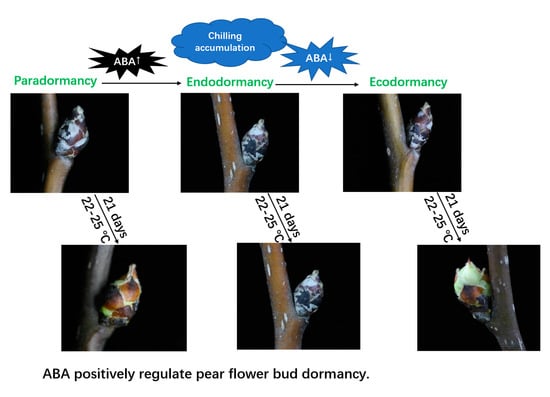
Abscisic Acid (ABA ) Promotes the Induction and Maintenance of Pear (Pyrus pyrifolia White Pear Group) Flower Bud Endodormancy
Abstract
:1. Introduction
2. Results
2.1. Daily Temperatures, Bud-Break Percentages, and ABA Content during the Natural Flower Bud Dormancy Cycle
2.2. Identification of ABA Metabolism- and Signaling-Related Genes in Pear
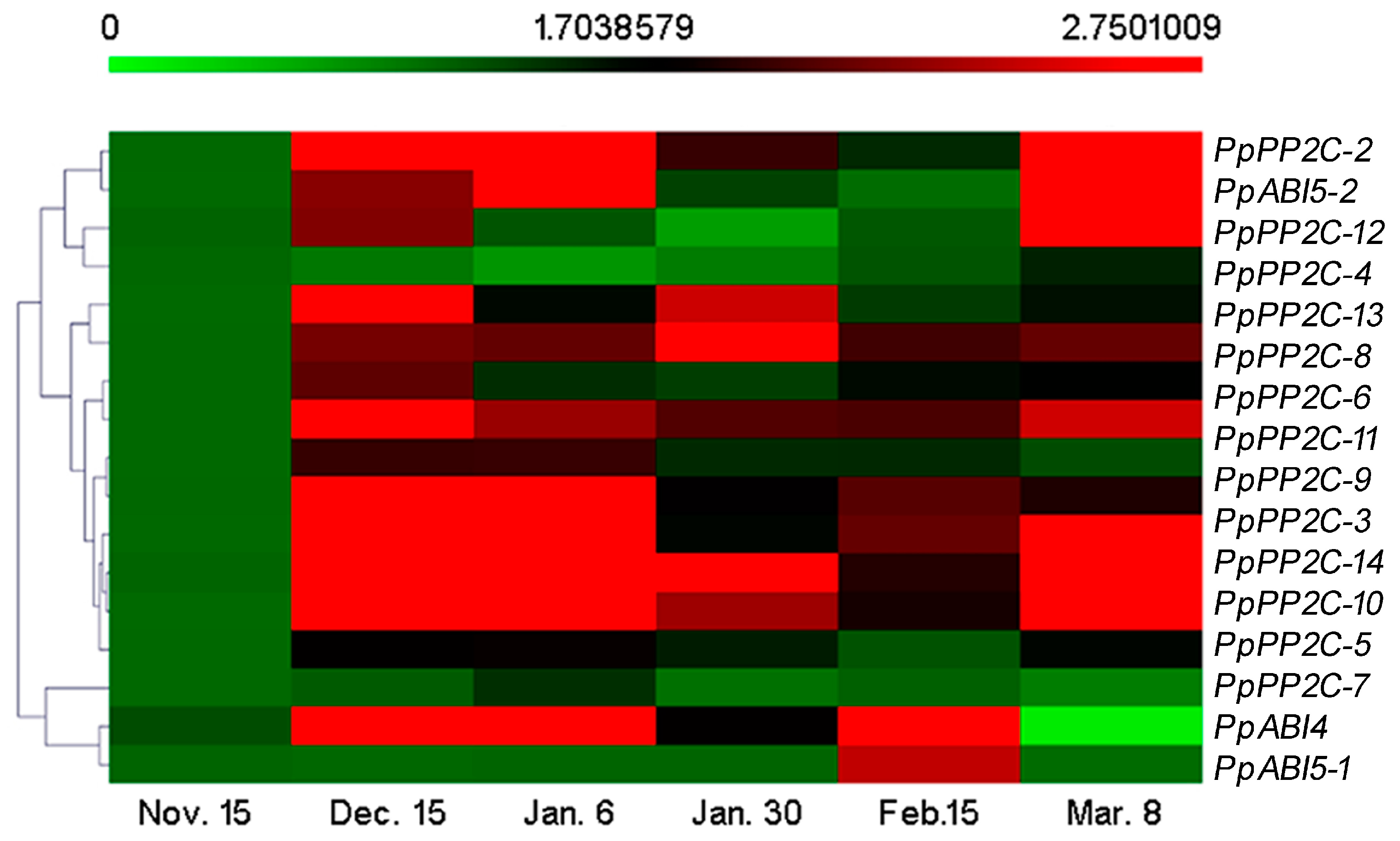
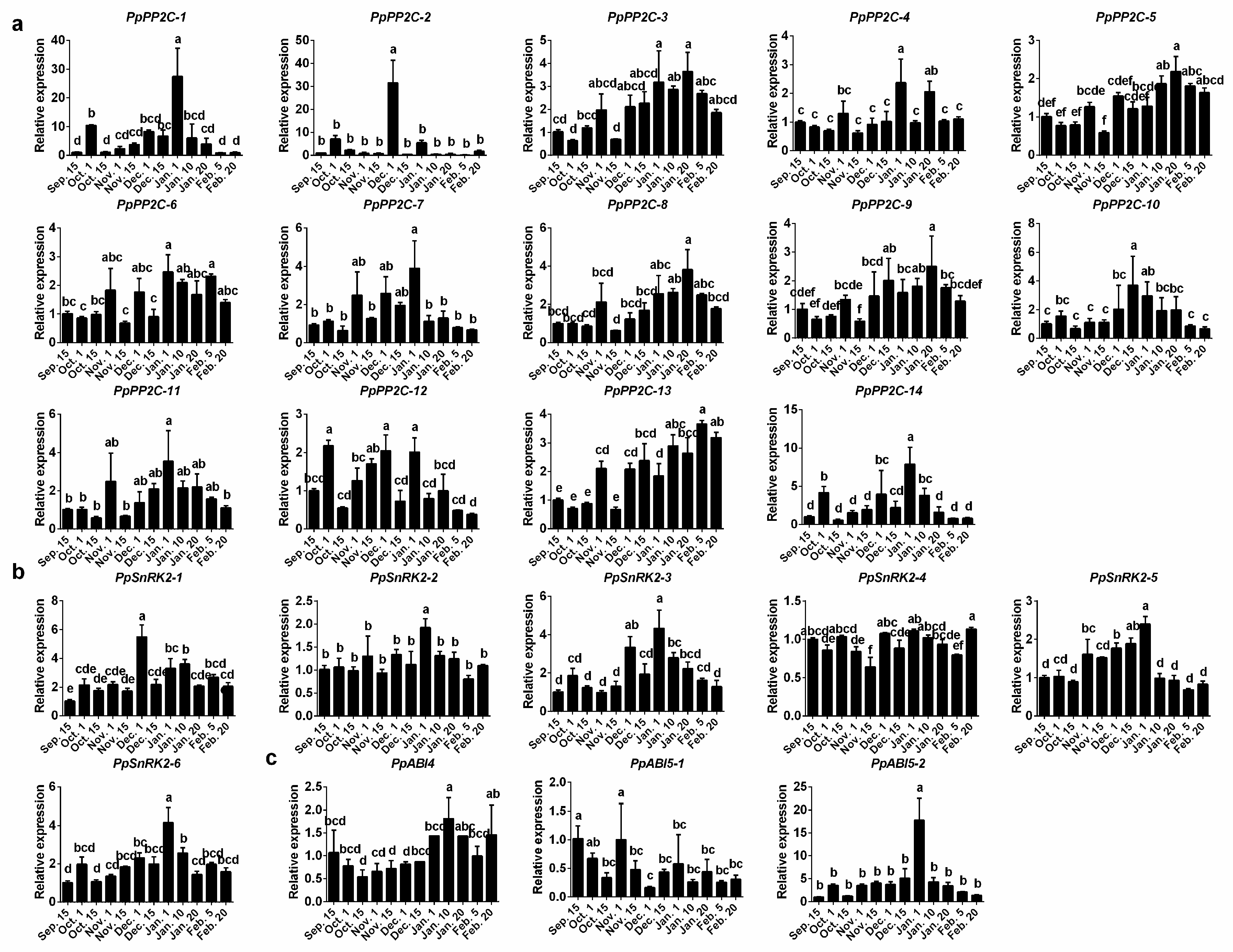
2.3. Expression Patterns of ABA Synthesis, Catabolism, and Signaling Genes during Bud Dormancy
2.4. Effects of Exogenous ABA on the Endodormancy-Related Induction of Pear Flower Buds
2.5. Effects of Exogenous ABA on Pear ABA Metabolism- and Signaling-Related Gene Expression Levels
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Measurement of Lateral Flower Bud’ Dormancy Status
4.3. Daily Temperature during Winter
4.4. ABA Treatments
4.5. Identification of Genes Related to ABA Biosynthesis, Metabolism, and Signaling
4.6. Determination of ABA Concentration
4.7. RNA Isolation and Quantitative Real-Time RT-PCR Analysis
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| ABA | Abscisic acid |
| NCED | 9-cis-Epoxycarotenoid dioxygenase |
| CYP707A | ABA 8′-hydroxylase |
