Candidate Genes and Molecular Markers Correlated to Physiological Traits for Heat Tolerance in Fine Fescue Cultivars
Abstract
:1. Introduction
2. Results
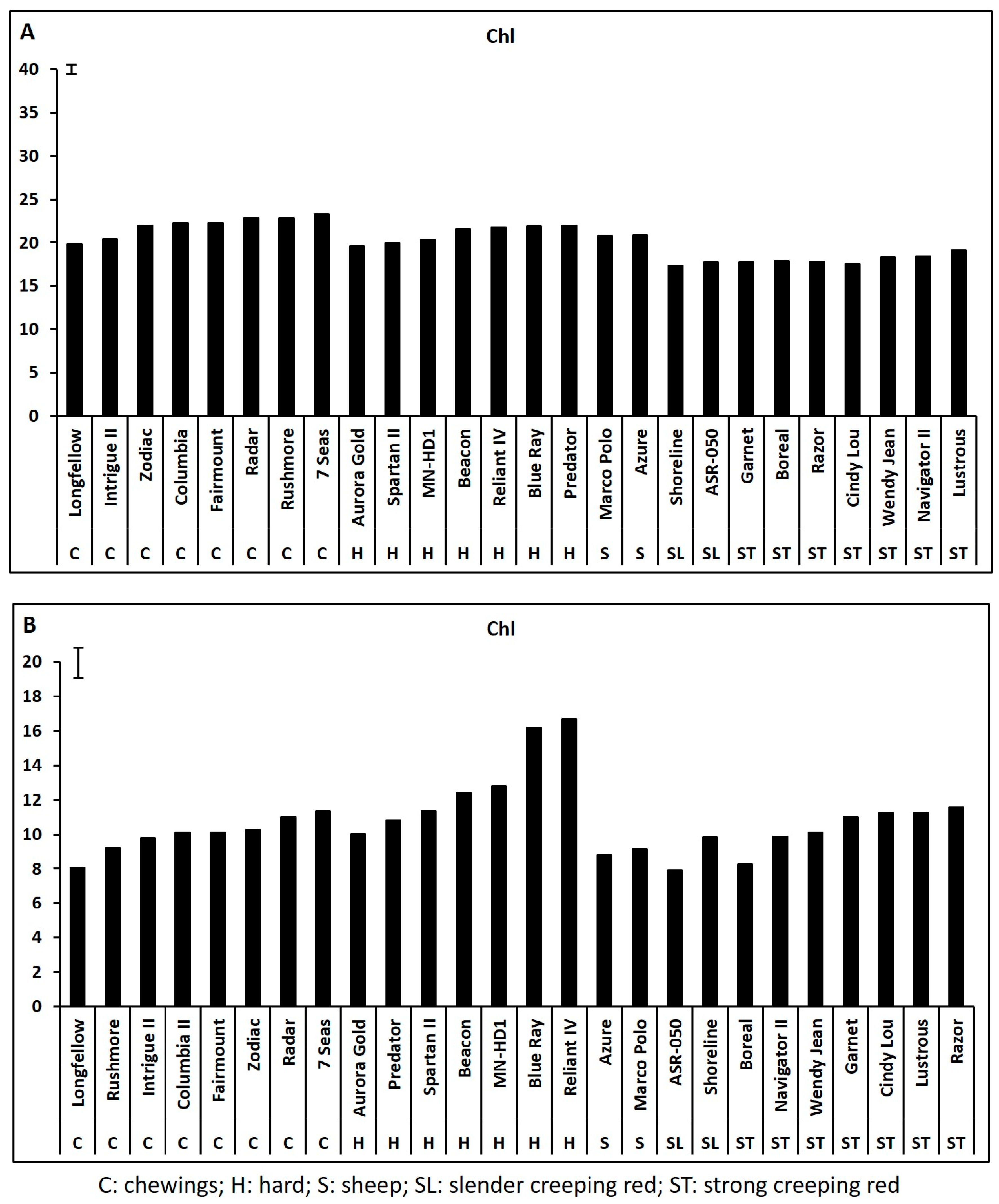
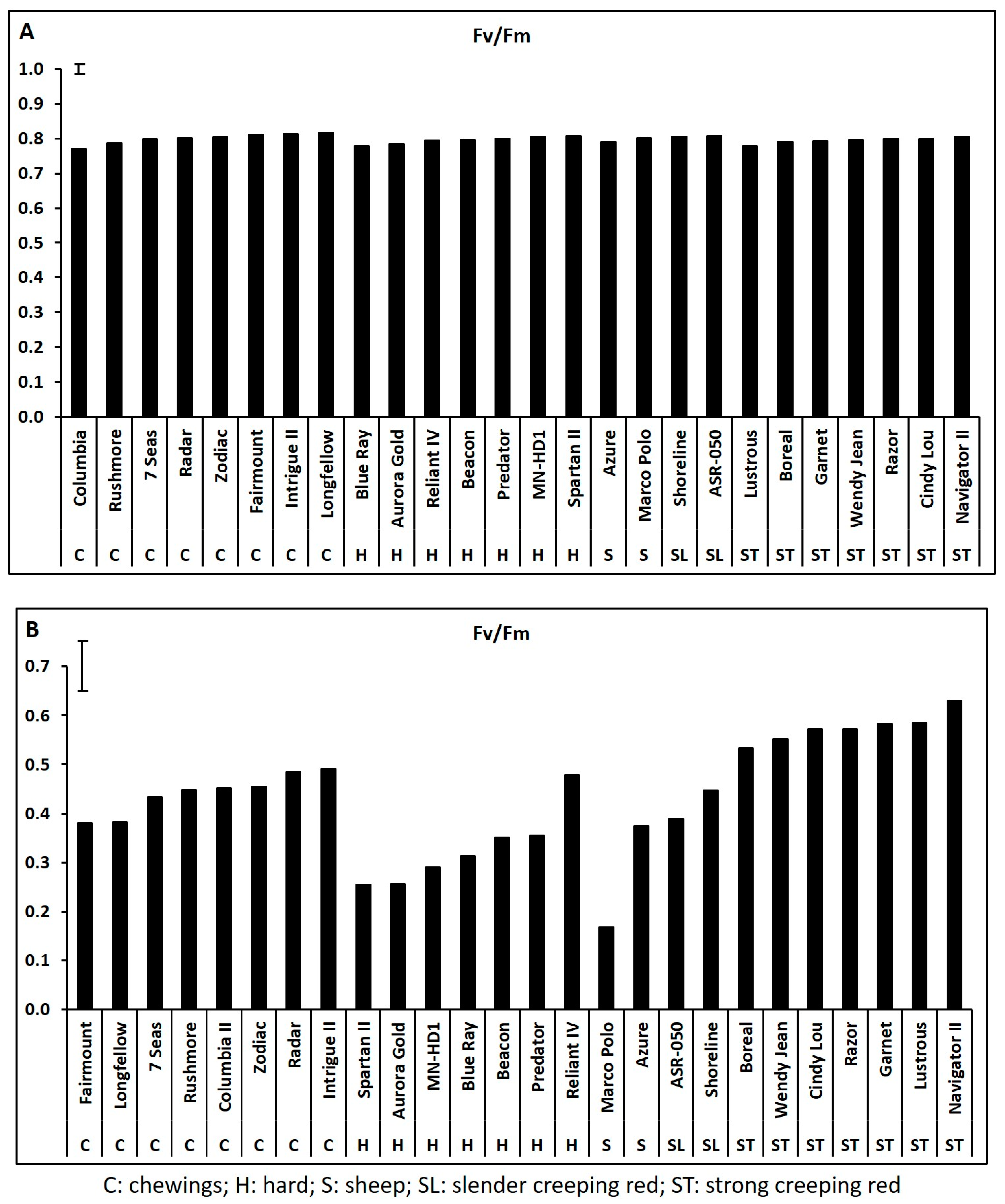
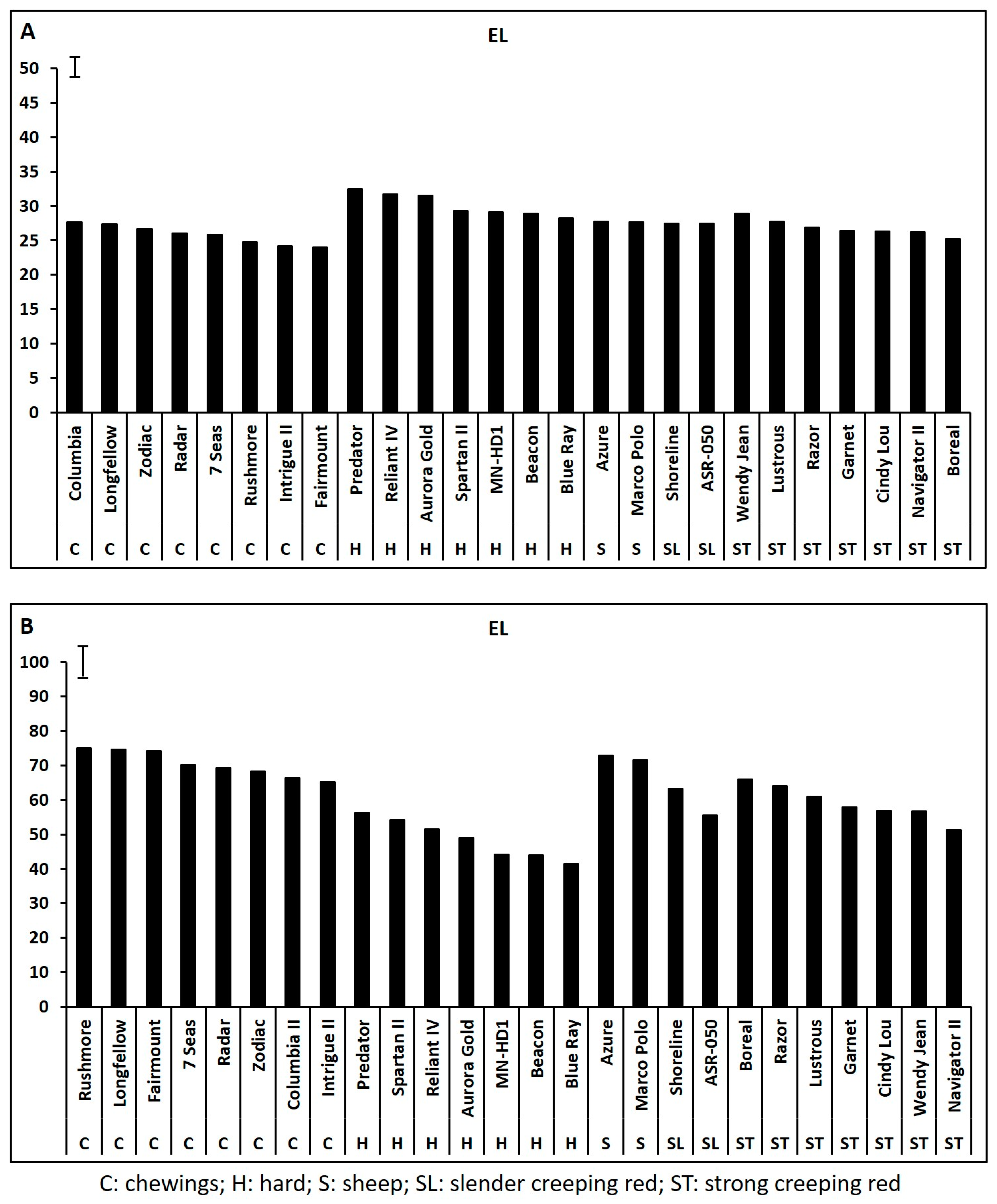
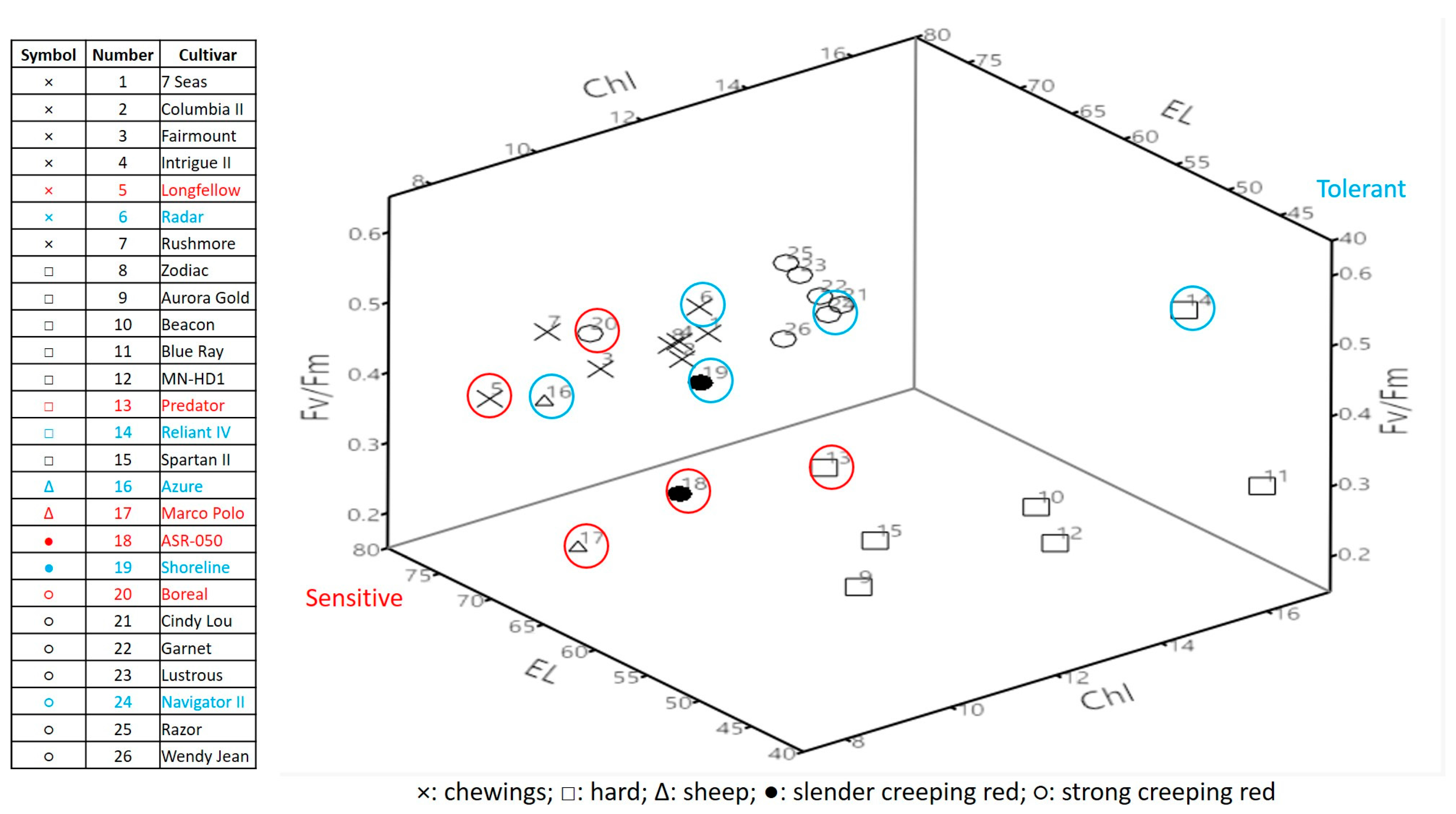
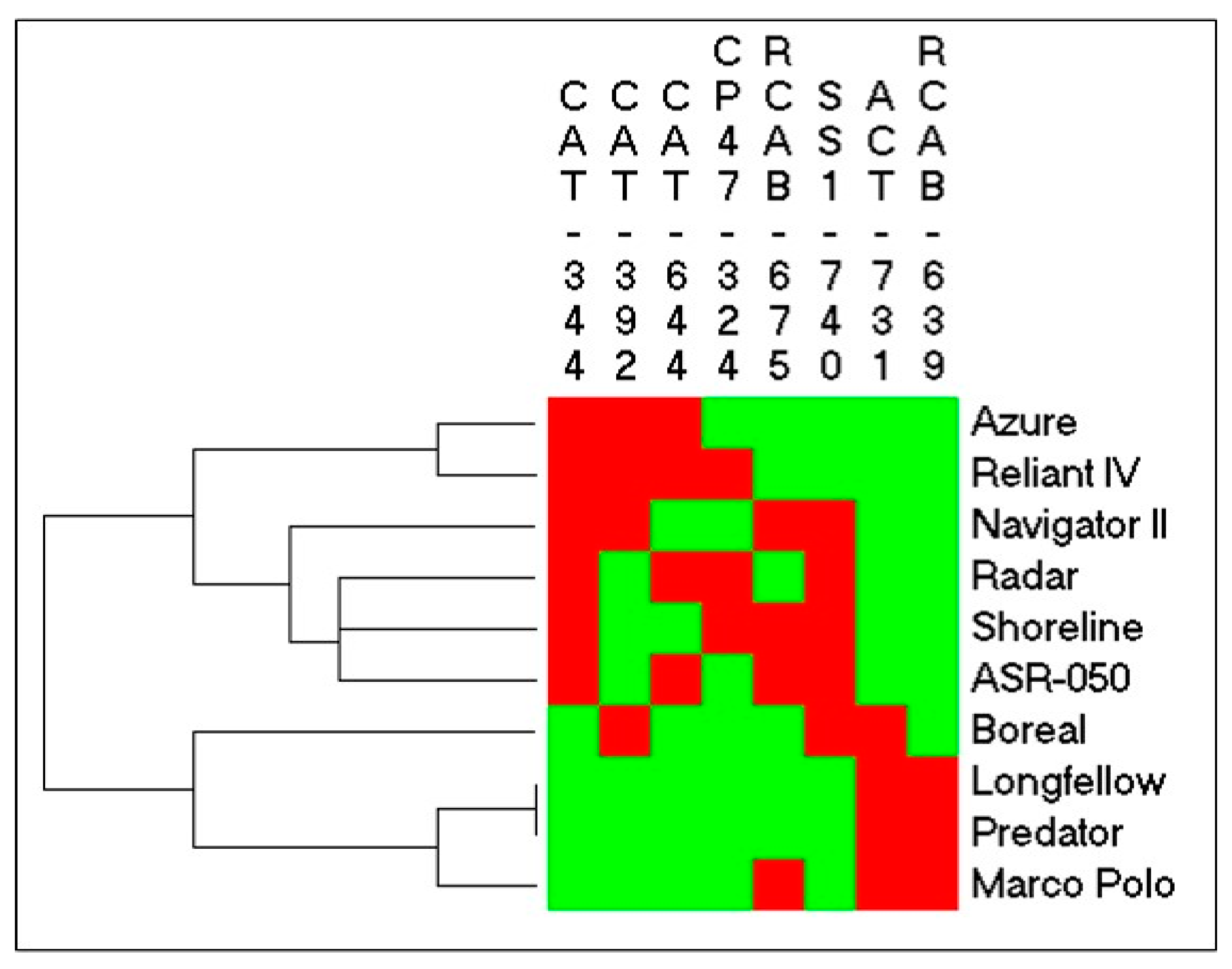
2.1. Genetic Variations in Physiological Traits Associated with Heat Tolerance
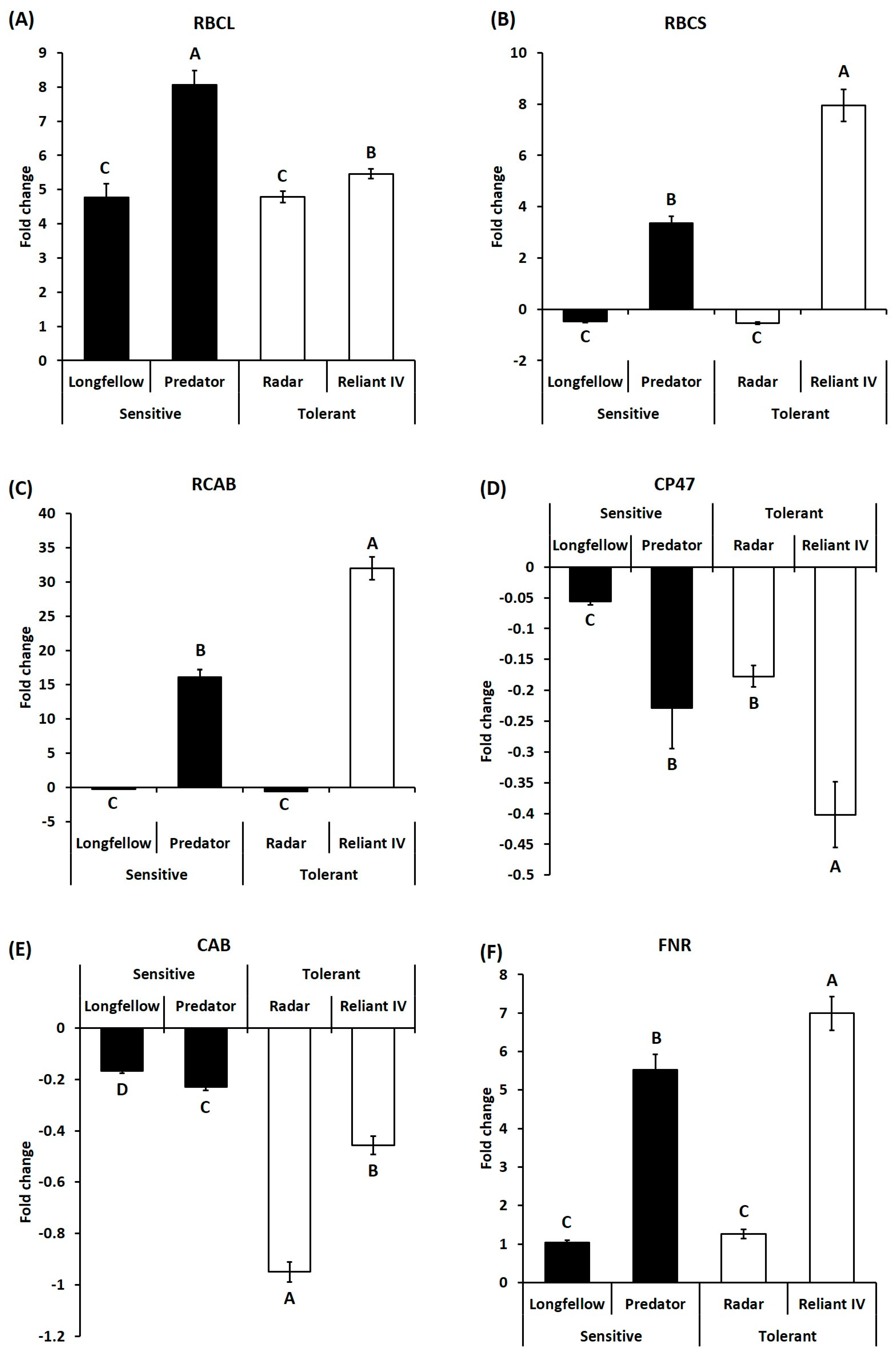
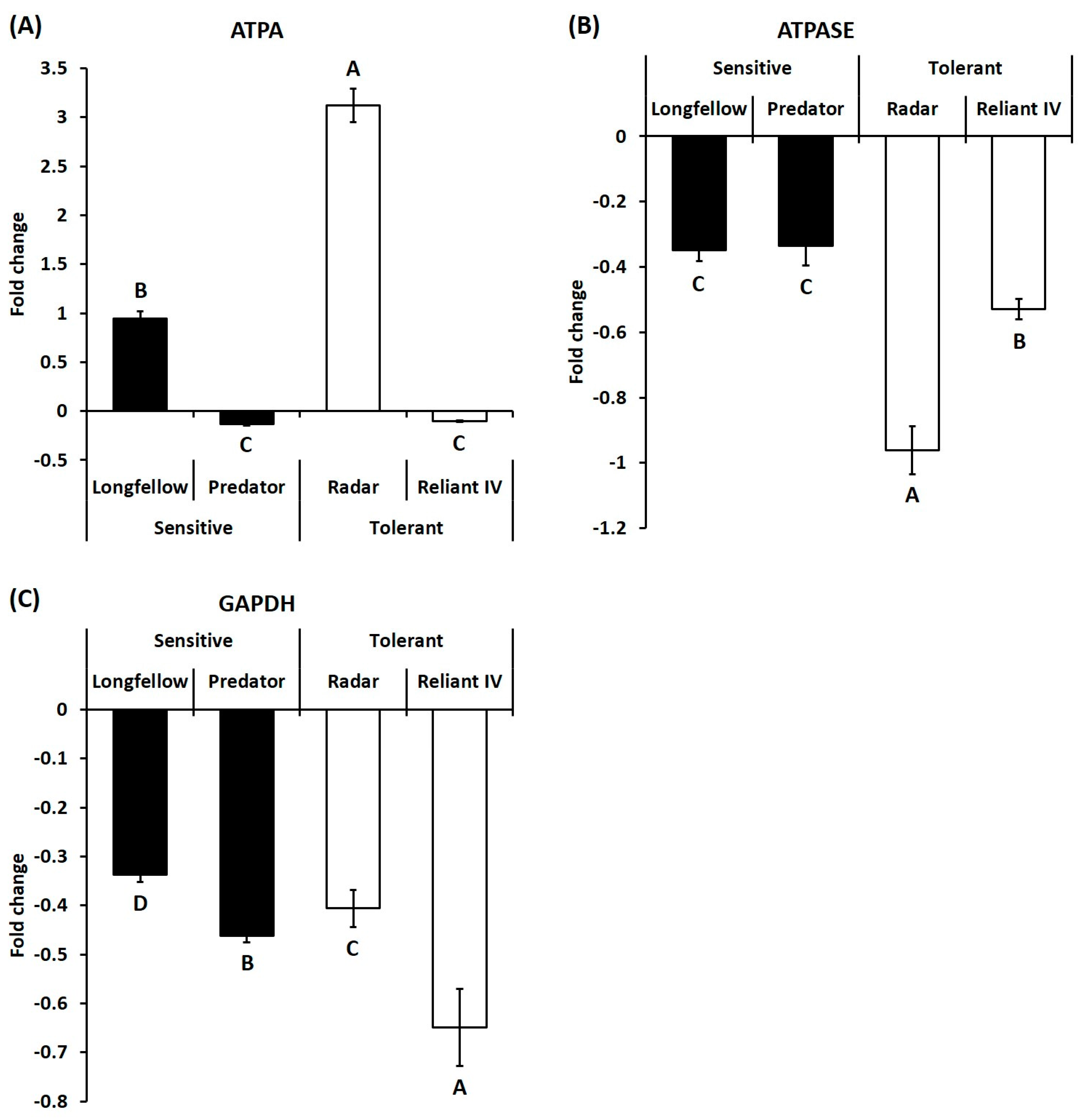
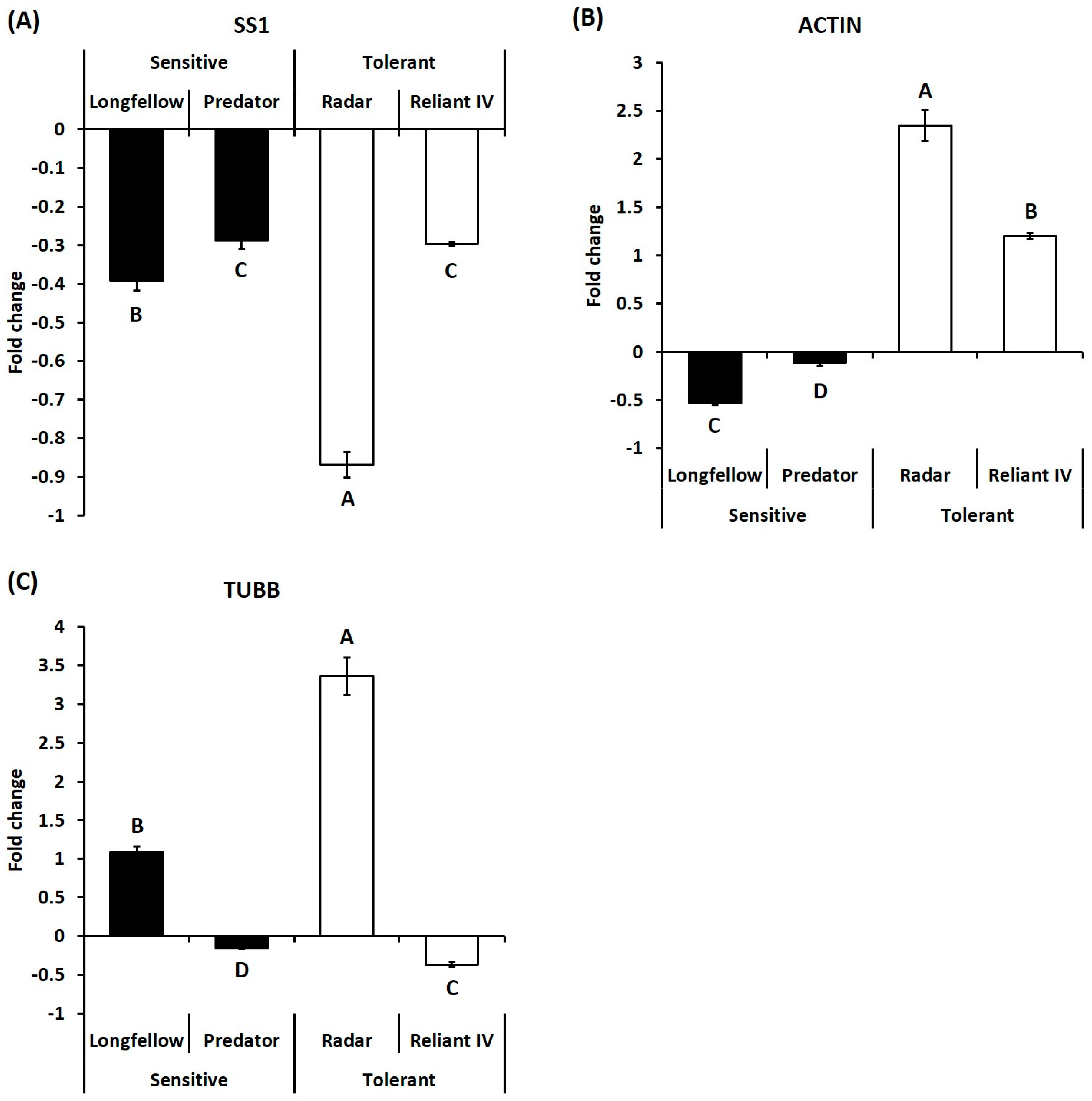
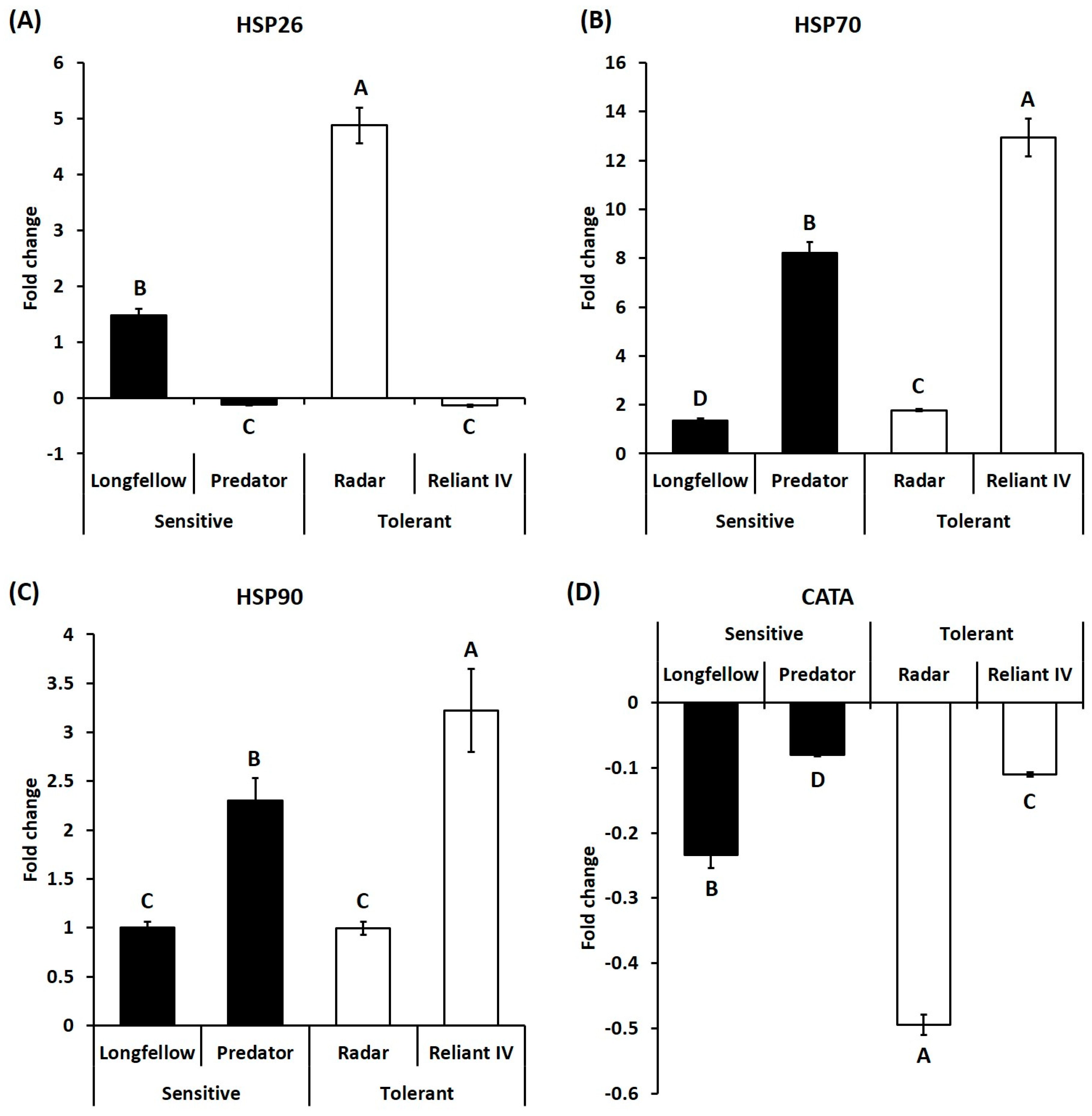
2.2. Differential Expression of Candidate Genes in Heat-Sensitive and Heat-Tolerant Cultivars
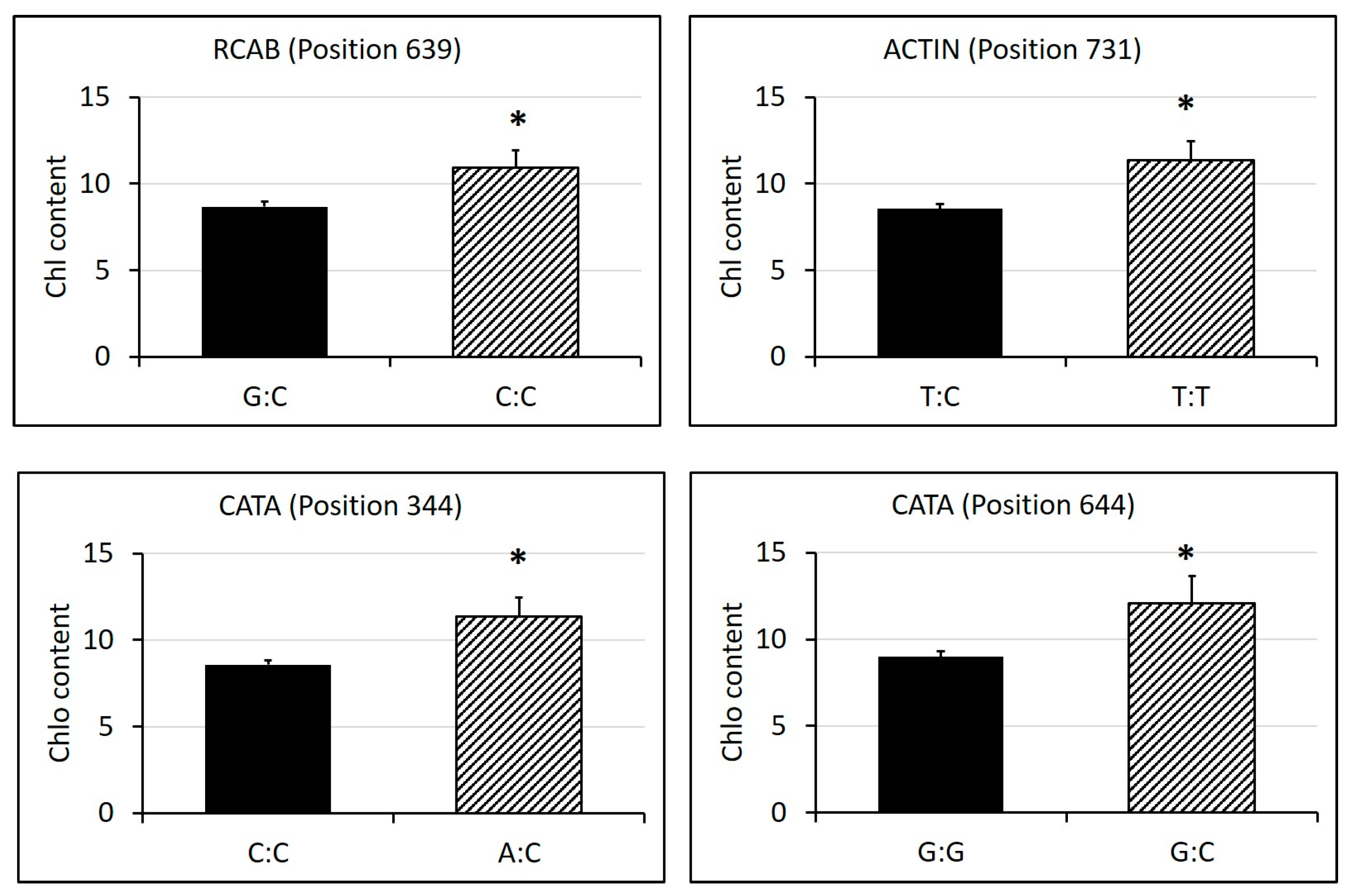
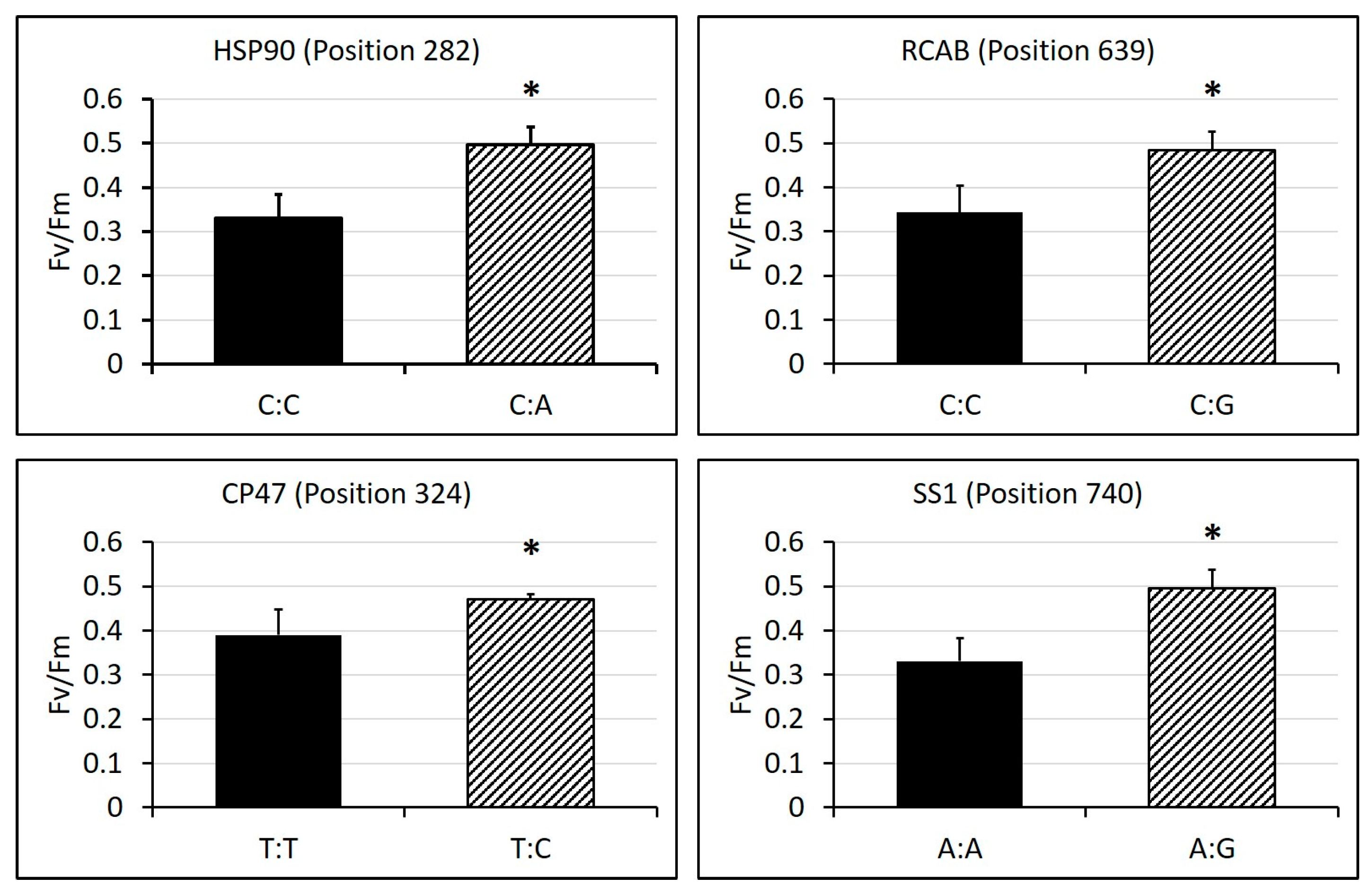
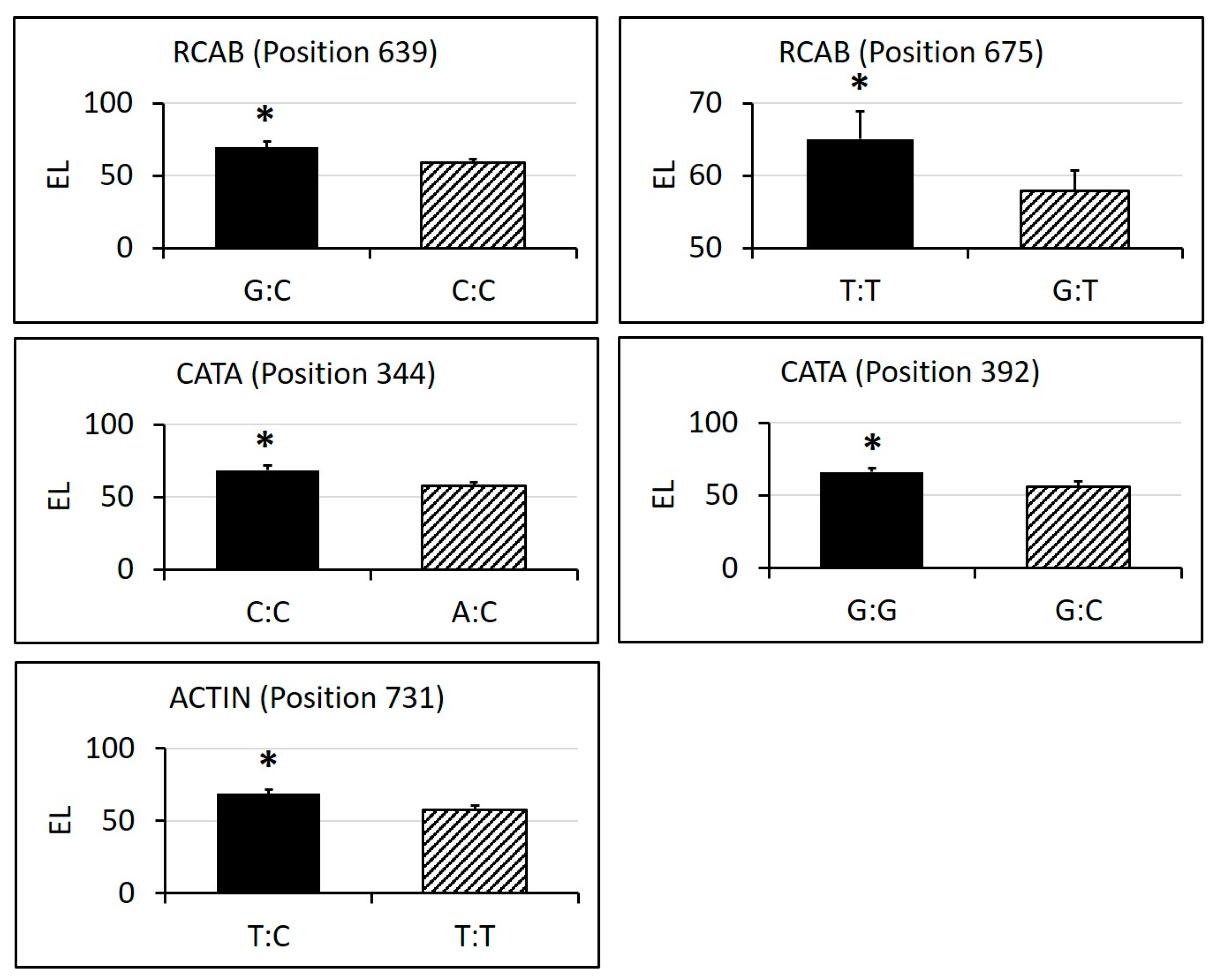
2.3. Identification of SNPs in Candidate Gene Sequences and Correlations with Heat Tolerance
3. Materials and Methods
3.1. Plant Materials and Growth Conditions
3.2. Temperature Treatments
3.3. Physiological Analysis
3.4. Gene Expression Analysis
3.5. SNP Identification
3.6. Statistical Analysis
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Wahid, A.; Gelani, S.; Ashraf, M.; Foolad, M. Heat tolerance in plants: An overview. Environ. Exp. Bot. 2007, 61, 199–223. [Google Scholar] [CrossRef]
- Liu, X.; Huang, B. Heat stress injury in relation to membrane lipid peroxidation in creeping bentgrass. Crop Sci. 2000, 40, 503–510. [Google Scholar] [CrossRef]
- Jiang, Y.; Huang, B. Drought and heat stress injury to two cool-season turfgrasses in relation to antioxidant metabolism and lipid peroxidation. Crop Sci. 2001, 41, 436–442. [Google Scholar] [CrossRef]
- Jiang, Y.; Huang, B. Physiological responses to heat stress alone or in combination with drought: A comparison between tall fescue and perennial ryegrass. HortScience 2001, 36, 682–686. [Google Scholar]
