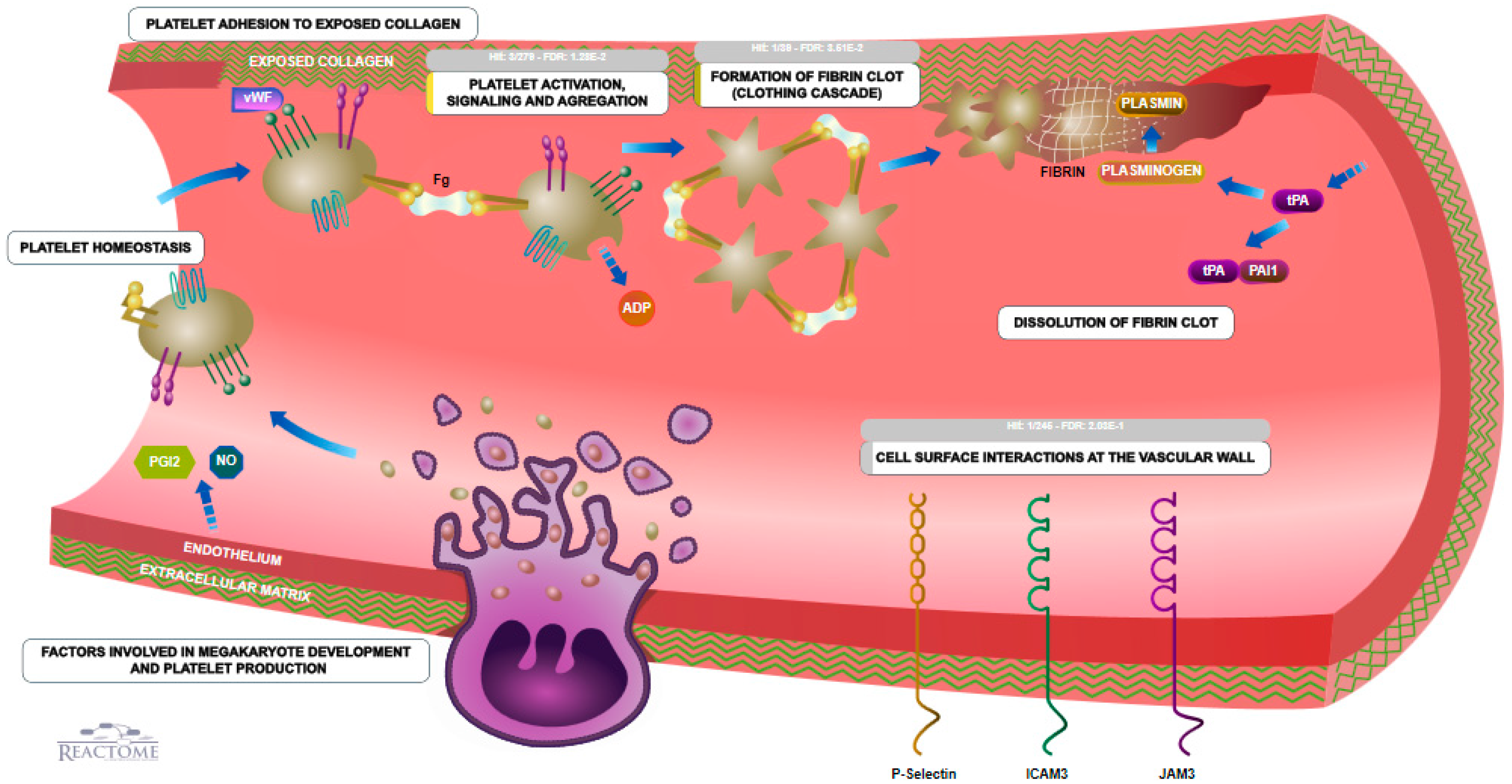
3.5. Coagulation
3.6. Biological Pathways
3.7. Limitations
4. Materials and Methods
4.1. Inclusion and Exclusion Criteria
4.2. Simulated Flight in the Pressure Chamber
4.3. Collection and Isolation of Serum Samples
4.4. Depletion of High-Abundant Proteins
4.5. Sample Preparation for 2DE Electrophoresis
4.6. Two-Dimensional Gel Electrophoresis (First and Second Dimension)
4.7. Identification of Relevant Protein Spots
4.8. MS Identification of Proteins
4.9. Bioinformatic Analysis of Proteins
4.10. Hierarchical Cluster Analysis
4.11. Protein Interaction and Network Identification
4.12. Reactome
4.13. KEGG
4.14. WebGestalt
5. Conclusions
Author Contributions
Conflicts of Interest
References
- Ahmad, Y.; Sharma, N.K.; Ahmad, M.F.; Sharma, M.; Garg, I.; Bhargava, K. Proteomic identification of novel differentiation plasma protein markers in hypobaric hypoxia-induced rat model. PLoS ONE 2014, 9, e98027. [Google Scholar]
- Ahmad, Y.; Sharma, N.K.; Ahmad, M.F.; Sharma, M.; Garg, I.; Bhargava, K. An insight into the changes in human plasma proteome on adaptation to hypobaric hypoxia. PLoS ONE 2013, 8, e67548. [Google Scholar]
- Jessen, T.H.; Weber, R.E.; Fermi, G.; Tame, J.; Braunitzer, G. Adaptation of bird hemoglobins to high altitudes: Demonstration of molecular mechanism by protein engineering. Proc. Natl. Acad. Sci. USA 1991, 88, 6519–6522. [Google Scholar] [CrossRef] [PubMed]
- Storz, J.F.; Runck, A.M.; Sabatino, S.J.; Kelly, J.K.; Ferrand, N.; Moriyama, H.; Weber, R.E.; Fagoe, A. Evolutionary and functional insights into the mechanism underlying high-altitude adaptation of deer mouse hemoglobin. Proc. Natl. Acad. Sci. USA 2009, 106, 14450–14455. [Google Scholar] [CrossRef] [PubMed]
- Padhy, G.; Sethy, N.K.; Ganju, L.; Bhargava, K. Abundance of plasma antioxidant proteins confers tolerance to acute hypobaric hypoxia exposure. High Alt. Med. Biol. 2013, 14, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Fountoulakis, M. Proteomics: Current technologies and applications in neurological disorders and toxicology. Amino Acids 2001, 21, 363–381. [Google Scholar] [CrossRef] [PubMed]
- Seillier-Moiseiwitsch, F.; Trost, D.C.; Moiseiwitsch, J. Statistical methods for proteomics. Methods Mol. Biol. 2002, 184, 51–80. [Google Scholar]
- Kim, H.; Eliuk, S.; Deshane, J.; Meleth, S.; Sanderson, T.; Pinner, A.; Robinson, G.; Wilson, L.; Kirk, M.; Barnes, S. Biological Aging; Humana Press Inc.: Clifton, NJ, USA, 2005; pp. 349–391. [Google Scholar]
