Natural Antisense Transcripts: Molecular Mechanisms and Implications in Breast Cancers
Abstract
:1. Introduction
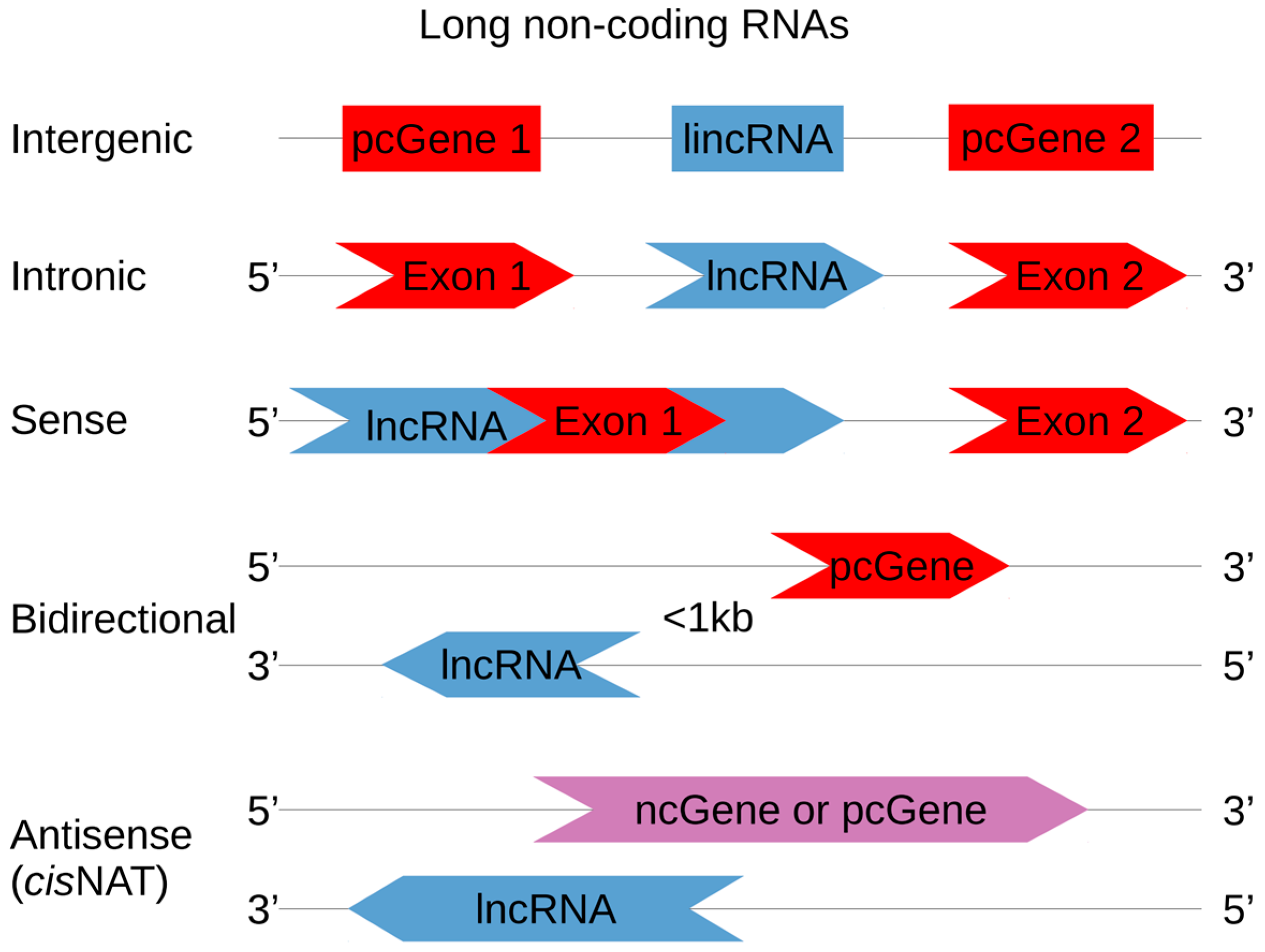
2. Generic Definition of NATs
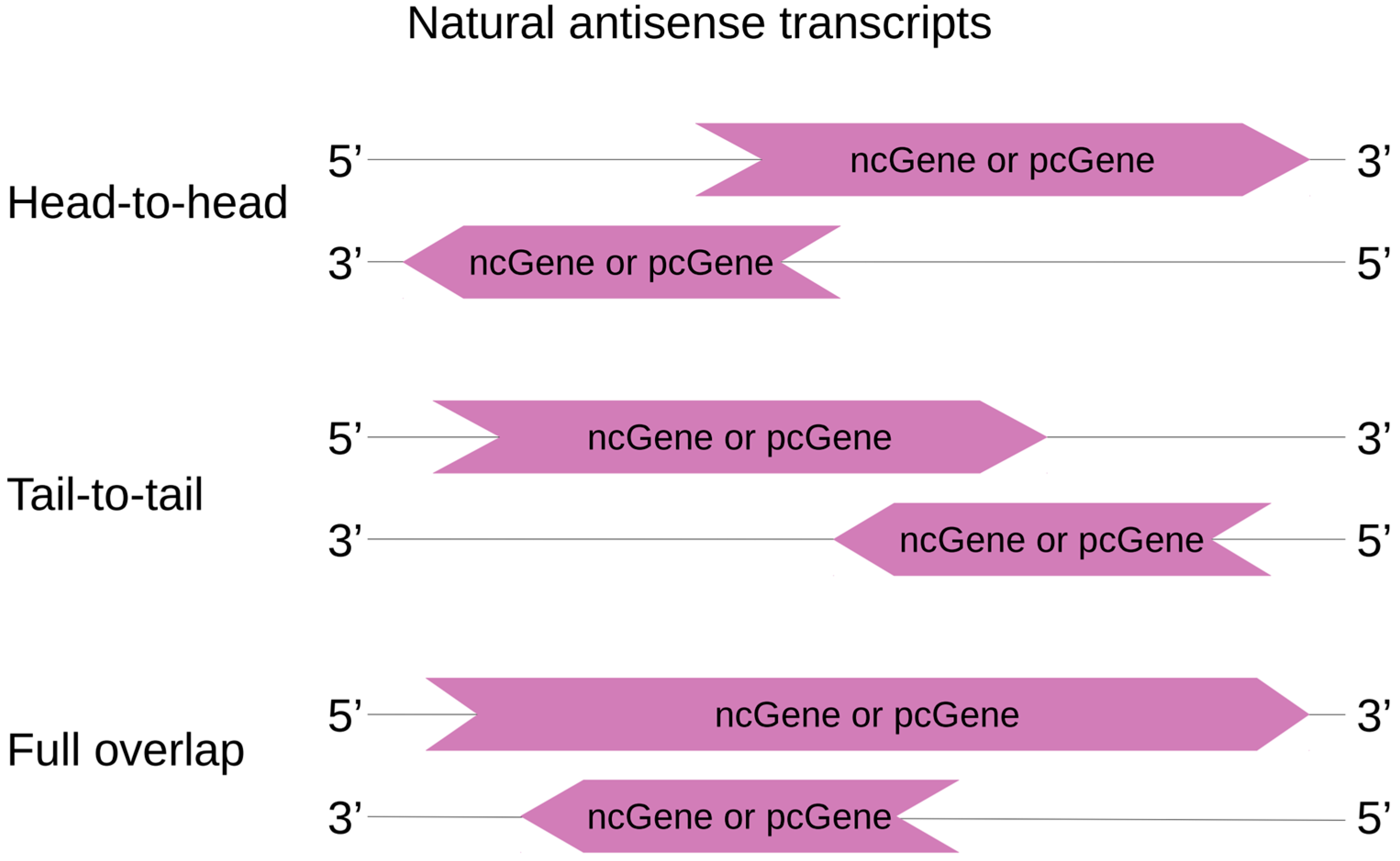
3. NAT: Structure, Localization, and Expression Regulation
4. NAT: Role, Function and Mechanism of Action
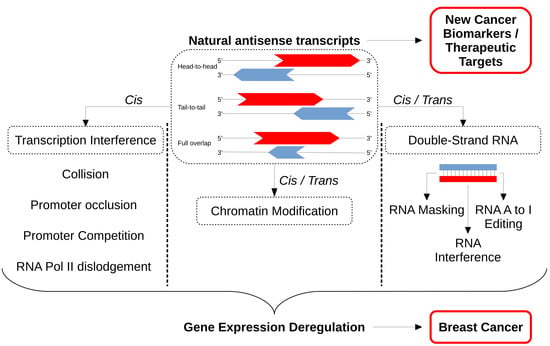
4.1. Action in Cis or Trans
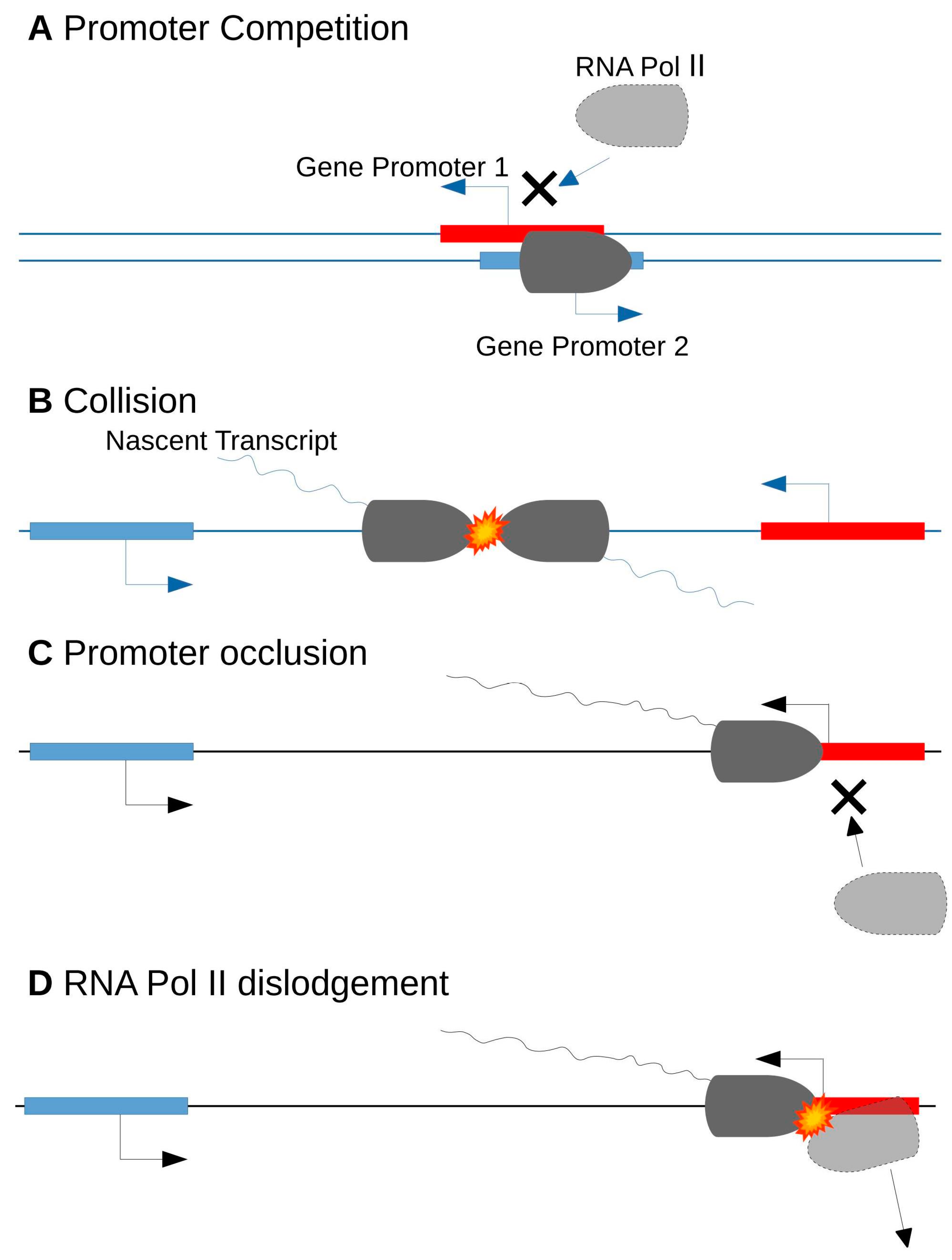
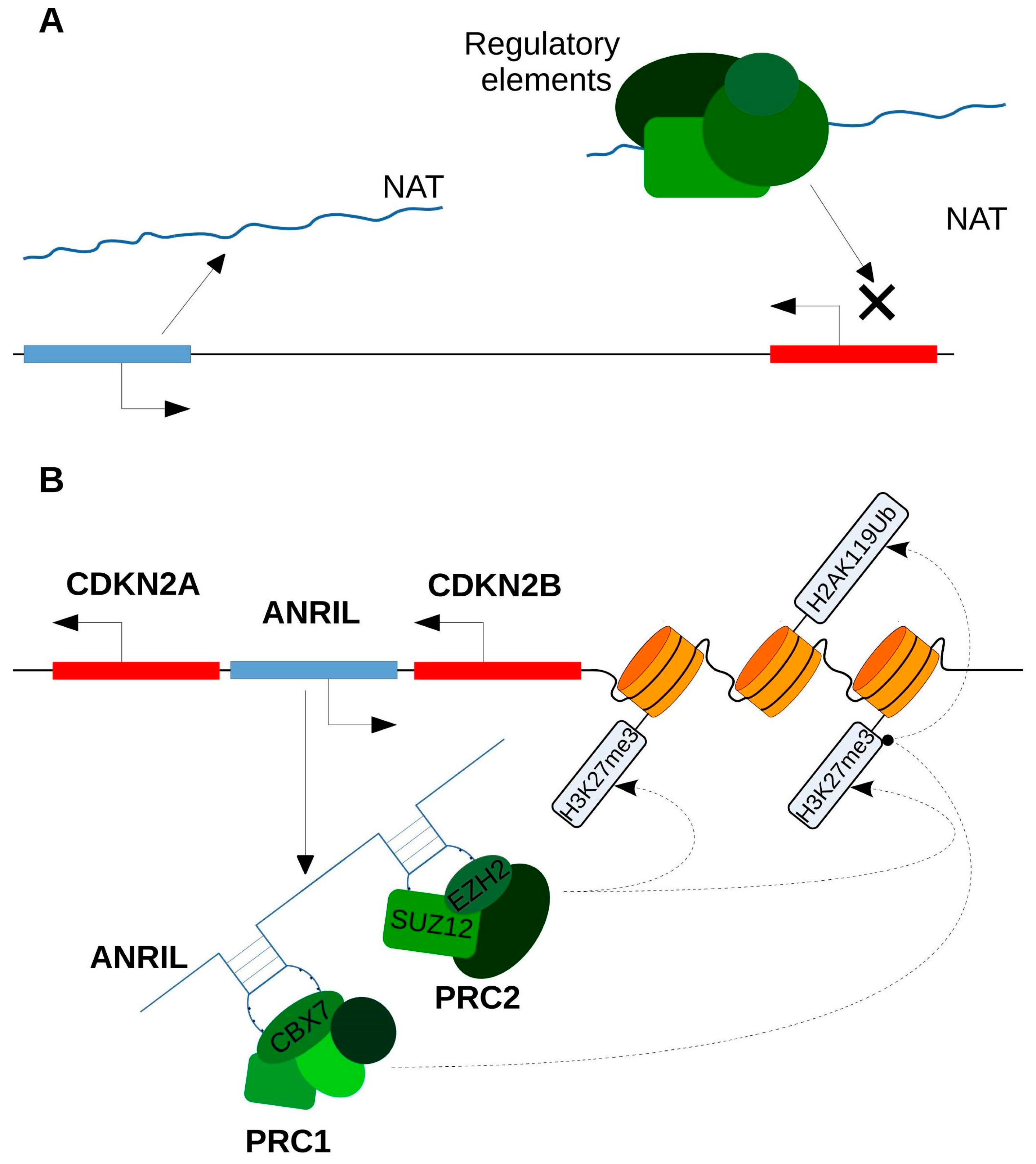
4.2. Transcriptional Interference
4.3. Chromatin Modification
4.3.1. Double-Stranded RNA/RNA Masking
4.3.2. Double-Stranded RNA/RNA A to I Editing
4.3.3. Double-Stranded RNA/RNA Interference
5. NATs in Breast Cancer
5.1. NATs as Cancer Biomarkers
5.2. NATs as Therapeutic Targets
6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| NAT | Natural antisense transcripts |
| nc | Non-coding |
| pc | Protein-coding |
| ds | Double-stranded |
| pcGene | Protein-coding gene |
| lncRNA | Long non-coding RNA |
| lincRNA | Long intergenic non-coding RNA |
| ncRNA | Non-coding RNA |
| mRNA | Messenger RNA |
| RNA Pol II | RNA polymerase II |
| PRC | Polycomb Repressive Complex |
| ASO | Antisense oligonucleotide |
References
- Carninci, P.; Kasukawa, T.; Katayama, S.; Gough, J.; Frith, M.C.; Maeda, N.; Oyama, R.; Ravasi, T.; Lenhard, B.; Wells, C.; et al. The transcriptional landscape of the mammalian genome. Science 2005, 309, 1559–1563. [Google Scholar] [CrossRef] [PubMed]
- Katayama, S.; Tomaru, Y.; Kasukawa, T.; Waki, K.; Nakanishi, M.; Nakamura, M.; Nishida, H.; Yap, C.C.; Suzuki, M.; Kawai, J.; et al. Antisense transcription in the mammalian transcriptome. Science 2005, 309, 1564–1566. [Google Scholar] [CrossRef] [PubMed]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef] [PubMed]
- ENCODE Project Consortium; Birney, E.; Stamatoyannopoulos, J.A.; Dutta, A.; Gingeras, T.R.; Margulies, E.H.; Weng, Z.; Snyder, M.; Dermitzakis, E.T.; Thurman, R.E.; et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 2007, 447, 799–816. [Google Scholar] [CrossRef] [PubMed]
- Nishizawa, M.; Ikeya, Y.; Okumura, T.; Kimura, T. Post-transcriptional inducible gene regulation by natural antisense RNA. Front. Biosci. 2015, 20, 1–36. [Google Scholar] [CrossRef]
- Kapranov, P.; Cheng, J.; Dike, S.; Nix, D.A.; Duttagupta, R.; Willingham, A.T.; Stadler, P.F.; Hertel, J.; Hackermüller, J.; Hofacker, I.L.; et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 2007, 316, 1484–1488. [Google Scholar] [CrossRef] [PubMed]
- Taft, R.J.; Pang, K.C.; Mercer, T.R.; Dinger, M.; Mattick, J.S. Non-coding RNAs: Regulators of disease. J. Pathol. 2010, 220, 126–139. [Google Scholar] [CrossRef] [PubMed]
- Palazzo, A.F.; Lee, E.S. Non-coding RNA: What is functional and what is junk? Front. Genet. 2015, 6, 2. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J.S.; Makunin, I.V. Non-coding RNA. Hum. Mol. Genet. 2006, 15, R17–R29. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Kellis, M.; Wold, B.; Snyder, M.P.; Bernstein, B.E.; Kundaje, A.; Marinov, G.K.; Ward, L.D.; Birney, E.; Crawford, G.E.; Dekker, J.; et al. Defining functional DNA elements in the human genome. Proc. Natl. Acad. Sci. USA 2014, 111, 6131–6138. [Google Scholar] [CrossRef] [PubMed]
- Frankish, A.; Uszczynska, B.; Ritchie, G.R.S.; Gonzalez, J.M.; Pervouchine, D.; Petryszak, R.; Mudge, J.M.; Fonseca, N.; Brazma, A.; Guigo, R.; et al. Comparison of GENCODE and RefSeq gene annotation and the impact of reference geneset on variant effect prediction. BMC Genom. 2015, 16 (Suppl. 8), S2. [Google Scholar] [CrossRef] [PubMed]
- Jalali, S.; Gandhi, S.; Scaria, V. Navigating the dynamic landscape of long noncoding RNA and protein-coding gene annotations in GENCODE. Hum. Genom. 2016, 10, 35. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Li, H.; Fang, S.; Kang, Y.; Wu, W.; Hao, Y.; Li, Z.; Bu, D.; Sun, N.; Zhang, M.Q.; et al. NONCODE 2016: An informative and valuable data source of long non-coding RNAs. Nucleic Acids Res. 2016, 44, D203–D208. [Google Scholar] [CrossRef] [PubMed]
- The RNAcentral Consortium. RNAcentral: A comprehensive database of non-coding RNA sequences. Nucleic Acids Res. 2017, 45, D128–D134. [Google Scholar] [CrossRef]
- Volders, P.-J.; Helsens, K.; Wang, X.; Menten, B.; Martens, L.; Gevaert, K.; Vandesompele, J.; Mestdagh, P. LNCipedia: A database for annotated human lncRNA transcript sequences and structures. Nucleic Acids Res. 2013, 41, D246–D251. [Google Scholar] [CrossRef] [PubMed]
- Wilusz, J.E.; Sunwoo, H.; Spector, D.L. Long noncoding RNAs: Functional surprises from the RNA world. Genes Dev. 2009, 23, 1494–1504. [Google Scholar] [CrossRef] [PubMed]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Bajic, V.B.; Zhang, Z. On the classification of long non-coding RNAs. RNA Biol. 2013, 10, 925–933. [Google Scholar] [CrossRef] [PubMed]
- Nakaya, H.I.; Amaral, P.P.; Louro, R.; Lopes, A.; Fachel, A.A.; Moreira, Y.B.; El-Jundi, T.A.; da Silva, A.M.; Reis, E.M.; Verjovski-Almeida, S. Genome mapping and expression analyses of human intronic noncoding RNAs reveal tissue-specific patterns and enrichment in genes related to regulation of transcription. Genome Biol. 2007, 8, R43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- St Laurent, G.; Savva, Y.A.; Kapranov, P. Dark matter RNA: An intelligent scaffold for the dynamic regulation of the nuclear information landscape. Front. Genet. 2012, 3, 57. [Google Scholar] [CrossRef] [PubMed]
- Gardner, E.J.; Nizami, Z.F.; Talbot, C.C., Jr.; Gall, J.G. Stable intronic sequence RNA (sisRNA), a new class of noncoding RNA from the oocyte nucleus of Xenopus tropicalis. Genes Dev. 2012, 26, 2550–2559. [Google Scholar] [CrossRef] [PubMed]
- Guttman, M.; Amit, I.; Garber, M.; French, C.; Lin, M.F.; Feldser, D.; Huarte, M.; Zuk, O.; Carey, B.W.; Cassady, J.P.; et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009, 458, 223–237. [Google Scholar] [CrossRef] [PubMed]
- Ponting, C.P.; Oliver, P.L.; Reik, W. Evolution and functions of long noncoding RNAs. Cell 2009, 136, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Amaral, P.P.; Clark, M.B.; Gascoigne, D.K.; Dinger, M.E.; Mattick, J.S. lncRNAdb: A reference database for long noncoding RNAs. Nucleic Acids Res. 2011, 39, D146–D151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- St Laurent, G.; Shtokalo, D.; Dong, B.; Tackett, M.R.; Fan, X.; Lazorthes, S.; Nicolas, E.; Sang, N.; Triche, T.J.; McCaffrey, T.A.; et al. VlincRNAs controlled by retroviral elements are a hallmark of pluripotency and cancer. Genome Biol. 2013, 14, R73. [Google Scholar] [CrossRef] [PubMed]
- Dinger, M.E.; Pang, K.C.; Mercer, T.R.; Mattick, J.S. Differentiating protein-coding and noncoding RNA: Challenges and ambiguities. PLoS Comput. Biol. 2008, 4, e1000176. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-L.; Carmichael, G.G. Long noncoding RNAs in mammalian cells: What, where, and why? Wiley Interdiscip. Rev. RNA 2010, 1, 2–21. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.-H.; Abdelmohsen, K.; Gorospe, M. Posttranscriptional gene regulation by long noncoding RNA. J. Mol. Biol. 2013, 425, 3723–3730. [Google Scholar] [CrossRef] [PubMed]