- Huang, B.; Liu, X.; Xu, Q. Supraoptimal soil temperatures induced oxidative stress in leaves of creeping bentgrass cultivars differing in heat tolerance. Crop Sci. 2001, 41, 430–435. [Google Scholar] [CrossRef]
- Du, H.; Wang, Z.; Huang, B. Differential responses of warm-season and cool-season turfgrass species to heat stress associated with antioxidant enzyme activity. J. Am. Soc. Hortic. Sci. 2009, 134, 417–422. [Google Scholar]
- Wang, J.; Cui, L.; Wang, Y.; Li, J. Growth, lipid peroxidation and photosynthesis in two tall fescue cultivars differing in heat tolerance. Biol. Plant. 2009, 53, 237–242. [Google Scholar] [CrossRef]
- Yin, H.; Chen, Q.; Yi, M. Effects of short-term heat stress on oxidative damage and responses of antioxidant system in Lilium longiflorum. Plant Growth Regul. 2008, 54, 45–54. [Google Scholar] [CrossRef]
- Xu, Y.; Zhan, C.; Huang, B. Heat shock proteins in association with heat tolerance in grasses. Int. J. Proteom. 2011, 2011, 529648. [Google Scholar] [CrossRef] [PubMed]
- Sari-Gorla, M.; Krajewski, P.; di Fonzo, N.; Villa, M.; Frova, C. Genetic analysis of drought tolerance in maize by molecular markers. II. Plant height and flowering. Theor. Appl. Genet. 1999, 99, 289–295. [Google Scholar] [CrossRef]
- Forster, B.; Ellis, R.; Thomas, W.; Newton, A.; Tuberosa, R.; This, D.; El-Enein, R.; Bahri, M.; Ben Salem, M. The development and application of molecular markers for abiotic stress tolerance in barley. J. Exp. Bot. 2000, 51, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Rizhsky, L.; Liang, H.; Mittler, R. The combined effect of drought stress and heat shock on gene expression in tobacco. Plant Physiol. 2002, 130, 1143–1151. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.G.; Ahsan, N.; Lee, S.H.; Kang, K.Y.; Bahk, J.D.; Lee, I.J.; Lee, B.H. A proteomic approach in analyzing heat-responsive proteins in rice leaves. Proteomics 2007, 7, 3369–3383. [Google Scholar] [CrossRef] [PubMed]