- Wilkins, M.R.; Pasquali, C.; Appel, R.D.; Ou, K.; Golaz, O.; Sanchez, J.C.; Yan, J.X.; Gooley, A.A.; Hughes, G.; Humphery, S.I.; et al. From proteins to proteomes: Large scale protein identification by two-dimensional electrophoresis and amino acid analysis. Biotechnology 1996, 14, 61–65. [Google Scholar] [CrossRef] [PubMed]
- Kelder, T.; Conklin, B.R.; Evelo, C.T.; Pico, A.R. Finding the right questions: Exploratory pathway analysis to enhance biological discovery in large datasets. PLoS ONE 2010, 8, e1000472. [Google Scholar] [CrossRef] [PubMed]
- Aerospace Medical Association. Cabin cruising altitudes for regular transport aircraft. Aviat. Space Environ. Med. 2008, 79, 433–439. [Google Scholar]
- Meyer, M.F.; Mikolajczak, S.; Luers, J.C.; Lotfipour, S.; Beutner, D.; Jumah, M.D. Characterizing the passive opening of the eustachian tube in a hypo-/hyperbaric pressure chamber. Laryngorhinootologie 2013, 92, 600–606. [Google Scholar] [PubMed]
- Mikolajczak, S. Characterizing the active opening of the eustachian tube in a hypo-/hyperbaric pressure chamber. Otol. Neurotol. 2014, 36, 70–75. [Google Scholar]
- Schumacker, P.T.; Chandel, N.; Agusti, A.G. Oxygen conformance of cellular respiration in hepatocytes. Am. J. Physiol. 1993, 265, L395–L402. [Google Scholar] [PubMed]
- Langen, H.; Röder, D.; Juranville, J.F.; Fountoulakis, M. Effect of protein application mode and acrylamide concentration on the resolution of protein spots separated by two-dimensional gel electrophoresis. Electrophoresis 1997, 18, 2085–2090. [Google Scholar] [CrossRef] [PubMed]
- Garbis, S.; Lubec, G.; Fountoulakis, M. Limitations of current proteomics technologies. J. Chromatogr. A 2005, 1077, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Futcher, B.; Latter, G.I.; Monardo, P.; Mclaughlin, C.S.; Garrels, J.I. A sampling of the yeast proteome. Mol. Cell Biol. 1999, 19, 7357–7368. [Google Scholar] [CrossRef] [PubMed]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry–based shotgun proteomics. Nat. Protocols 2016, 11, 2301–2319. [Google Scholar] [CrossRef] [PubMed]
- Godoy, L.D.; Olsen, J.V.; Cox, J.; Nielsen, M.L.; Hubner, N.C.; Walther, T.C.; Mann, M. Comprehensive massspectrometry- based proteome quantification of haploid versus diploid yeast. Nature 2011, 455, 1251–1254. [Google Scholar] [CrossRef] [PubMed]