- Vance, K.W.; Ponting, C.P. Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet. 2014, 30, 348–355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dinger, M.E.; Amaral, P.P.; Mercer, T.R.; Pang, K.C.; Bruce, S.J.; Gardiner, B.B.; Askarian-Amiri, M.E.; Ru, K.; Soldà, G.; Simons, C.; et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008, 18, 1433–1445. [Google Scholar] [CrossRef] [PubMed]
- Maenner, S.; Müller, M.; Becker, P.B. Roles of long, non-coding RNA in chromosome-wide transcription regulation: Lessons from two dosage compensation systems. Biochimie 2012, 94, 1490–1498. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, M.; Kitagawa, K.; Kotake, Y.; Niida, H.; Ohhata, T. Cell cycle regulation by long non-coding RNAs. Cell. Mol. Life Sci. 2013, 70, 4785–4794. [Google Scholar] [CrossRef] [PubMed]
- Grammatikakis, I.; Panda, A.C.; Abdelmohsen, K.; Gorospe, M. Long noncoding RNAs(lncRNAs) and the molecular hallmarks of aging. Aging 2014, 6, 992–1009. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Dammert, M.A.; Hoppe, S.; Bierhoff, H.; Grummt, I. Heat shock represses rRNA synthesis by inactivation of TIF-IA and lncRNA-dependent changes in nucleosome positioning. Nucleic Acids Res. 2016, 44, 8144–8152. [Google Scholar] [CrossRef] [PubMed]
- Angrand, P.-O.; Vennin, C.; Le Bourhis, X.; Adriaenssens, E. The role of long non-coding RNAs in genome formatting and expression. Front. Genet. 2015, 6, 165. [Google Scholar] [CrossRef] [PubMed]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Tsuiji, H.; Yoshimoto, R.; Hasegawa, Y.; Furuno, M.; Yoshida, M.; Nakagawa, S. Competition between a noncoding exon and introns: Gomafu contains tandem UACUAAC repeats and associates with splicing factor-1. Genes Cells 2011, 16, 479–490. [Google Scholar] [CrossRef] [PubMed]
- Balbin, O.A.; Malik, R.; Dhanasekaran, S.M.; Prensner, J.R.; Cao, X.; Wu, Y.-M.; Robinson, D.; Wang, R.; Chen, G.; Beer, D.G.; et al. The landscape of antisense gene expression in human cancers. Genome Res. 2015, 25, 1068–1079. [Google Scholar] [CrossRef] [PubMed]
- Li, J.-T.; Zhang, Y.; Kong, L.; Liu, Q.-R.; Wei, L. Trans-natural antisense transcripts including noncoding RNAs in 10 species: Implications for expression regulation. Nucleic Acids Res. 2008, 36, 4833–4844. [Google Scholar] [CrossRef] [PubMed]
- Vanhée-Brossollet, C.; Vaquero, C. Do natural antisense transcripts make sense in eukaryotes? Gene 1998, 211, 1–9. [Google Scholar] [CrossRef]
- Seila, A.C.; Calabrese, J.M.; Levine, S.S.; Yeo, G.W.; Rahl, P.B.; Flynn, R.A.; Young, R.A.; Sharp, P.A. Divergent transcription from active promoters. Science 2008, 322, 1849–1851. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Zhang, L.; Luo, W.; Zhang, X. Characteristics of antisense transcript promoters and the regulation of their activity. Int. J. Mol. Sci. 2016, 17, 9. [Google Scholar] [CrossRef] [PubMed]
- Patton, J.; Block, S.; Coombs, C.; Martin, M.E. Identification of functional elements in the murine Gabp alpha/ATP synthase coupling factor 6 bi-directional promoter. Gene 2006, 369, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Schuettengruber, B.; Doetzlhofer, A.; Kroboth, K.; Wintersberger, E.; Seiser, C. Alternate activation of two divergently transcribed mouse genes from a bidirectional promoter is linked to changes in histone modification. J. Biol. Chem. 2003, 278, 1784–1793. [Google Scholar] [CrossRef] [PubMed]
- Grinchuk, O.V.; Motakis, E.; Yenamandra, S.P.; Ow, G.S.; Jenjaroenpun, P.; Tang, Z.; Yarmishyn, A.A.; Ivshina, A.V.; Kuznetsov, V.A. Sense-antisense gene-pairs in breast cancer and associated pathological pathways. Oncotarget 2015, 6, 42197–42221. [Google Scholar] [CrossRef] [PubMed]
- Whitehouse, I.; Rando, O.J.; Delrow, J.; Tsukiyama, T. Chromatin remodelling at promoters suppresses antisense transcription. Nature 2007, 450, 1031–1035. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.; Xu, Z.; Clauder-Münster, S.; Steinmetz, L.M.; Buratowski, S. Set3 HDAC mediates effects of overlapping noncoding transcription on gene induction kinetics. Cell 2012, 150, 1158–1169. [Google Scholar] [CrossRef] [PubMed]
- Giannakakis, A.; Zhang, J.; Jenjaroenpun, P.; Nama, S.; Zainolabidin, N.; Aau, M.Y.; Yarmishyn, A.A.; Vaz, C.; Ivshina, A.V.; Grinchuk, O.V.; et al. Contrasting expression patterns of coding and noncoding parts of the human genome upon oxidative stress. Sci. Rep. 2015, 5, 9737. [Google Scholar] [CrossRef] [PubMed]
- Wenric, S.; ElGuendi, S.; Caberg, J.-H.; Bezzaou, W.; Fasquelle, C.; Charloteaux, B.; Karim, L.; Hennuy, B.; Frères, P.; Collignon, J.; et al. Transcriptome-wide analysis of natural antisense transcripts shows their potential role in breast cancer. Sci. Rep. 2017, 7, 17452. [Google Scholar] [CrossRef] [PubMed]
- Pelechano, V.; Steinmetz, L.M. Gene regulation by antisense transcription. Nat. Rev. Genet. 2013, 14, 880–893. [Google Scholar] [CrossRef] [PubMed]
- Rosikiewicz, W.; Makałowska, I. Biological functions of natural antisense transcripts. Acta Biochim. Pol. 2016, 63, 665–673. [Google Scholar] [CrossRef] [PubMed]
- Makalowska, I.; Lin, C.-F.; Makalowski, W. Overlapping genes in vertebrate genomes. Comput. Biol. Chem. 2005, 29, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Shearwin, K.E.; Callen, B.P.; Egan, J.B. Transcriptional interference--a crash course. Trends Genet. 2005, 21, 339–345. [Google Scholar] [CrossRef] [PubMed]
- Stojic, L.; Niemczyk, M.; Orjalo, A.; Ito, Y.; Ruijter, A.E.M.; Uribe-Lewis, S.; Joseph, N.; Weston, S.; Menon, S.; Odom, D.T.; et al. Transcriptional silencing of long noncoding RNA GNG12-AS1 uncouples its transcriptional and product-related functions. Nat. Commun. 2016, 7, 10406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oeder, S.; Mages, J.; Flicek, P.; Lang, R. Uncovering information on expression of natural antisense transcripts in Affymetrix MOE430 datasets. BMC Genom. 2007, 8, 200. [Google Scholar] [CrossRef] [PubMed]
- Grigoriadis, A.; Oliver, G.R.; Tanney, A.; Kendrick, H.; Smalley, M.J.; Jat, P.; Neville, A.M. Identification of differentially expressed sense and antisense transcript pairs in breast epithelial tissues. BMC Genom. 2009, 10, 324. [Google Scholar] [CrossRef] [PubMed]
- Conley, A.B.; Jordan, I.K. Epigenetic regulation of human cis-natural antisense transcripts. Nucleic Acids Res. 2012, 40, 1438–1445. [Google Scholar] [CrossRef] [PubMed]
- Ling, M.H.T.; Ban, Y.; Wen, H.; Wang, S.M.; Ge, S.X. Conserved expression of natural antisense transcripts in mammals. BMC Genom. 2013, 14, 243. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Sun, H.; Wang, H. Long noncoding RNAs in DNA methylation: New players stepping into the old game. Cell Biosci. 2016, 6, 45. [Google Scholar] [CrossRef] [PubMed]
- Khorkova, O.; Myers, A.J.; Hsiao, J.; Wahlestedt, C. Natural antisense transcripts. Hum. Mol. Genet. 2014, 23, R54–R63. [Google Scholar] [CrossRef] [PubMed]
- Davidovich, C.; Cech, T.R. The recruitment of chromatin modifiers by long noncoding RNAs: Lessons from PRC2. RNA 2015, 21, 2007–2022. [Google Scholar] [CrossRef] [PubMed]