- Qin, D.; Wu, H.; Peng, H.; Yao, Y.; Ni, Z.; Li, Z.; Zhou, C.; Sun, Q. Heat stress-responsive transcriptome analysis in heat susceptible and tolerant wheat (Triticum aestivum L.) by using wheat genome array. BMC Genom. 2008, 9, 432. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Huang, B. Root proteomic responses to heat stress in two Agrostis grass species contrasting in heat tolerance. J. Exp. Bot. 2008, 59, 4183–4194. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Huang, B. Differential proteomic response to heat stress in thermal Agrostis scabra and heat-sensitive Agrostis stolonifera. Physiol. Plant. 2010, 139, 192–204. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Gianfagna, T.; Huang, B. Proteomic changes associated with expression of a gene (ipt) controlling cytokinin synthesis for improving heat tolerance in a perennial grass species. J. Exp. Bot. 2010, 61, 3273–3289. [Google Scholar] [CrossRef] [PubMed]
- Thudi, M.; Upadhyaya, H.D.; Rathore, A.; Gaur, P.M.; Krishnamurthy, L.; Roorkiwal, M.; Nayak, S.N.; Chaturvedi, S.K.; Basu, P.S.; Gangarao, N. Genetic dissection of drought and heat tolerance in chickpea through genome-wide and candidate gene-based association mapping approaches. PLoS ONE 2014, 9, e96758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Mian, M.R.; Chekhovskiy, K.; So, S.; Kupfer, D.; Lai, H.; Roe, B.A. Differential gene expression in Festuca under heat stress conditions. J. Exp. Bot. 2005, 56, 897–907. [Google Scholar] [CrossRef] [PubMed]
- Bita, C.E.; Zenoni, S.; Vriezen, W.H.; Mariani, C.; Pezzotti, M.; Gerats, T. Temperature stress differentially modulates transcription in meiotic anthers of heat-tolerant and heat-sensitive tomato plants. BMC Genom. 2011, 12, 384. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.-W.; Vierling, E. Mutants of Arabidopsis thaliana defective in the acquisition of tolerance to high temperature stress. Proc. Natl. Acad. Sci. USA 2000, 97, 4392–4397. [Google Scholar] [CrossRef] [PubMed]