- Picotti, P.; Bodenmiller, B.; Mueller, L.N.; Domon, B.; Aebersold, R. Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell 2009, 138, 795–806. [Google Scholar] [CrossRef] [PubMed]
- Wasinger, V.C.; Cordwell, S.J.; Cerpam, P.A.; Wilkins, M.R.; Duncan, M.W.; Harris, R.; Williams, K.L.; Humphery, S.I. Progress with gene-product mapping of the Mollicutes: Mycoplasma genitalium. Electrophoresis 1995, 16, 1090–1094. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Luo, G.; Ni, B. Progress in mass spectrometry-based proteomics in hypoxia-related diseases and high-altitude medicine. OMICS 2017, 21, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Rigbolt, K.T.; Vanselow, J.T.; Blagoev, B. GProX, a user-friendly platform for bioinformatics analysis and visualization of quantitative proteomics data. Mol. Cell Proteom. 2011, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Jiang, B.H.; Rue, E.; Wang, G.L.; Roe, R.; Semenza, G.L. Dimerization, DNA binding, and transactivation properties of hypoxia-inducible factor 1. J. Biol. Chem. 1996, 271, 17771–17778. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.L.; Jiang, B.H.; Rue, E.A.; Semenza, G.L. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. USA 1995, 92, 5510–5514. [Google Scholar] [CrossRef] [PubMed]
- Krzeszowiak, J.; Zawadzki, M.; Markiewicz, G.I.; Kawalec, A.; Pawlas, K. The influence of 9-day trekking in the Alps on the level of oxidative stress parameters and blood parameters in native lowlanders. Ann. Agric. Environ. Med. 2014, 21, 585–589. [Google Scholar] [CrossRef] [PubMed]
- Wadley, A.J.; Svendsen, I.S.; Gleeson, M. Heightened Exercise-Induced Oxidative Stress at Simulated Moderate Level Altitude vs. Sea Level in Trained Cyclists. Int. J. Sport Nutr. Exerc. Metab. 2017, 27, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Jones, S. Clusterin. Int. J. Biochem. Cell Biol. 2002, 34, 427–431. [Google Scholar] [CrossRef]
- Troakes, C.; Smyth, R.; Noor, F.; Maekawa, S.; Killick, R.; King, A.; Al-Sarraj, S. Clusterin expression is upregulated following acute head injury and localizes to astrocytes in old head injury. Neuropathology 2017, 37, 12–24. [Google Scholar] [CrossRef] [PubMed]
- Koltai, T. Clusterin: A key player in cancer chemoresistance and its inhibition. Onco. Targets Ther. 2014, 7, 447–456. [Google Scholar] [CrossRef] [PubMed]