- Meseure, D.D.; Vacher, S.; Alsibai, K.D.; Nicolas, A.; Chemlali, W.; Caly, M.; Lidereau, R.; Pasmant, E.; Callens, C.; Bieche, I. Expression of ANRIL-Polycomb Complexes-CDKN2A/B/ARF Genes in Breast Tumors: Identification of a Two-Gene (EZH2/CBX7) Signature with Independent Prognostic Value. Mol. Cancer Res. 2016, 14, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Kotake, Y.; Nakagawa, T.; Kitagawa, K.; Suzuki, S.; Liu, N.; Kitagawa, M.; Xiong, Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene 2011, 30, 1956–1962. [Google Scholar] [CrossRef] [PubMed]
- Aguilo, F.; Zhou, M.-M.; Walsh, M.J. Long noncoding RNA, polycomb, and the ghosts haunting INK4b-ARF-INK4a expression. Cancer Res. 2011, 71, 5365–5369. [Google Scholar] [CrossRef] [PubMed]
- Congrains, A.; Kamide, K.; Ohishi, M.; Rakugi, H. ANRIL: Molecular mechanisms and implications in human health. Int. J. Mol. Sci. 2013, 14, 1278–1292. [Google Scholar] [CrossRef] [PubMed]
- Deng, Q.; Sun, H.; He, B.; Pan, Y.; Gao, T.; Chen, J.; Ying, H.; Liu, X.; Wang, F.; Xu, Y.; et al. Prognostic value of long non-coding RNA HOTAIR in various cancers. PLoS ONE 2014, 9, e110059. [Google Scholar] [CrossRef] [PubMed]
- Bhan, A.; Mandal, S.S. LncRNA HOTAIR: A master regulator of chromatin dynamics and cancer. Biochim. Biophys. Acta 2015, 1856, 151–164. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Wang, X.; Fu, C.; Wang, X.; Zou, J.; Hua, H.; Bi, Z. Long noncoding RNA FGFR3-AS1 promotes osteosarcoma growth through regulating its natural antisense transcript FGFR3. Mol. Biol. Rep. 2016, 43, 427–436. [Google Scholar] [CrossRef] [PubMed]
- Zong, X.; Nakagawa, S.; Freier, S.M.; Fei, J.; Ha, T.; Prasanth, S.G.; Prasanth, K.V. Natural antisense RNA promotes 3′ end processing and maturation of MALAT1 lncRNA. Nucleic Acids Res. 2016, 44, 2898–2908. [Google Scholar] [CrossRef] [PubMed]
- Beltran, M.; Puig, I.; Peña, C.; García, J.M.; Alvarez, A.B.; Peña, R.; Bonilla, F.; de Herreros, A.G. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev. 2008, 22, 756–769. [Google Scholar] [CrossRef] [PubMed]
- Nishikura, K. Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 2010, 79, 321–349. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Carmichael, G.G. The fate of dsRNA in the nucleus: A p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell 2001, 106, 465–475. [Google Scholar] [CrossRef]
- Levanon, E.Y.; Eisenberg, E.; Yelin, R.; Nemzer, S.; Hallegger, M.; Shemesh, R.; Fligelman, Z.Y.; Shoshan, A.; Pollock, S.R.; Sztybel, D.; et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol. 2004, 22, 1001–1005. [Google Scholar] [CrossRef] [PubMed]
- Ramaswami, G.; Li, J.B. RADAR: A rigorously annotated database of A-to-I RNA editing. Nucleic Acids Res. 2014, 42, D109–D113. [Google Scholar] [CrossRef] [PubMed]
- Neeman, Y.; Dahary, D.; Levanon, E.Y.; Sorek, R.; Eisenberg, E. Is there any sense in antisense editing? Trends Genet. 2005, 21, 544–547. [Google Scholar] [CrossRef] [PubMed]
- Salameh, A.; Lee, A.K.; Cardó-Vila, M.; Nunes, D.N.; Efstathiou, E.; Staquicini, F.I.; Dobroff, A.S.; Marchiò, S.; Navone, N.M.; Hosoya, H.; et al. PRUNE2 is a human prostate cancer suppressor regulated by the intronic long noncoding RNA PCA3. Proc. Natl. Acad. Sci. USA 2015, 112, 8403–8408. [Google Scholar] [CrossRef] [PubMed]
- Hannon, G.J. RNA interference. Nature 2002, 418, 244–251. [Google Scholar] [CrossRef] [PubMed]
- Czech, B.; Hannon, G.J. Small RNA sorting: Matchmaking for Argonautes. Nat. Rev. Genet. 2011, 12, 19–31. [Google Scholar] [CrossRef] [PubMed]
- Polikepahad, S.; Corry, D.B. Profiling of T helper cell-derived small RNAs reveals unique antisense transcripts and differential association of miRNAs with argonaute proteins 1 and 2. Nucleic Acids Res. 2013, 41, 1164–1177. [Google Scholar] [CrossRef] [PubMed]
- Werner, A.; Cockell, S.; Falconer, J.; Carlile, M.; Alnumeir, S.; Robinson, J. Contribution of natural antisense transcription to an endogenous siRNA signature in human cells. BMC Genom. 2014, 15, 19. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Meng, Y.; Zuo, Z.; Xue, J.; Wang, H. NATpipe: An integrative pipeline for systematical discovery of natural antisense transcripts (NATs) and phase-distributed nat-siRNAs from de novo assembled transcriptomes. Sci. Rep. 2016, 6, 21666. [Google Scholar] [CrossRef] [PubMed]
- Beckedorff, F.C.; Ayupe, A.C.; Crocci-Souza, R.; Amaral, M.S.; Nakaya, H.I.; Soltys, D.T.; Menck, C.F.M.; Reis, E.M.; Verjovski-Almeida, S. The intronic long noncoding RNA ANRASSF1 recruits PRC2 to the RASSF1A promoter, reducing the expression of RASSF1A and increasing cell proliferation. PLoS Genet. 2013, 9, e1003705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sang, Y.; Tang, J.; Li, S.; Li, L.; Tang, X.; Cheng, C.; Luo, Y.; Qian, X.; Deng, L.-M.; Liu, L.; et al. LncRNA PANDAR regulates the G1/S transition of breast cancer cells by suppressing p16(INK4A) expression. Sci. Rep. 2016, 6, 22366. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Lv, F.; Liang, D.; Yang, Q.; Zhang, B.; Lin, H.; Wang, X.; Qian, G.; Xu, J.; You, W. HOTAIR may regulate proliferation, apoptosis, migration and invasion of MCF-7 cells through regulating the P53/Akt/JNK signaling pathway. Biomed. Pharmacother. 2017, 90, 555–561. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Li, Y.; Yang, B. Downregulated long non-coding RNA MEG3 in breast cancer regulates proliferation, migration and invasion by depending on p53’s transcriptional activity. Biochem. Biophys. Res. Commun. 2016, 478, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.-Y.; Yu, M.-S.; Li, X.; Zhang, Z.; Han, C.-R.; Yan, B. Overexpression of long non-coding RNA MEG3 suppresses breast cancer cell proliferation, invasion, and angiogenesis through AKT pathway. Tumour. Biol. 2017, 39. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Yang, M.; Jiang, R.; An, N.; Wang, X.; Liu, B. Long non-coding RNA HOTAIR regulates the proliferation, self-renewal capacity, tumor formation and migration of the cancer stem-like cell (CSC) subpopulation enriched from breast cancer cells. PLoS ONE 2017, 12, e0170860. [Google Scholar] [CrossRef] [PubMed]
- Jene-Sanz, A.; Váraljai, R.; Vilkova, A.V.; Khramtsova, G.F.; Khramtsov, A.I.; Olopade, O.I.; Lopez-Bigas, N.; Benevolenskaya, E.V. Expression of polycomb targets predicts breast cancer prognosis. Mol. Cell. Biol. 2013, 33, 3951–3961. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-M.; Liu, Y.; Wei, H.-Y.; Lv, K.-Z.; Fu, P. Linc-ROR induces epithelial-mesenchymal transition and contributes to drug resistance and invasion of breast cancer cells. Tumour. Biol. 2016, 37, 10861–10870. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Ye, X.-L.; Xu, J.; Cao, M.-G.; Fang, Z.-Y.; Li, L.-Y.; Guan, G.-H.; Liu, Q.; Qian, Y.-H.; Xie, D. The lncRNA H19 mediates breast cancer cell plasticity during EMT and MET plasticity by differentially sponging miR-200b/c and let-7b. Sci. Signal. 2017, 10. [Google Scholar] [CrossRef] [PubMed]
- Hou, P.; Zhao, Y.; Li, Z.; Yao, R.; Ma, M.; Gao, Y.; Zhao, L.; Zhang, Y.; Huang, B.; Lu, J. LincRNA-ROR induces epithelial-to-mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell Death Dis. 2014, 5, e1287. [Google Scholar] [CrossRef] [PubMed]