- Yamanouchi, U.; Yano, M.; Lin, H.; Ashikari, M.; Yamada, K. A rice spotted leaf gene, spl7, encodes a heat stress transcription factor protein. Proc. Natl. Acad. Sci. USA 2002, 99, 7530–7535. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Zhu, Q.; Xu, J.; Huang, B. Cloning and characterization of a gene, encoding expansin proteins inducible by heat stress and hormones in creeping bentgrass. Crop Sci. 2011, 51, 333–341. [Google Scholar] [CrossRef]
- Wang, X.; Huang, W.; Liu, J.; Yang, Z.; Huang, B. Molecular regulation and physiological functions of a novel fahsfa2c cloned from tall fescue conferring plant tolerance to heat stress. Plant Biotechnol. J. 2017, 15, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Jespersen, D.; Belanger, F.C.; Huang, B. Candidate genes and molecular markers associated with heat tolerance in colonial bentgrass. PLoS ONE 2017, 12, e0171183. [Google Scholar] [CrossRef] [PubMed]
- Jiang, G.-L. Molecular markers and marker-assisted breeding in plants. In Plant Breeding from Laboratories to Fields; Andersen, S.B., Ed.; InTech: Rijeka, Croatia, 2013; pp. 45–83. [Google Scholar]
- Rostoks, N.; Mudie, S.; Cardle, L.; Russell, J.; Ramsay, L.; Booth, A.; Svensson, J.T.; Wanamaker, S.I.; Walia, H.; Rodriguez, E.M. Genome-wide SNP discovery and linkage analysis in barley based on genes responsive to abiotic stress. Mol. Genet. Genom. 2005, 274, 515–527. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Wei, X.; Sang, T.; Zhao, Q.; Feng, Q.; Zhao, Y.; Li, C.; Zhu, C.; Lu, T.; Zhang, Z. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 2010, 42, 961–967. [Google Scholar] [CrossRef] [PubMed]
- Turgeon, A. Turfgrass Management, 6th ed.; Prentice Hall: Englewood Cliffs, NJ, USA, 2002. [Google Scholar]
- Morris, K. National Turfgrass Evaluation Program; NTEP: Beltsville, MD, USA, 2001. [Google Scholar]