| RCAR/PYR/PYL | Regulatory component of aba receptor/pyrabactin resistance/pyrabactin resistance like |
| PP2C | Protein phosphatase 2C |
| SnRK2 | Snf1-Related kinase 2 |
| ABI | Abscisic acid insensitive |
| GGDP | Geranylgeranyl diphosphate |
| CTAB | Cetyltrimethyl Ammonium Bromide |
References
- Faust, M.; Erez, A.; Rowland, L.J.; Wang, S.Y.; Norman, H.A. Bud dormancy in perennial fruit trees: Physiological basis for dormancy induction, maintenance, and release. HortScience 1997, 32, 623–629. [Google Scholar]
- Lang, G.A.; Early, J.D.; Martin, G.C.; Darnell, R.L. Endo-, para-, and ecodormancy: Physiological terminology and classification for dormancy research. HortScience 1987, 22, 371–377. [Google Scholar]
- Heide, O.M.; Prestrud, A.K. Low temperature, but not photoperiod, controls growth cessation and dormancy induction and release in apple and pear. Tree Physiol. 2005, 25, 109. [Google Scholar] [CrossRef] [PubMed]
- Tamura, F.; Tanabe, K.; Itai, A. Regulation of endodormancy in Japanese pear. Acta Hortic. 2002, 587, 325–336. [Google Scholar] [CrossRef]
- Kretzschmar, A.A.; Brighenti, L.M.; Rufato, L.; Pelizza, T.R.; Silveira, F.N.; Miquelutti, D.J.; Faoro, I.D. Chilling requirement for dormancy bud break in European pear. Acta Hortic. 2011, 909, 85–88. [Google Scholar] [CrossRef]