- Zou, Q.; Zhou, E.; Xu, F.; Zhang, D.; Yi, W.; Yao, J. A TP73-AS1/miR-200a/ZEB1 regulating loop promotes breast cancer cell invasion and migration. J. Cell. Biochem. 2017. [CrossRef] [PubMed]
- Chi, Y.; Huang, S.; Yuan, L.; Liu, M.; Huang, N.; Zhou, S.; Zhou, B.; Wu, J. Role of BC040587 as a predictor of poor outcome in breast cancer. Cancer Cell Int. 2014, 14, 123. [Google Scholar] [CrossRef] [PubMed]
- Xing, Z.; Lin, C.; Yang, L. Unraveling the therapeutic potential of the LncRNA-dependent noncanonical Hedgehog pathway in cancer. Mol. Cell. Oncol. 2015, 2, e998900. [Google Scholar] [CrossRef] [PubMed]
- Xing, Z.; Park, P.K.; Lin, C.; Yang, L. LncRNA BCAR4 wires up signaling transduction in breast cancer. RNA Biol. 2015, 12, 681–689. [Google Scholar] [CrossRef] [PubMed]
- Booy, E.P.; McRae, E.K.; Koul, A.; Lin, F.; McKenna, S.A. The long non-coding RNA BC200 (BCYRN1) is critical for cancer cell survival and proliferation. Mol. Cancer 2017, 16, 109. [Google Scholar] [CrossRef] [PubMed]
- Redis, R.S.; Sieuwerts, A.M.; Look, M.P.; Tudoran, O.; Ivan, C.; Spizzo, R.; Zhang, X.; de Weerd, V.; Shimizu, M.; Ling, H.; et al. CCAT2, a novel long non-coding RNA in breast cancer: Expression study and clinical correlations. Oncotarget 2013, 4, 1748–1762. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Zhao, Y.; Wu, X.; Song, G. Upregulation of CCAT2 promotes cell proliferation by repressing the P15 in breast cancer. Biomed. Pharmacother. 2017, 91, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.-J.; Li, Y.; Wu, Y.-Z.; Wang, Y.; Nian, W.-Q.; Wang, L.-L.; Li, L.-C.; Luo, H.-L.; Wang, D.-L. Long non-coding RNA CCAT2 promotes the breast cancer growth and metastasis by regulating TGF-beta signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 706–714. [Google Scholar] [PubMed]
- Cai, Y.; He, J.; Zhang, D. Long noncoding RNA CCAT2 promotes breast tumor growth by regulating the Wnt signaling pathway. OncoTargets Ther. 2015, 8, 2657–2664. [Google Scholar] [CrossRef]
- Sarrafzadeh, S.; Geranpayeh, L.; Tasharrofi, B.; Soudyab, M.; Nikpayam, E.; Iranpour, M.; Mirfakhraie, R.; Gharesouran, J.; Ghafouri-Fard, S.; Ghafouri-Fard, S. Expression study and clinical correlations of MYC and CCAT2 in breast cancer patients. Iran. Biomed. J. 2017, 21, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Huan, J.; Xing, L.; Lin, Q.; Xui, H.; Qin, X. Long noncoding RNA CRNDE activates Wnt/beta-catenin signaling pathway through acting as a molecular sponge of microRNA-136 in human breast cancer. Am. J. Transl. Res. 2017, 9, 1977–1989. [Google Scholar] [PubMed]
- Sha, S.; Yuan, D.; Liu, Y.; Han, B.; Zhong, N. Targeting long non-coding RNA DANCR inhibits triple negative breast cancer progression. Biol. Open 2017, 6, 1310–1316. [Google Scholar] [CrossRef] [PubMed]
- Miano, V.; Ferrero, G.; Reineri, S.; Caizzi, L.; Annaratone, L.; Ricci, L.; Cutrupi, S.; Castellano, I.; Cordero, F.; de Bortoli, M. Luminal long non-coding RNAs regulated by estrogen receptor alpha in a ligand-independent manner show functional roles in breast cancer. Oncotarget 2015, 7, 3201–3216. [Google Scholar] [CrossRef] [PubMed]
- Niknafs, Y.S.; Han, S.; Ma, T.; Speers, C.; Zhang, C.; Wilder-Romans, K.; Iyer, M.K.; Pitchiaya, S.; Malik, R.; Hosono, Y.; et al. The lncRNA landscape of breast cancer reveals a role for DSCAM-AS1 in breast cancer progression. Nat. Commun. 2016, 7, 12791. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Lv, S.-X.; Lv, L.; Liu, Y.-H.; Dong, S.-Y.; Yao, Z.-H.; Dai, X.-X.; Zhang, X.-H.; Wang, O.-C. Identification of lncRNA FAM83H-AS1 as a novel prognostic marker in luminal subtype breast cancer. OncoTargets Ther. 2016, 9, 7039–7045. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Song, W.-Q.; Sun, P.; Jin, L.; Dai, H.-Y. LncRNA-GAS5 induces PTEN expression through inhibiting miR-103 in endometrial cancer cells. J. Biomed. Sci. 2015, 22, 100. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Li, Z. Long non-coding RNA growth arrest-specific transcript 5 in tumor biology. Oncol. Lett. 2015, 10, 1953–1958. [Google Scholar] [CrossRef] [PubMed]
- Pickard, M.R.; Williams, G.T. The hormone response element mimic sequence of GAS5 lncRNA is sufficient to induce apoptosis in breast cancer cells. Oncotarget 2016, 7, 10104–10116. [Google Scholar] [CrossRef] [PubMed]
- Mourtada-Maarabouni, M.; Pickard, M.R.; Hedge, V.L.; Farzaneh, F.; Williams, G.T. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene 2009, 28, 195–208. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zhai, L.; Wang, H.; Liu, C.; Zhang, J.; Chen, W.; Wei, Q. Downregulation of LncRNA GAS5 causes trastuzumab resistance in breast cancer. Oncotarget 2016, 7, 27778–27786. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Ma, P.; Liu, S.-M.; Zhou, X. Circulating long noncoding RNA GAS5 as a potential biomarker in breast cancer for assessing the surgical effects. Tumour. Biol. 2016, 37, 6847–6854. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, Y.; Li, Y.; Ren, K.; Li, X.; Han, X.; Wang, J. Long non-coding RNA H19 promotes the proliferation and invasion of breast cancer through upregulating DNMT1 expression by sponging miR-152. J. Biochem. Mol. Toxicol. 2017, 31. [Google Scholar] [CrossRef] [PubMed]
- Si, X.; Zang, R.; Zhang, E.; Liu, Y.; Shi, X.; Zhang, E.; Shao, L.; Li, A.; Yang, N.; Han, X.; et al. LncRNA H19 confers chemoresistance in ERalpha-positive breast cancer through epigenetic silencing of the pro-apoptotic gene BIK. Oncotarget 2016, 7, 81452–81462. [Google Scholar] [CrossRef] [PubMed]
- Vennin, C.; Spruyt, N.; Dahmani, F.; Julien, S.; Bertucci, F.; Finetti, P.; Chassat, T.; Bourette, R.P.; Le Bourhis, X.; Adriaenssens, E. H19 non coding RNA-derived miR-675 enhances tumorigenesis and metastasis of breast cancer cells by downregulating c-Cbl and Cbl-b. Oncotarget 2015, 6, 29209–29223. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Luo, Z.; Zhang, Y.; Zhang, L.; Wu, L.; Liu, L.; Yang, J.; Song, X.; Liu, J. Circulating lncRNA H19 in plasma as a novel biomarker for breast cancer. Cancer Biomark. 2016, 17, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.-Z.; Liu, Y.-R.; Xu, X.-E.; Jin, X.; Hu, X.; Yu, K.-D.; Shao, Z.-M. Transcriptome analysis of triple-negative breast cancer reveals an integrated mRNA-lncRNA signature with predictive and prognostic value. Cancer Res. 2016, 76, 2105–2114. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Xing, L.-Q.; Liu, Y.-J. A three-long noncoding RNA signature as a diagnostic biomarker for differentiating between triple-negative and non-triple-negative breast cancers. Medicine 2017, 96, e6222. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Ma, J.; Cheng, Y.; Zhang, H.; Luo, W.; Zhang, H. HMMR antisense RNA 1, a novel long noncoding RNA, regulates the progression of basal-like breast cancer cells. Breast Cancer 2016, 8, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Bhan, A.; Hussain, I.; Ansari, K.I.; Kasiri, S.; Bashyal, A.; Mandal, S.S. Antisense transcript long noncoding RNA (lncRNA) HOTAIR is transcriptionally induced by estradiol. J. Mol. Biol. 2013, 425, 3707–3722. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.-C.; Hung, T.; Argani, P.; Rinn, J.L.; et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010, 464, 1071–1076. [Google Scholar] [CrossRef] [PubMed]