- Wang, J.; Juliani, H.R.; Jespersen, D.; Huang, B. Differential profiles of membrane proteins, fatty acids, and sterols associated with genetic variations in heat tolerance for a perennial grass species, hard fescue (Festuca trachyphylla). Environ. Exp. Bot. 2017, 140, 65–75. [Google Scholar] [CrossRef]
- Hoagland, D.R.; Arnon, D.I. The Water-Culture Method for Growing Plants without Soil; University of California College of Agriculture: Davis, CA, USA, 1938. [Google Scholar]
- Hiscox, J.T.; Israelstam, G. A method for the extraction of chlorophyll from leaf tissue without maceration. Can. J. Bot. 1979, 57, 1332–1334. [Google Scholar] [CrossRef]
- Arnon, D.I. Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 1949, 24, 1. [Google Scholar] [CrossRef] [PubMed]
- Blum, A.; Ebercon, A. Cell-membrane stability as a measure of drought and heat tolerance in wheat. Crop Sci. 1981, 21, 43–47. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Weckx, S.; Del-Favero, J.; Rademakers, R.; Claes, L.; Cruts, M.; De Jonghe, P.; Van Broeckhoven, C.; De Rijk, P. NovoSNP, a novel computational tool for sequence variation discovery. Genome Res. 2005, 15, 436–442. [Google Scholar] [CrossRef] [PubMed]
- Caraux, G.; Pinloche, S. Permutmatrix: A graphical environment to arrange gene expression profiles in optimal linear order. Bioinformatics 2004, 21, 1280–1281. [Google Scholar] [CrossRef] [PubMed]
- Marcum, K.B. Cell membrane thermostability and whole-plant heat tolerance of Kentucky bluegrass. Crop Sci. 1998, 38, 1214–1218. [Google Scholar] [CrossRef]
- Jespersen, D.; Meyer, W.; Huang, B. Physiological traits and genetic variations associated with drought and heat tolerance in creeping bentgrass. Int. Turfgrass Soc. Res. J. 2013, 12, 459–464. [Google Scholar]
- Rachmilevitch, S.; DaCosta, M.; Huang, B. Physiological and biochemical indicators for stress tolerance. In Plant-Environment Interactions, 3rd ed.; CRC Press: Boca Raton, FL, USA, 2006; pp. 321–355. [Google Scholar]
- Nilsen, E.; Orcutt, D. The Physiology of Plants under Deficit. Abiotic Factors; John Wiley & Sons: New York, NY, USA, 1996. [Google Scholar]