- Sansanwal, P.; Li, L.; Sarwal, M.M. Inhibition of intracellular clusterin attenuates cell death in nephropathic cystinosis. J. Am. Soc. Nephrol. 2015, 26, 612–625. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.C.; Tsai, P.; Sun, H.Y.; Hsu, M.C.; Lee, J.C.; Wu, I.C.; Tsao, C.W.; Chang, T.T.; Young, K.C. Apolipoprotein J, a glucose-upregulated molecular chaperone, stabilizes core and NS5A to promote infectious hepatitis C virus virion production. J. Hepatol. 2014, 61, 984–993. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Park, S.Y.; Shin, E.; Lee, S.H.; Kim, Y.S.; Lee, D.H.; Roh, G.S.; Kim, H.J.; Kang, S.S.; Cho, G.J.; et al. Hypoxia inducible factor-1α directly regulates nuclear clusterin transcription by interacting with hypoxia response elements in the clusterin promoter. Mol. Cells 2014, 37, 178–186. [Google Scholar] [CrossRef] [PubMed]
- Nizet, V.; Johnsonm, R.S. Interdependence of hypoxic and innate immune responses. Nat. Rev. Immunol. 2009, 9, 609–617. [Google Scholar] [CrossRef] [PubMed]
- Rolfs, A.; Kvietikova, I.; Gassmann, M.; Wenger, R.H. Oxygen-regulated transferrin expression is mediated by hypoxia-inducible factor-1. J. Biol. Chem. 1997, 272, 20055–20062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forsythe, J.A.; Jiang, B.H.; Iyer, N.V.; Agani, F.; Leung, S.W.; Koos, R.D.; Semenza, G.L. Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor 1. Mol. Cell Biol. 1996, 16, 4604–4613. [Google Scholar] [CrossRef] [PubMed]
- Iyer, N.V.; Kotch, L.E.; Agani, F.; Leung, S.W.; Laughner, E.; Wenger, R.H.; Gassmann, M.; Gearhart, J.D.; Lawler, A.M.; Yu, A.Y.; et al. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1α. Genes Dev. 1998, 12, 149–162. [Google Scholar] [CrossRef] [PubMed]
- Patel, N.; Gonsalves, C.S.; Malik, P.; Kalra, V.K. Placenta growth factor augments endothelin-1 and endothelin-B receptor expression via hypoxia-inducible factor-1α. Blood 2008, 112, 856–865. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, M.; Sakata, M.; Takeda, T.; Yamamoto, T.; Okamoto, Y.; Sawada, K.; Kimura, A.; Minekawa, R.; Tahara, M.; Tasaka, K.; et al. Induction of glucose transporter 1 expression through hypoxia-inducible factor 1α under hypoxic conditions in trophoblast-derived cells. J. Endocrinol. 2004, 183, 145–154. [Google Scholar] [CrossRef] [PubMed]
- León-Velarde, F.; Mejía, O. Gene expression in chronic high altitude diseases. High Alt. Med. Biol. 2008, 9, 130–139. [Google Scholar]
- Zhou, S.; Wu, H.; Zeng, C.; Xiong, X.; Tang, S.; Tang, Z.; Sun, X. Apolipoprotein E protects astrocytes from hypoxia and glutamate-induced apoptosis. FEBS Lett. 2013, 587, 254–258. [Google Scholar] [CrossRef] [PubMed]