- Kogo, R.; Shimamura, T.; Mimori, K.; Kawahara, K.; Imoto, S.; Sudo, T.; Tanaka, F.; Shibata, K.; Suzuki, A.; Komune, S.; et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011, 71, 6320–6326. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.-H.; Abdelmohsen, K.; Kim, J.; Yang, X.; Martindale, J.L.; Tominaga-Yamanaka, K.; White, E.J.; Orjalo, A.V.; Rinn, J.L.; Kreft, S.G.; et al. Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nat. Commun. 2013, 4, 2939. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.-J.; Zhang, Y.; Luo, Z.-L.; Liu, L.; Yang, J.; Wu, L.-C.; Yu, S.-S.; Liu, J.-B. Long non-coding RNA HOTAIR in plasma as a potential biomarker for breast cancer diagnosis. Nan Fang Yi Ke Da Xue Xue Bao 2016, 36, 488–492. [Google Scholar] [PubMed]
- Ozes, A.R.; Wang, Y.; Zong, X.; Fang, F.; Pilrose, J.; Nephew, K.P. Therapeutic targeting using tumor specific peptides inhibits long non-coding RNA HOTAIR activity in ovarian and breast cancer. Sci. Rep. 2017, 7, 894. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.Q.D.; Dostie, J. Reciprocal regulation of chromatin state and architecture by HOTAIRM1 contributes to temporal collinear HOXA gene activation. Nucleic Acids Res. 2017, 45, 1091–1104. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Malouf, G.G.; Chen, Y.; Zhang, J.; Yao, H.; Valero, V.; Weinstein, J.N.; Spano, J.-P.; Meric-Bernstam, F.; Khayat, D.; et al. Comprehensive analysis of long non-coding RNAs in human breast cancer clinical subtypes. Oncotarget 2014, 5, 9864–9876. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Wang, J.; Wu, F.; Song, Y.; Zhao, S.; Zhang, Q. Long non-coding RNA HOXA-AS2 promotes proliferation and invasion of breast cancer by acting as a miR-520c-3p sponge. Oncotarget 2017, 8, 46090–46103. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Jia, G.; Qu, Y.; Du, Q.; Liu, B.; Liu, B. Long Non-Coding RNA (LncRNA) HOXA11-AS Promotes Breast Cancer Invasion and Metastasis by Regulating Epithelial-Mesenchymal Transition. Med. Sci. Monit. 2017, 23, 3393–3403. [Google Scholar] [CrossRef] [PubMed]
- Li, W.-X.; Sha, R.-L.; Bao, J.-Q.; Luan, W.; Su, R.-L.; Sun, S.-R. Expression of long non-coding RNA linc-ITGB1 in breast cancer and its influence on prognosis and survival. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 3397–3401. [Google Scholar] [PubMed]
- Yan, M.; Zhang, L.; Li, G.; Xiao, S.; Dai, J.; Cen, X. Long noncoding RNA linc-ITGB1 promotes cell migration and invasion in human breast cancer. Biotechnol. Appl. Biochem. 2017, 64, 5–13. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Li, C.; Chen, J.; Zhang, K.; Chu, X.; Wang, R.; Chen, L. The Emerging Roles of Long Noncoding RNA ROR (lincRNA-ROR) and its Possible Mechanisms in Human Cancers. Cell. Physiol. Biochem. 2016, 40, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Katsaros, D.; Loo, L.W.M.; Hernandez, B.Y.; Chong, C.; Canuto, E.M.; Biglia, N.; Lu, L.; Risch, H.; Chu, W.-M.; et al. Prognostic and predictive values of long non-coding RNA LINC00472 in breast cancer. Oncotarget 2015, 6, 8579–8592. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Wang, Z.; Loo, L.W.M.; Ni, Y.; Jia, W.; Fei, P.; Risch, H.A.; Katsaros, D.; Yu, H. LINC00472 expression is regulated by promoter methylation and associated with disease-free survival in patients with grade 2 breast cancer. Breast Cancer Res. Treat. 2015, 154, 473–482. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.M.; Boczek, N.J.; Berres, M.W.; Ma, X.; Smith, D.I. LSINCT5 is over expressed in breast and ovarian cancer and affects cellular proliferation. RNA Biol. 2011, 8, 496–505. [Google Scholar] [CrossRef] [PubMed]
- Jadaliha, M.; Zong, X.; Malakar, P.; Ray, T.; Singh, D.K.; Freier, S.M.; Jensen, T.; Prasanth, S.G.; Karni, R.; Ray, P.S.; et al. Functional and prognostic significance of long non-coding RNA MALAT1 as a metastasis driver in ER negative lymph node negative breast cancer. Oncotarget 2016, 7, 40418–40436. [Google Scholar] [CrossRef] [PubMed]
- Gutschner, T.; Hammerle, M.; Diederichs, S. MALAT1—A paradigm for long noncoding RNA function in cancer. J. Mol. Med. 2013, 91, 791–801. [Google Scholar] [CrossRef] [PubMed]
- Huang, N.-S.; Chi, Y.-Y.; Xue, J.-Y.; Liu, M.-Y.; Huang, S.; Mo, M.; Zhou, S.-L.; Wu, J. Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 (MALAT1) interacts with estrogen receptor and predicted poor survival in breast cancer. Oncotarget 2016, 7, 37957–37965. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Fan, R.; Chen, L.; Qian, H. Clinical Significance of Long Non-coding RNA MALAT1 Expression in Tissue and Serum of Breast Cancer. Ann. Clin. Lab. Sci. 2016, 46, 418–424. [Google Scholar] [PubMed]
- Zhang, W.; Shi, S.; Jiang, J.; Li, X.; Lu, H.; Ren, F. LncRNA MEG3 inhibits cell epithelial-mesenchymal transition by sponging miR-421 targeting E-cadherin in breast cancer. Biomed. Pharmacother. 2017, 91, 312–319. [Google Scholar] [CrossRef] [PubMed]
- Mondal, T.; Subhash, S.; Vaid, R.; Enroth, S.; Uday, S.; Reinius, B.; Mitra, S.; Mohammed, A.; James, A.R.; Hoberg, E.; et al. MEG3 long noncoding RNA regulates the TGF-β pathway genes through formation of RNA-DNA triplex structures. Nat. Commun. 2015, 6, 7743. [Google Scholar] [CrossRef] [PubMed]
- Nie, F.-Q.; Ma, S.; Xie, M.; Liu, Y.-W.; De, W.; Liu, X.-H. Decreased long noncoding RNA MIR31HG is correlated with poor prognosis and contributes to cell proliferation in gastric cancer. Tumour. Biol. 2016, 37, 7693–7701. [Google Scholar] [CrossRef] [PubMed]
- Augoff, K.; McCue, B.; Plow, E.F.; Sossey-Alaoui, K. miR-31 and its host gene lncRNA LOC554202 are regulated by promoter hypermethylation in triple-negative breast cancer. Mol. Cancer 2012, 11, 5. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Lu, B.; Wang, J.; Wang, J.; Shi, Y.; Lian, Y.; Zhu, Y.; Wang, J.; Fan, Y.; Wang, Z.; et al. Long non-coding RNA Loc554202 induces apoptosis in colorectal cancer cells via the caspase cleavage cascades. J. Exp. Clin. Cancer Res. 2015, 34, 100. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zhang, Z.; Liu, X.; Cheng, X.; Zhang, Y.; Han, X.; Zhang, Y.; Liu, S.; Yang, J.; Xu, B.; et al. The FOXN3-NEAT1-SIN3A repressor complex promotes progression of hormonally responsive breast cancer. J. Clin. Investig. 2017, 127, 3421–3440. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Wu, W.-B.; Wang, Z.-W.; Wang, X.-H. lncRNA NEAT1 is closely related with progression of breast cancer via promoting proliferation and EMT. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 1020–1026. [Google Scholar] [PubMed]
- Hirose, T.; Virnicchi, G.; Tanigawa, A.; Naganuma, T.; Li, R.; Kimura, H.; Yokoi, T.; Nakagawa, S.; Benard, M.; Fox, A.H.; et al. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol. Biol. Cell 2014, 25, 169–183. [Google Scholar] [CrossRef] [PubMed]