- Ashraf, M.; Harris, P. Photosynthesis under stressful environments: An overview. Photosynthetica 2013, 51, 163–190. [Google Scholar] [CrossRef]
- Portis, A.R. Rubisco activase-Rubisco’s catalytic chaperone. Photosynth. Res. 2003, 75, 11–27. [Google Scholar] [CrossRef] [PubMed]
- Avigad, G.; Milner, Y. UDP-glucose: Fructose transglucosylase from sugar beet roots. Methods Enzymol. 1966, 8, 341–345. [Google Scholar]
- Gupta, A.K.; Kaur, N. Sugar signalling and gene expression in relation to carbohydrate metabolism under abiotic stresses in plants. J. Biosci. 2005, 30, 761–776. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Burgess, P.; Huang, B. Root antioxidant mechanisms in relation to root thermotolerance in perennial grass species contrasting in heat tolerance. PLoS ONE 2015, 10, e0138268. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Huang, B. Differential responses to heat stress in activities and isozymes of four antioxidant enzymes for two cultivars of Kentucky bluegrass contrasting in heat tolerance. J. Am. Soc. Hortic. Sci. 2010, 135, 116–124. [Google Scholar]
- Wang, W.; Vinocur, B.; Shoseyov, O.; Altman, A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 2004, 9, 244–252. [Google Scholar] [CrossRef] [PubMed]
- Milioni, D.; Hatzopoulos, P. Genomic organization of hsp90 gene family in Arabidopsis. Plant Mol. Biol. 1997, 35, 955–961. [Google Scholar] [CrossRef] [PubMed]
- Volkmann, D.; Baluška, F. Actin cytoskeleton in plants: From transport networks to signaling networks. Microsc. Res. Tech. 1999, 47, 135–154. [Google Scholar] [CrossRef]
- Staiger, C.J. Signaling to the actin cytoskeleton in plants. Annu. Rev. Plant Biol. 2000, 51, 257–288. [Google Scholar] [CrossRef] [PubMed]
- Shimmen, T.; Yokota, E. Cytoplasmic streaming in plants. Curr. Opin. Cell Biol. 2004, 16, 68–72. [Google Scholar] [CrossRef] [PubMed]