- Sharma, N.K.; Sethy, N.K.; Bhargava, K. Comparative proteome analysis reveals differential regulation of glycolytic and antioxidant enzymes in cortex and hippocampus exposed to short-term hypobaric hypoxia. J. Proteom. 2013, 79, 277–298. [Google Scholar] [CrossRef] [PubMed]
- Arnaud, J.; Quilici, J.C.; Gutierrez, N.; Beard, J.; Vergnes, H. Methaemoglobin and erythrocyte reducing systems in high-altitude natives. Ann. Hum. Biol. 1979, 6, 585–592. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, T.; Ahmad, S.; Gupta, N.; Sahu, A.; Ahmad, Y.; Nair, V.; Chatterjee, T.; Bajaj, N.; Sengupta, S.; Ganju, L.; et al. Altered expression of platelet proteins and calpain activity mediate hypoxia-induced prothrombotic phenotype. Blood 2014, 123, 1250–1260. [Google Scholar] [CrossRef] [PubMed]
- Ninivaggi, M.; de Laat, M.; Lancé, M.M.D.; Kicken, C.H.; Pelkmans, L.; Bloemen, S.; Dirks, M.L.; van Loon, L.J.; Govers-Riemslag, J.W.; Lindhout, T.; et al. Hypoxia induces a prothrombotic state independently of the physical activity. PLoS ONE 2015, 10, e0141797. [Google Scholar] [CrossRef] [PubMed]
- Toff, W.D.; Jones, C.I.; Ford, I.; Pearse, R.J.; Watson, H.G.; Watt, S.J.; Ross, J.A.; Gradwell, D.P.; Batchelor, A.J.; Abrams, K.R.; et al. Effect of hypobaric hypoxia, simulating conditions during long-haul air travel, on coagulation, fibrinolysis, platelet function, and endothelial activation. JAMA 2006, 295, 2251–2261. [Google Scholar] [CrossRef] [PubMed]
- Schreijer, A.J.; Hoylaerts, M.F.; Meijers, J.C.M.; Lijnen, H.R.; Middeldorp, S.; Büller, H.R.; Reitsma, P.H.; Rosendaal, F.R.; Cannegieter, S.C. Explanations for coagulation activation after air travel. J. Thromb. Haemost. 2010, 8, 971–978. [Google Scholar] [CrossRef] [PubMed]
- Schobersberger, W.; Hoffmann, G.; Gunga, H.-C. Interaction of hypoxia and haemostasis—Hypoxia as a prothrombotic factor at high altitude? Wien. Med. Wochenschr. 2005, 155, 157–162. [Google Scholar] [CrossRef] [PubMed]
- Schaber, M.; Leichtfried, V.; Fries, D.; Wille, M.; Gatterer, H.; Faulhaber, M.; Würtinger, P.; Schobersberger, W. Influence of acute normobaric hypoxia on hemostasis in volunteers with and without acute mountain sickness. BioMed Res. Int. 2015, 2015, 593938–593939. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; Bhargava, K.; Ahmad, Y. The meta-analytical paradigm in an in silico hybrid: Pathways and networks perturbed during exposure to varying degrees of hypobaric hypoxia. Proteom. Clin. Appl. 2017, 11, 1600160. [Google Scholar] [CrossRef] [PubMed]