- Qian, K.; Liu, G.; Tang, Z.; Hu, Y.; Fang, Y.; Chen, Z.; Xu, X. The long non-coding RNA NEAT1 interacted with miR-101 modulates breast cancer growth by targeting EZH2. Arch. Biochem. Biophys. 2017, 615, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Shao, C.; Wu, Q.-J.; Chen, G.; Zhou, J.; Yang, B.; Li, H.; Gou, L.-T.; Zhang, Y.; Wang, Y.; et al. NEAT1 scaffolds RNA-binding proteins and the Microprocessor to globally enhance pri-miRNA processing. Nat. Struct. Mol. Biol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Ke, H.; Zhao, L.; Feng, X.; Xu, H.; Zou, L.; Yang, Q.; Su, X.; Peng, L.; Jiao, B. NEAT1 is Required for Survival of Breast Cancer Cells Through FUS and miR-548. Gene Regul. Syst. Biol. 2016, 10, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Iranpour, M.; Soudyab, M.; Geranpayeh, L.; Mirfakhraie, R.; Azargashb, E.; Movafagh, A.; Ghafouri-Fard, S. Expression analysis of four long noncoding RNAs in breast cancer. Tumour. Biol. 2016, 37, 2933–2940. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Luo, J.; Jiao, S. Comprehensive characterization of cancer subtype associated long non-coding RNAs and their clinical implications. Sci. Rep. 2014, 4, 6591. [Google Scholar] [CrossRef] [PubMed]
- Colombo, T.; Farina, L.; Macino, G.; Paci, P. PVT1: A rising star among oncogenic long noncoding RNAs. BioMed Res. Int. 2015, 2015, 304208. [Google Scholar] [CrossRef] [PubMed]
- Tseng, Y.-Y.; Bagchi, A. The PVT1-MYC duet in cancer. Mol. Cell. Oncol. 2015, 2, e974467. [Google Scholar] [CrossRef] [PubMed]
- Sarver, A.L.; Murray, C.D.; Temiz, N.A.; Tseng, Y.-Y.; Bagchi, A. MYC and PVT1 synergize to regulate RSPO1 levels in breast cancer. Cell Cycle 2016, 15, 881–885. [Google Scholar] [CrossRef] [PubMed]
- Tseng, Y.-Y.; Moriarity, B.S.; Gong, W.; Akiyama, R.; Tiwari, A.; Kawakami, H.; Ronning, P.; Reuland, B.; Guenther, K.; Beadnell, T.C.; et al. PVT1 dependence in cancer with MYC copy-number increase. Nature 2014, 512, 82–86. [Google Scholar] [CrossRef] [PubMed]
- Shahryari, A.; Jazi, M.S.; Samaei, N.M.; Mowla, S.J. Long non-coding RNA SOX2OT: Expression signature, splicing patterns, and emerging roles in pluripotency and tumorigenesis. Front. Genet. 2015, 6, 196. [Google Scholar] [CrossRef] [PubMed]
- Askarian-Amiri, M.E.; Seyfoddin, V.; Smart, C.E.; Wang, J.; Kim, J.E.; Hansji, H.; Baguley, B.C.; Finlay, G.J.; Leung, E.Y. Emerging role of long non-coding RNA SOX2OT in SOX2 regulation in breast cancer. PLoS ONE 2014, 9, e102140. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Li, J.; Liu, Y.; Ding, J.; Fan, Y.; Tian, Y.; Wang, L.; Lian, Y.; Wang, K.; Shu, Y. The long noncoding RNA SPRY4-IT1 increases the proliferation of human breast cancer cells by upregulating ZNF703 expression. Mol. Cancer 2015, 14, 51. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Chen, Y.; Chen, Z.; He, A.; Xie, H.; Zhang, Q.; Cai, Z.; Liu, Y.; Huang, W. SPRY4-IT1: A novel oncogenic long non-coding RNA in human cancers. Tumour. Biol. 2017, 39, 1010428317711406. [Google Scholar] [CrossRef] [PubMed]
- Cusanelli, E.; Chartrand, P. Telomeric repeat-containing RNA TERRA: A noncoding RNA connecting telomere biology to genome integrity. Front. Genet. 2015, 6, 143. [Google Scholar] [CrossRef] [PubMed]
- Azzalin, C.M.; Reichenbach, P.; Khoriauli, L.; Giulotto, E.; Lingner, J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 2007, 318, 798–801. [Google Scholar] [CrossRef] [PubMed]
- Luke, B.; Lingner, J. TERRA: Telomeric repeat-containing RNA. EMBO J. 2009, 28, 2503–2510. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.; Norseen, J.; Wiedmer, A.; Riethman, H.; Lieberman, P.M. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol. Cell 2009, 35, 403–413. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Xu, F.; Zhang, D.; Yi, W.; Chen, X.; Chen, G.; Zhou, E. TP73-AS1 promotes breast cancer cell proliferation through miR-200a-mediated TFAM inhibition. J. Cell. Biochem. 2017. [CrossRef] [PubMed]
- Gumireddy, K.; Li, A.; Yan, J.; Setoyama, T.; Johannes, G.J.; Orom, U.A.; Tchou, J.; Liu, Q.; Zhang, L.; Speicher, D.W.; et al. Identification of a long non-coding RNA-associated RNP complex regulating metastasis at the translational step. EMBO J. 2013, 32, 2672–2684. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Zhou, N.; Watabe, K.; Lu, Z.; Wu, F.; Xu, M.; Mo, Y.-Y. Long non-coding RNA UCA1 promotes breast tumor growth by suppression of p27 (Kip1). Cell Death Dis. 2014, 5, e1008. [Google Scholar] [CrossRef] [PubMed]
- Tuo, Y.-L.; Li, X.-M.; Luo, J. Long noncoding RNA UCA1 modulates breast cancer cell growth and apoptosis through decreasing tumor suppressive miR-143. Eur. Rev. Med. Pharmacol. Sci. 2015, 19, 3403–3411. [Google Scholar] [PubMed]
- Xiao, C.; Wu, C.-H.; Hu, H.-Z. LncRNA UCA1 promotes epithelial-mesenchymal transition (EMT) of breast cancer cells via enhancing Wnt/beta-catenin signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 2819–2824. [Google Scholar] [PubMed]
- Li, X.; Wu, Y.; Liu, A.; Tang, X. Long non-coding RNA UCA1 enhances tamoxifen resistance in breast cancer cells through a miR-18a-HIF1alpha feedback regulatory loop. Tumour. Biol. 2016, 37, 14733–14743. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Luo, J. Long Non-Coding RNA (lncRNA) Urothelial Carcinoma-Associated 1 (UCA1) Enhances Tamoxifen Resistance in Breast Cancer Cells via Inhibiting mTOR Signaling Pathway. Med. Sci. Monit. 2016, 22, 3860–3867. [Google Scholar] [CrossRef] [PubMed]
- Tian, F.-M.; Meng, F.-Q.; Wang, X.-B. Overexpression of long-noncoding RNA ZFAS1 decreases survival in human NSCLC patients. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 5126–5131. [Google Scholar] [PubMed]
- Askarian-Amiri, M.E.; Crawford, J.; French, J.D.; Smart, C.E.; Smith, M.A.; Clark, M.B.; Ru, K.; Mercer, T.R.; Thompson, E.R.; Lakhani, S.R.; et al. SNORD-host RNA Zfas1 is a regulator of mammary development and a potential marker for breast cancer. RNA 2011, 17, 878–891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Xing, C. Upregulation of long noncoding RNA ZFAS1 predicts poor prognosis and prompts invasion and metastasis in colorectal cancer. Pathol. Res. Pract. 2016, 212, 690–695. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Wang, F.; Chen, H.; Tan, Q.; Qiu, S.; Chen, S.; Jing, W.; Yu, M.; Liang, C.; Ye, S.; et al. Increased expression of long-noncoding RNA ZFAS1 is associated with epithelial-mesenchymal transition of gastric cancer. Aging 2016, 8, 2023–2038. [Google Scholar] [CrossRef] [PubMed]
- Hansji, H.; Leung, E.Y.; Baguley, B.C.; Finlay, G.J.; Cameron-Smith, D.; Figueiredo, V.C.; Askarian-Amiri, M.E. ZFAS1: A long noncoding RNA associated with ribosomes in breast cancer cells. Biol. Direct 2016, 11, 62. [Google Scholar] [CrossRef] [PubMed]
- Paik, S.; Shak, S.; Tang, G.; Kim, C.; Baker, J.; Cronin, M.; Baehner, F.L.; Walker, M.G.; Watson, D.; Park, T.; et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N. Engl. J. Med. 2004, 351, 2817–2826. [Google Scholar] [CrossRef] [PubMed]
- Gnant, M.; Sestak, I.; Filipits, M.; Dowsett, M.; Balic, M.; Lopez-Knowles, E.; Greil, R.; Dubsky, P.; Stoeger, H.; Rudas, M.; et al. Identifying clinically relevant prognostic subgroups of postmenopausal women with node-positive hormone receptor-positive early-stage breast cancer treated with endocrine therapy: A combined analysis of ABCSG-8 and ATAC using the PAM50 risk of recurrence. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2015, 26, 1685–1691. [Google Scholar] [CrossRef] [PubMed]