- Sangwan, V.; Örvar, B.L.; Beyerly, J.; Hirt, H.; Dhindsa, R.S. Opposite changes in membrane fluidity mimic cold and heat stress activation of distinct plant map kinase pathways. Plant J. 2002, 31, 629–638. [Google Scholar] [CrossRef] [PubMed]
- Marguet, D.; Lenne, P.F.; Rigneault, H.; He, H.T. Dynamics in the plasma membrane: How to combine fluidity and order. EMBO J. 2006, 25, 3446–3457. [Google Scholar] [CrossRef] [PubMed]
| Gene | Accession | Primer Sequence | |
|---|---|---|---|
| RBCL | GO796655 | Forward | GTTCAGTTCAGTCAAGTGGTAG |
| Reverse | GGAGTTCAGATCAGGCAAAG | ||
| RBCS | GO852616 | Forward | GATACTACGACGGCAGGTA |
| Reverse | CATAGGCGTCAGGGTACT | ||
| RCAB | GO876625 | Forward | GGCTATCTTGAAGTGGGTAAAT |
| Reverse | GTTGGCTTTGGAGGGATAAA | ||
| CP47 | GO847696 | Forward | CAGTAGTATCGCTGCTGTATTT |
| Reverse | AGTATCCTGATCCCACTGATAA | ||
| CAB | GO884317 | Forward | TGAGGCTGTCTGGTTCAA |
| Reverse | ACCCATGAGCACAACCT | ||
| FNR | GO803921 | Forward | GACTTCGCTGTGAGCAG |
| Reverse | GTTGTCCTTCTTGAGCATTTC | ||
| ATPA | GO855374 | Forward | GCTTCCCTGAATACCCAAAG |
| Reverse | CTTTCAACCCTTTCAACCAATC | ||
| ATPASE | GO869342 | Forward | ACACTGTGCCTTGCTTAC |
| Reverse | GGGCGAACAGGATCTTTAATA | ||
| GAPDH | GO789276 | Forward | CCGCACTGAACATCGTG |
| Reverse | TGATAACGAGGTCCACCA | ||
| SS1 | GO874751 | Forward | CATCAAAGGACAAGGAGGAG |
| Reverse | CATTACGGACACGGTTCAT | ||
| ACTIN | GO839904 | Forward | TGTTCTCAGTGGAGGTTCTA |
| Reverse | CCTTTCAGGTGGTGCAATA | ||
| TUBB | GO795142 | Forward | CATGTAAGGAGATCCGTGTG |
| Reverse | CCTAGTGGTATTGCGTTGATAA | ||
| HSP26 | DT690825 | Forward | GACGACAAGGAGGTGAAGA |
| Reverse | GGATGACGAGCGTGTAGT | ||
| HSP70 | GO882325 | Forward | ACAAGATCACCATCACCAAC |
| Reverse | CTTGGCATCAACCTTCTTCT | ||
| HSP90 | GO882272 | Forward | ACTACGTCACCAGGATGAA |
| Reverse | TGACCTCATAGCCCTTCTT | ||
| CATA | GO877704 | Forward | TGATGGATGGCTGGATCT |
| Reverse | GATCTAGGCATGGTCTCTTATTG | ||
| EF1A | HO060093 | Forward | CAGATCAATGAGCCCAAGAG |