- Liao, W.T.; Liu, B.; Chen, J.; Cui, J.H.; Gao, Y.X.; Liu, F.Y.; Xu, G.; Sun, B.D.; Zhang, E.L.; Yuan, Z.B.; et al. Metabolite modulation in human plasma in the early phase of acclimatization to hypobaric hypoxia. Sci. Rep. 2016, 6, 22589. [Google Scholar] [CrossRef] [PubMed]
- Bendz, B.; Rostrup, M.; Sevre, K.; Andersen, T.O.; Sandset, P.M. Association between acute hypobaric hypoxia and activation of coagulation in human beings. Lancet 2000, 356, 1657–1658. [Google Scholar] [CrossRef]
- Mikolajczak, S.; Meyer, M.F.; Felsch, M.; Jumah, M.D.; Huttenbrink, K.B.; Grosheva, M.; Luers, J.C.; Beutner, D. Is there a difference between active opening of the Eustachian tube in a hypobaric surrounding compared to a hyperbaric surrounding? Undersea Hyperb Med. 2015, 42, 593–599. [Google Scholar] [PubMed]
- Klose, J.; Kobalz, U. Two-dimensional electrophoresis of proteins: An updated protocol and implications for a functional analysis of the genome. Electrophoresis 1995, 16, 1034–1059. [Google Scholar] [CrossRef] [PubMed]
- Key, M.A. Tutorial in displaying mass spectrometry-based proteomic data using heat maps. BMC Bioinformatics 2012, 13, S10. [Google Scholar] [CrossRef] [PubMed]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nature Methods 2016, 13, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010, 38, W214–W220. [Google Scholar]
- Mostafavi, S.; Ray, D.; Warde, F.D.; Grouios, C.; Morris, Q. GeneMANIA: A real-time multiple association network integration algorithm for predicting gene function. Genome Biol. 2008, 9, S4. [Google Scholar] [CrossRef] [PubMed]
- Joshi-Tope, G.; Gillespie, M.; Vastrik, I.; D’Eustachio, P.; Schmidt, E.; de Bono, B.; Jassal, B.; Gopinath, G.R.; Wu, G.R.; Matthews, L.; et al. Reactome: A knowledgebase of biological pathways. Nucleic Acids Res. 2005, 33, D428–D432. [Google Scholar]
- Wenger, R.H. Cellular adaptation to hypoxia: O2-sensing protein hydroxylases, hypoxia-inducible transcription factors, and O2-regulated gene expression. FASEB J. 2002, 16, 1151–1162. [Google Scholar] [CrossRef] [PubMed]
- Chandel, N.S.; Budinger, G.R.; Schumacker, P.T. Molecular oxygen modulates cytochrome c oxidase function. J. Biol. Chem. 1996, 271, 18672–18677. [Google Scholar] [CrossRef] [PubMed]

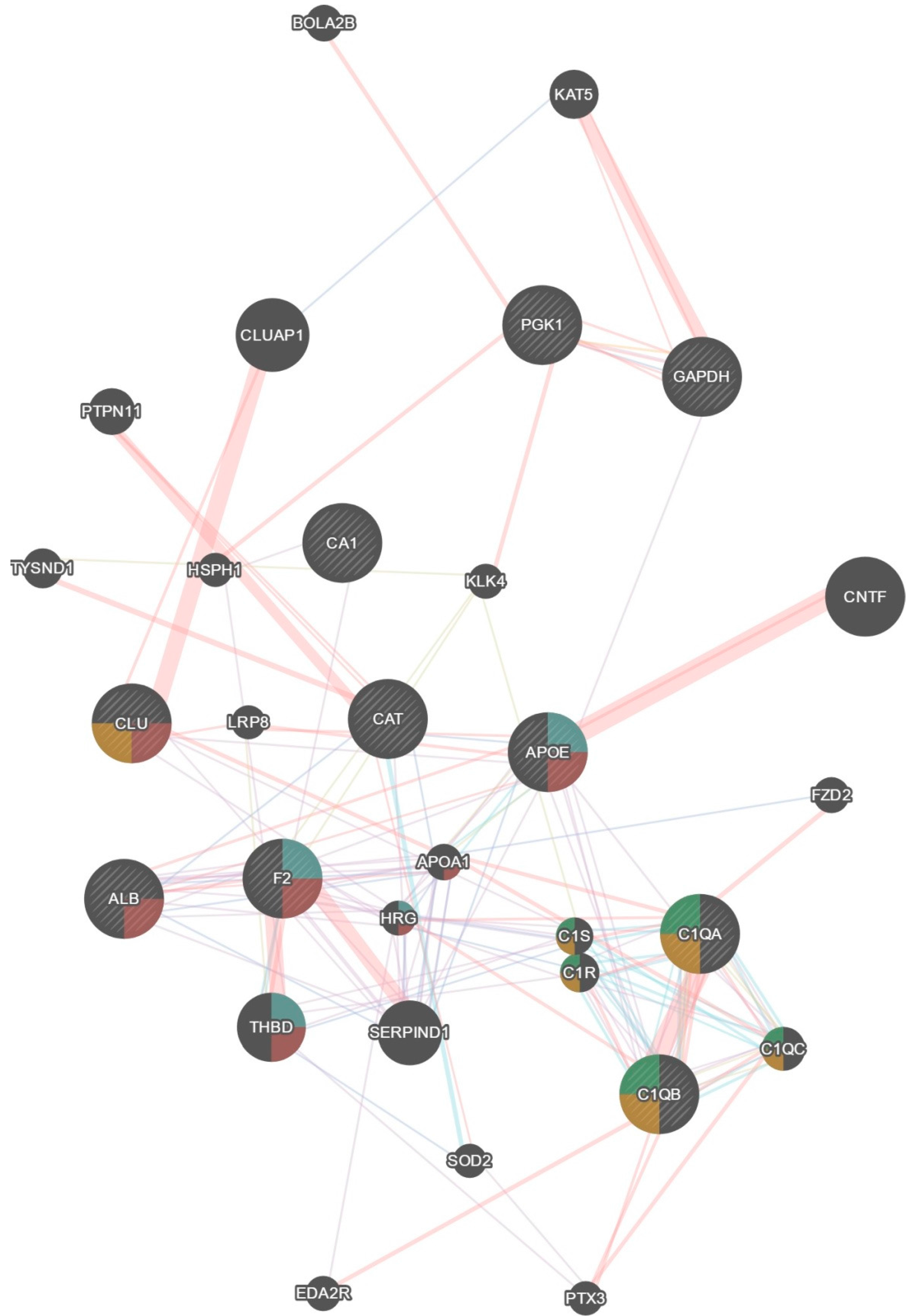
| Original Dataset Proteins | GeneMania Predicted Proteins |
|---|---|