- Mook, S.; Schmidt, M.K.; Viale, G.; Pruneri, G.; Eekhout, I.; Floore, A.; Glas, A.M.; Bogaerts, J.; Cardoso, F.; Piccart-Gebhart, M.J.; et al. The 70-gene prognosis-signature predicts disease outcome in breast cancer patients with 1-3 positive lymph nodes in an independent validation study. Breast Cancer Res. Treat. 2009, 116, 295–302. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Wang, Q.; Zhan, Y.; Chen, X.; Yu, Q.; Zhang, J.; Wang, Y.; Xu, X.-J.; Zhu, L. Transcriptome sequencing uncovers a three-long noncoding RNA signature in predicting breast cancer survival. Sci. Rep. 2016, 6, 27931. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Zhong, L.; Xu, W.; Sun, Y.; Zhang, Z.; Zhao, H.; Yang, L.; Sun, J. Discovery of potential prognostic long non-coding RNA biomarkers for predicting the risk of tumor recurrence of breast cancer patients. Sci. Rep. 2016, 6, 31038. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Li, J.; Koirala, P.; Ding, X.; Chen, B.; Wang, Y.; Wang, Z.; Wang, C.; Zhang, X.; Mo, Y.-Y. Long non-coding RNAs as prognostic markers in human breast cancer. Oncotarget 2016, 7, 20584–20596. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-R.; Jiang, Y.-Z.; Xu, X.-E.; Hu, X.; Yu, K.-D.; Shao, Z.-M. Comprehensive Transcriptome Profiling Reveals Multigene Signatures in Triple-Negative Breast Cancer. Clin. Cancer Res. 2016, 22, 1653–1662. [Google Scholar] [CrossRef] [PubMed]
- Wahlestedt, C. Targeting long non-coding RNA to therapeutically upregulate gene expression. Nat. Rev. Drug Discov. 2013, 12, 433–446. [Google Scholar] [CrossRef] [PubMed]
- MacLeod, A.R.; Crooke, S.T. RNA Therapeutics in Oncology: Advances, Challenges, and Future Directions. J. Clin. Pharmacol. 2017, 57, S43–S59. [Google Scholar] [CrossRef] [PubMed]
- Modarresi, F.; Faghihi, M.A.; Lopez-Toledano, M.A.; Fatemi, R.P.; Magistri, M.; Brothers, S.P.; van der Brug, M.P.; Wahlestedt, C. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat. Biotechnol. 2012, 30, 453–459. [Google Scholar] [CrossRef] [PubMed]
| lncRNA in Breast Cancer | Access Number | Type | pcGene | Alteration/Role in Breast Cancer | Mechanism of Action | Therapeutic Utility | Ref. |
|---|---|---|---|---|---|---|---|
| ANRASSF1 | ENSG00000281358 | NAT (nc|pc) | RASSF1 | Upregulation/Oncogenic | Binds to PRC2 and silences the tumor suppressor gene RASSF1A. | [84] | |
| ANRIL | ENSG00000240498 | NAT (nc|pc) | CDKN2A, CDKN2B | Upregulation/Oncogenic | ANRIL is the NAT of CDKN2B gene (p15); binds to components of PRC1 (CBX7) and PRC2 (SUZ12) to silence the INK4 locus by epigenetic mechanisms. | Overexpressed in a variety of cancers and diseases. | [63,64,65] |
| LINC00901 | ENSG00000242385 | NAT (nc|pc) | LSAMP | Downregulation/Tumor suppressor | Low expression is associated with low overall survival. | Potential prognostic marker. | [95] |
| BCAR4 | ENSG00000262117 | lincRNA | Upregulation/Oncogenic | Interaction with SNIP1 and PNUTS in Hedgehog canonical pathway leads to a resistance to cancer treatments with SMO inhibitors. | Responsible for the acquisition of resistance to treatments and upregulation of non-canonical hedgehog pathway. | [96,97] | |
| BCYRN1 | ENSG00000236824 | NAT (nc|nc) | Upregulation/Oncogenic | BCYRN1 expression is associated with cell proliferation. | Knockdown BCYRN1 impacts viability of actively proliferating cells through growth arrest and apoptosis. Potential therapeutic target for various cancers. | [98] | |
| CCAT2 | ENSG00000280997 | NAT (nc|nc) | Upregulation/Oncogenic | Downregulates p15 through interaction with EZH2. Regulates TGF-β and Wnt signaling pathways. Promotes cell proliferation, invasion, tumor growth and metastasis. | Potential prognosis biomarker and therapeutic target. | [99,100,101,102,103] | |
| CRNDE | ENSG00000245694 | lincRNA | Upregulation/Oncogenic | Molecular sponge of miRNA-136 in breast cancer, activating Wnt/β-catenin. | Associated with unfavorable prognosis. | [104] | |
| DANCR | ENSG00000226950 | lincRNA | Upregulation/Oncogenic | Participates in cell proliferation and invasion. | Associated with a worse prognosis in TNBC. | [105] | |
| DSCAM-AS1 | ENSG00000235123 | NAT (nc|pc) | DSCAM | Upregulation/Oncogenic | Expression induced by estrogen stimulation. Positive correlation with genes associated with cancer aggression, tamoxifen resistance, and metastasis. | Biomarker for luminal subtype. | [106,107] |
| FAM83H-AS1 | ENSG00000282685 | lincRNA | Upregulation/Oncogenic | Most upregulated in luminal subtype of breast cancer. | Prognostic marker of luminal subtype. | [108] | |
| GAS5 | ENSG00000234741 | NAT (nc|pc)/bidirectional lncRNA/lincRNA (multiple transcripts) | ZBTB37 | Downregulation/Tumor suppressor | Required for decoy of glucocorticoid receptor (GR), inhibits transcriptional induction by GR, stops growth and triggers apoptosis, induces PTEN through miR-103 inhibition. | Responsible for the acquisition of trastuzumab resistance. Potential circulating biomarker. | [109,110,111,112,113,114] |
| H19 | ENSG00000130600 | lincRNA | Upregulation/Oncogenic | Mediates breast cancer cell plasticity, invasion, and proliferation by sponging several miR (miR-200b/c, let-7b, miR-152), silences pro-apoptotic gene BIK through epigenetic modifications, precursor of miR-675 (pro-tumoral and pro-metastatic). | Upregulated in cancer. Potential circulating biomarker for early screening and prognosis monitoring in breast cancer. | [92,115,116,117,118] | |
| HIF1A-AS2 | ENSG00000258667 | NAT (nc|pc) | HIF1A | Upregulation/Oncogenic | Involved in cell proliferation and invasion, contributes to chemotherapy resistance. | In TNBC, biomarker for detection, prognosis and prediction for recurrence and response to taxane chemotherapy. | [119,120] |
| HMMR-AS1 | ENSG00000251018 | NAT (nc|pc) | HMMR | Upregulation/Oncogenic | Involved in cell proliferation and invasion. | Positive correlation with HMMR, BRCA1, BRCA2 (oncogenic), biomarker and potential target in basal-like breast cancer. | [121] |
| HOTAIR | ENSG00000228630 | NAT (nc|pc)/lincRNA (multiple transcripts) | HOXC11 | Upregulation/Oncogenic | Guides epigenetic mechanisms to silence tumor suppressor genes through interaction with PRC2 and LSD1, involved in protein degradation by interaction with E3-ubiquitin ligases, tumor invasion, apoptosis and EMT. | Over-expressed in cancer, biomarker and potential therapeutic target. | [86,89,122,123,124,125,126,127] |
| HOTAIRM1 | ENSG00000233429 | NAT (nc|pc)/bidirectional lncRNA (multiple transcripts) | HOXA1, HOXA2 | Upregulation/Oncogenic | Modulates gene expression in HoxA gene cluster by interacting with PRC1 and PRC2 complexes. High positive correlation with HOXA1 expression. | Increased in basal-like subtype breast cancer. | [128,129] |
| HOXA-AS2 | ENSG00000253552 | NAT (nc|pc) | HOXA3, HOXA4 | Upregulation/Oncogenic | Acts as an endogenous sponge of miR-520c-3p and indirectly controls the expression of miR520c-3p target genes (TGFBR2 and RELA). | [130] | |