| Reverse | ATTCACACCAGTCTCAACAC | ||
| Gene | Reference Length (bp) | Sequencing Length (bp) | Total SNP Number (Q > 10) | Frequency (bp per SNP) |
|---|---|---|---|---|
| ACTIN | 1004 | 600 | 58 | 10.3 |
| ATPA | 1036 | 1008 | 9 | 112.0 |
| CATA | 1081 | 569 | 53 | 10.7 |
| CP47 | 1041 | 631 | 10 | 63.1 |
| HSP90 | 1056 | 924 | 58 | 15.9 |
| RCAB | 1210 | 361 | 11 | 32.8 |
| SS1 | 1193 | 491 | 17 | 28.9 |
| Gene | Accession | Primer Sequence | |
|---|---|---|---|
| ACTIN | GO839904 | Forward | GTGCTTGACTCTGGTGATG |
| Reverse | AGTGCTAAGAGAGGCCAA | ||
| ATPA | GO855374 | Forward | CGTGGTCAACGAGAACTTAT |
| Reverse | GCATCAAGTTCCATCTTTCTTT | ||
| CATA | GO877704 | Forward | CGACGAGGAGGACAAGTT |
| Reverse | TGAGCTCATGGCTGACC | ||
| CP47 | GO847696 | Forward | CTGGCTCGATGGCTTTATAC |
| Reverse | GCAGCGATACTACTGGAAAG | ||
| HSP90 | GO882272 | Forward | TGACTGGGAGGAGCATTT |
| Reverse | TAGTCTCTGCTGGACATGTAG | ||
| RCAB | GO876625 | Forward | TTGATGAACTTGGCGGATAA |
| Reverse | GATAACCCATCCTTCGTATACAT | ||
| SS1 | GO874751 | Forward | CATGTCCATCTACTTCCCATAC |
| Reverse | GCGATAGAGCTCAGCATTAC | ||
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Y.; Wang, J.; Bonos, S.A.; Meyer, W.A.; Huang, B. Candidate Genes and Molecular Markers Correlated to Physiological Traits for Heat Tolerance in Fine Fescue Cultivars. Int. J. Mol. Sci. 2018, 19, 116. https://doi.org/10.3390/ijms19010116
Xu Y, Wang J, Bonos SA, Meyer WA, Huang B. Candidate Genes and Molecular Markers Correlated to Physiological Traits for Heat Tolerance in Fine Fescue Cultivars. International Journal of Molecular Sciences. 2018; 19(1):116. https://doi.org/10.3390/ijms19010116
Chicago/Turabian StyleXu, Yi, Jinyu Wang, Stacy A. Bonos, William A. Meyer, and Bingru Huang. 2018. "Candidate Genes and Molecular Markers Correlated to Physiological Traits for Heat Tolerance in Fine Fescue Cultivars" International Journal of Molecular Sciences 19, no. 1: 116. https://doi.org/10.3390/ijms19010116