| Pgk1, Cat, Apoe, Clu, F2, Alb, C1qa, C1qb, Ca1, Gapdh | Bola2b, Eda2r, Ptx3, Sod2, Serpind1, C1qc, C1r, C1s, Thbd, Hrg, Apoa1, Fzd2, Lrp8, Cntf, Klk4, Hsph1, Tysnd1, Ptpn11, Cluap1, Kat5 |
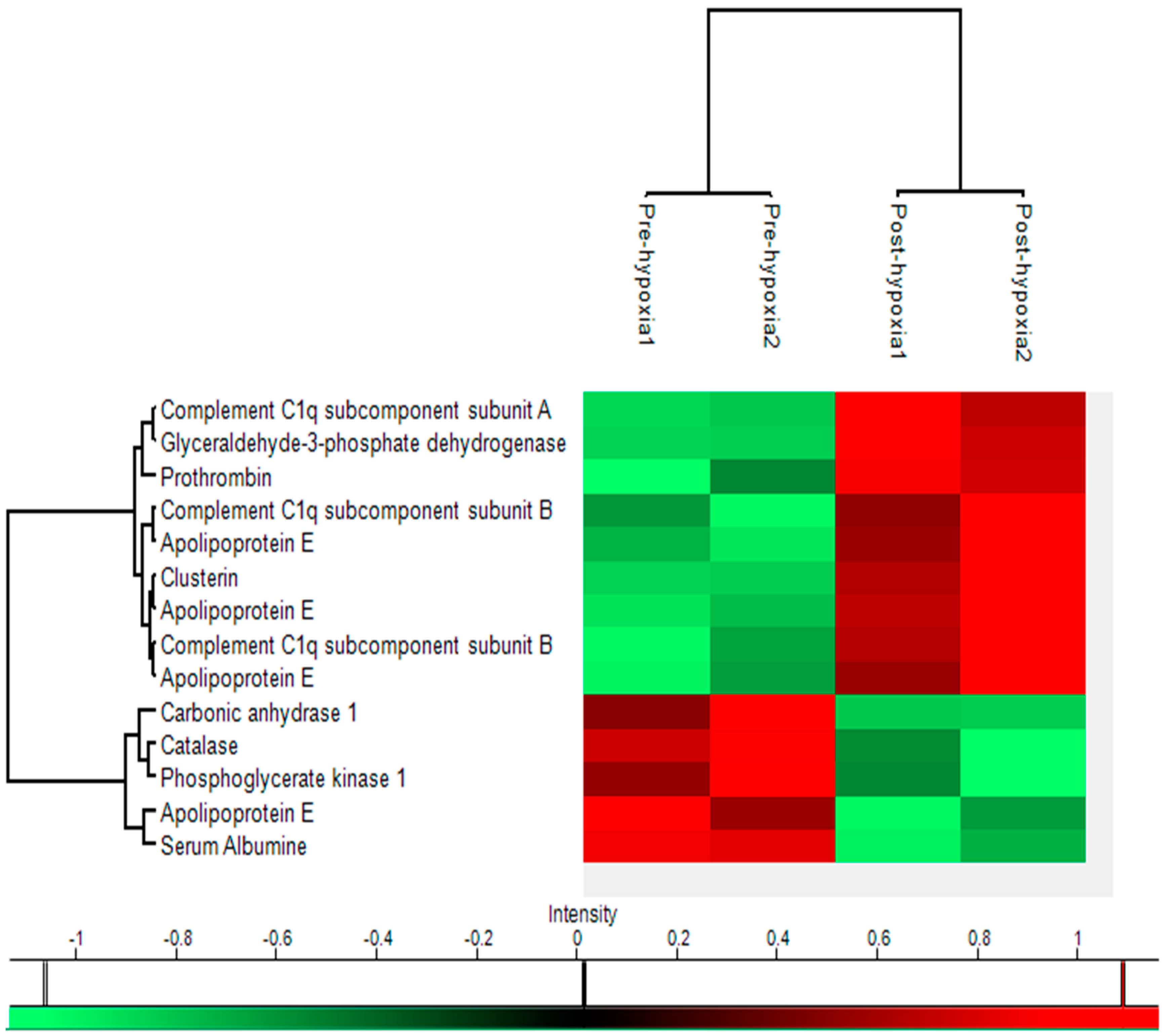
| Name | Uniprot key | Gene | Function | Location | Pre-hypoxia Mean | Post-hypoxia Mean | p-value | Fold-change | Molecular Weight | pI value | Sequence Coverage | Mascot Score |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Catalase | P04040 | CAT | Protection from toxic effects of hydrogen peroxide, aid to cellular growth of e.g. T-cells, B-cells, AML cells, Melanome cells, etc | Cytosol, endoplasmatic reticulum, mitochondria, peroxisome | 0.032 | 0.023 | 0.021 | −1.398 | 59,719 | 6.9 | 63% | 1941 |
| Carbonic anhydrase 1 | P00915 | CA1 | Bicarbonate transport, carbon metabolism | Cytosol, exosomes | 0.416 | 0.298 | 0.038 | −1.398 | 28,852 | 6.59 | 75% | 1176 |
| Apolipoprotein E | P02649 | APOE | Cholesterol metabolism, Lipid metabolism, Lipid transport, Steroid metabolism, Sterol metabolism, Transport protein | Chylomicron, HDL, VLDL | 0.056 | 0.043 | 0.029 | −1.294 | 36,132 | 5.65 | 69% | 1785 |
| Serum Albumine | P02768 | ALB | Binding capacity for water, Ca2+, Na+, K+, 0 fatty acids, hormones, bilirubin and drugs, regulation of the colloidal osmotic pressure of blood, zinc transporter | Serum, extracellular | 0.474 | 0.381 | 0.007 | −1.245 | 69,321 | 5.92 | 66% | 2262 |
| Phosphoglycerate kinase 1 | P00558 | PGK1 | Gluconeogenesis, glycolysis, epithelial cell differentiation | Cytoplasm | 0.021 | 0.017 | 0.033 | −1.203 | 44,586 | 8.3 | 70% | 1539 |
| Complement C1q subcomponent subunit B | P02746 | C1QB | Complement activation, innate Immune system, Inner ear development, Proteolysis | Blood microparticle, collagen, complement C1 complex, exosomes | 0.157 | 0.193 | 0.022 | 1.23 | 26,704 | 8.83 | 39% | 686 |
| Clusterin | P10909 | CLU | Apoptosis, complement system, immune defence, innate immune system | Cytoplasma, cytoplasmic vescicles, endoplasmatic reticulum, membrane, mitochondria, microsomes, nucleus | 0.663 | 0.84 | 0.014 | 1.267 | 52,461 | 5.85 | 43% | 1407 |
| Complement C1q subcomponent subunit B | P02746 | C1QB | Complement activation, innate immune system, inner ear development, proteolysis | Cytosol, endoplasmatic reticulum, mitochondria, peroxisome | 0.095 | 0.132 | 0.047 | 1.38 | 26,704 | 8.83 | 41% | 656 |