| HOXA11-AS | ENSG00000240990 | NAT (nc|pc) | HOXA11 | Upregulation/Oncogenic | Promotes cell proliferation, invasion and metastasis by regulating EMT. | Biomarker for metastasis and prognosis in breast cancer. Blocked relation between HOXA11-AS and EMT may have therapeutic utility. | [131] |
| Lnc-ITGB1-6:7 | Lnc-ITGB1-6 (LNCipedia) | lincRNA | Upregulation/Oncogenic | Promotes cell proliferation, invasion and metastasis by regulating EMT. High linc-ITGB1 expression is associated with poor prognosis. | Biomarker in prognosis of breast cancer. | [132,133] | |
| Linc-RoR | ENSG00000258609 | lincRNA | Upregulation/Oncogenic | Induces EMT. Contributes to tumor growth, invasion, metastasis and drug resistance through endogenous competition with various miR (145, 205, 133, 34) and inhibition of p53 expression. | Upregulation is a marker in multi-drug resistance, chemotherapy tolerance. Potential therapeutic target for aggressive and metastatic breast cancer. | [91,134] | |
| LINC00472 | ENSG00000233237 | lincRNA | Downregulation/Tumor suppressor | Associated with tumor grade, estrogen receptor status and molecular subtype in breast cancer. Repressed by methylation of its promoter. | Potential prognosis and predictive biomarker. | [135,136] | |
| LSINCT5 | ENSG00000281560 | lincRNA | Upregulation/Oncogenic | Promotes cell proliferation. | [137] | ||
| MALAT1 | ENSG00000251562 | NAT (nc|nc) | Upregulation/Oncogenic | Plays a critical role in pre-mRNA alternative splicing. Regulates EMT gene expression. | Knockdown reduces cell growth, invasion, migration and differentiation into cystic tumors. Potential prognosis marker in ER− and prediction marker for endocrine treatment sensitivity in ER+. | [138,139,140,141] | |
| MEG3 | ENSG00000214548 | NAT (nc|nc) | Downregulation/Tumor suppressor | Represses MDM2, leading to p53 accumulation. Silences genomic loci of TGFβ-associated genes by interaction with PRC2. Represses AKT signaling pathway. Inhibits EMT by sponging miR-421. | Expression promotes apoptosis, inhibits proliferation and angiogenesis. | [87,88,142,143] | |
| MIR31HG (LOC554202) | ENSG00000171889 | Sense-overlapping lncRNA | Downregulation/Tumor suppressor | Host gene of miR-31. Silenced in TNBC by promoter hypermethylation. Inhibits invasion-metastasis cascade by targeting pro-metastasis genes (i.e., RhoA and WAVE3). | [144,145,146] | ||
| NEAT1 | ENSG00000245532 | lincRNA | Upregulation/Oncogenic | Modulates miRNA biogenesis by organizing key components of paraspeckles and regulates transcription through protein sequestration into paraspeckles. Promotes proliferation and EMT. In ER+, NEAT1 is indispensable for interaction between FOXN3 and SINA3 complex. Regulates EZH2 through miR-101. | Overexpression of miR-548ar-3p downregulates NEAT1 and results in inhibition of cell growth. | [147,148,149,150,151,152] | |
| PANDAR | ENSG00000281450 | lincRNA | Upregulation/Oncogenic | Represses p16INK4A expression through modulating the recruitment of Bmi1 to the p16INK4A promoter. Removes cycle arrest possibility during G1/S transition. | Potential therapeutic target. | [85] | |
| PTPRG-AS1 | ENSG00000241472 | NAT (nc|pc) | PTPRG, C3ORF14 | Upregulation/Oncogenic | Differentially expressed between ER+ and ER− subtypes. | [153,154] | |
| PVT1 | ENSG00000249859 | NAT (nc|pc) | TMEM75 | Upregulation/Oncogenic | Co-operation between c-Myc and PVT1. Enhances c-Myc stability through inhibiting its phosphorylation. | Due to synergy between c-Myc and PVT1, silencing PVT1 expression decreases cell proliferation and increases apoptosis. Potential therapeutic target. | [155,156,157,158] |
| SNHG17 | ENSG00000196756 | lincRNA | Differentially expressed between ER+ and ER− subtypes. Low expression associated with overall survival. Expression correlates with tumor grade. | [154] | |||
| SOX2-OT | ENSG00000242808 | NAT (nc|pc), sense-overlapping lncRNA, lincRNA | DNAJC19 | Upregulation/Oncogenic | Through positive effect on SOX2 expression, SOX2OT plays a key role in pluripotency and tumorigenesis. | Potential prognosis marker and therapeutic target. | [159,160] |
| SPRY4-IT1 | ENSG00000281881 | Sense-intronic lncRNA | Upregulation/Oncogenic | Upregulates ZNF703 involved in the activation of the mTor signaling pathway. Promotes cell proliferation and inhibits apoptosis. | SPRY4-IT1 positively correlates with tumor size and pathological stage. Prognostic marker and potential therapeutic target. | [161,162] | |
| TERRA (Telomeric repeat-containing RNA) | lncRNA | Misregulation | Transcribed from telomeric C-rich strand. Interacts with TRF1 and TRF2 to facilitate heterochromatin formation. Provides RNA template to aid telomerase function. | Potential therapeutic target to impair telomerase activity. | [163,164,165,166] | ||
| TP73-AS1 | ENSG00000227372 | NAT (nc|pc) | TP73 | Upregulation/Oncogenic | TP73-AS1/miR-200a/ZEB1 forms a regulating loop. TP73-AS1 competes with ZEB1 for binding to miR-200a. ZEB1 binds to TP73-AS1 promoter and activates its expression. Upregulation of TP73-AS1/ZEB1 promotes cell invasion and migration. | Potential therapeutic target. | [94,167] |
| treRNA | ENSG00000231265 | lincRNA | Upregulation/Oncogenic | Regulates translation through interaction with ribonucleoprotein complex, which will bind to the translation initiation factor (EIF4G1). Overexpressed in lymph-node metastasis. Promotes tumor invasion and metastasis. Regulates expression of metastasis promoting-gene Snail. Suppresses epithelial markers and translation of E-cadherin mRNA. | [168] | ||
| UCA1 | ENSG00000214049 | lincRNA | Upregulation/Oncogenic | Enhances chemotherapy resistance (tamoxifen) through mTor pathway inhibition and miR-18a downregulation. Promotes EMT through activating Wnt/β-catenin signaling. UCA1/hnRNP1 suppresses p27 protein level by competition. Downregulates tumor suppressor miR-143. | Potential urine biomarker. Knockdown reduces chemoresistance, cell migration and tumor size. | [169,170,171,172,173] | |
| ZFAS1 | ENSG00000177410 | NAT (nc|pc) | ZNFX1 | Downregulation in breast cancer/Upregulated in other cancers | Associated with ribosomes in breast cancer. Role in development and cell differentiation in mammary gland. | Potential biomarker. | [174,175,176,177,178] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Latgé, G.; Poulet, C.; Bours, V.; Josse, C.; Jerusalem, G. Natural Antisense Transcripts: Molecular Mechanisms and Implications in Breast Cancers. Int. J. Mol. Sci. 2018, 19, 123. https://doi.org/10.3390/ijms19010123
Latgé G, Poulet C, Bours V, Josse C, Jerusalem G. Natural Antisense Transcripts: Molecular Mechanisms and Implications in Breast Cancers. International Journal of Molecular Sciences. 2018; 19(1):123. https://doi.org/10.3390/ijms19010123
Chicago/Turabian StyleLatgé, Guillaume, Christophe Poulet, Vincent Bours, Claire Josse, and Guy Jerusalem. 2018. "Natural Antisense Transcripts: Molecular Mechanisms and Implications in Breast Cancers" International Journal of Molecular Sciences 19, no. 1: 123. https://doi.org/10.3390/ijms19010123