| Glyceraldehyde-3-phosphate dehydrogenase | P04406 | GAPDH | Oxidoreductase, transferase, apoptose, glycolysis, regulation of translation | Cytoplasm, cytoskeleton, membrane, nucleus | 0.019 | 0.027 | 0.004 | 1.393 | 36,030 | 8.57 | 66% | 1113 |
| Apolipoprotein E | P02649 | APOE | Cholesterol metabolism, lipid metabolism, lipid transport, steroid metabolism, sterol metabolism, transport protein | Chylomicron, HDL, VLDL | 0.026 | 0.037 | 0.005 | 1.399 | 36,132 | 5.65 | 74% | 1912 |
| Apolipoprotein E | P02649 | APOE | Cholesterol metabolism, lipid metabolism, lipid transport, steroid metabolism, sterol metabolism, transport protein | Chylomicron, HDL, VLDL | 0.085 | 0.121 | 0.037 | 1.419 | 36,132 | 5.65 | 65% | 1438 |
| Prothrombin | P00734 | F2 | Acute-phase protein, blood coagulation, haemostasis | Extracellular | 0.024 | 0.036 | 0.04 | 1.498 | 69,992 | 5.64 | 38% | 1487 |
| Apolipoprotein E | P02649 | APOE | Cholesterol metabolism, lipid metabolism, lipid transport, steroid metabolism, sterol metabolism, transport protein | Chylomicron, HDL, VLDL | 0.026 | 0.041 | 0.028 | 1.588 | 36,132 | 5.65 | 44% | 884 |
| Complement C1q subcomponent subunit A | P02745 | C1QA | Complement pathway, immune defence, innate immune system | Collagen, complement C1 complex, exosomes | 0.068 | 0.143 | 0.011 | 2.109 | 26,000 | 9.26 | 20% | 320 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hinkelbein, J.; Jansen, S.; Iovino, I.; Kruse, S.; Meyer, M.; Cirillo, F.; Drinhaus, H.; Hohn, A.; Klein, C.; Robertis, E.D.; et al. Thirty Minutes of Hypobaric Hypoxia Provokes Alterations of Immune Response, Haemostasis, and Metabolism Proteins in Human Serum. Int. J. Mol. Sci. 2017, 18, 1882. https://doi.org/10.3390/ijms18091882
Hinkelbein J, Jansen S, Iovino I, Kruse S, Meyer M, Cirillo F, Drinhaus H, Hohn A, Klein C, Robertis ED, et al. Thirty Minutes of Hypobaric Hypoxia Provokes Alterations of Immune Response, Haemostasis, and Metabolism Proteins in Human Serum. International Journal of Molecular Sciences. 2017; 18(9):1882. https://doi.org/10.3390/ijms18091882
Chicago/Turabian StyleHinkelbein, Jochen, Stefanie Jansen, Ivan Iovino, Sylvia Kruse, Moritz Meyer, Fabrizio Cirillo, Hendrik Drinhaus, Andreas Hohn, Corinna Klein, Edoardo De Robertis, and et al. 2017. "Thirty Minutes of Hypobaric Hypoxia Provokes Alterations of Immune Response, Haemostasis, and Metabolism Proteins in Human Serum" International Journal of Molecular Sciences 18, no. 9: 1882. https://doi.org/10.3390/ijms18091882