- Campoy, J.A.; Ruiz, D.; Egea, J. Dormancy in temperate fruit trees in a global warming context: A review. Sci. Hortic. 2011, 130, 357–372. [Google Scholar] [CrossRef]
- Karssen, C.M.; Swan, B.V.D.; Breekland, A.E.; Koornneef, M. Induction of dormancy during seed development by endogenous abscisic acid: Studies on abscisic acid deficient genotypes of Arabidopsis thaliana (L.) Heynh. Planta 1983, 157, 158. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, H.; Nishijima, T.; Yamato, Y.; Hamano, M.; Koshioka, M.; Miura, H. Involvement of abscisic acid in bulb dormancy of Allium wakegi Araki. II. A comparison between dormant and nondormant cultivars. Plant Growth Regul. 1999, 29, 195–200. [Google Scholar] [CrossRef]
- Seeley, S.D.; Powell, L.E.J. Seasonal changes of free and hydrolyzable abscisic acid in vegatative apple buds [Plant dormancy. J. Am. Soc. Hortic. Sci. 1981, 106, 405–409. [Google Scholar]
- Rodríguez, A.; Sánchez-Tamés, R. Dormancy and seasonal changes of plant growth regulators in hazel buds. Physiol. Plant. 1986, 66, 288–292. [Google Scholar] [CrossRef]
- Zheng, C.; Halaly, T.; Acheampong, A.K.; Takebayashi, Y.; Jikumaru, Y.; Kamiya, Y.; Or, E. Abscisic acid (ABA) regulates grape bud dormancy, and dormancy release stimuli may act through modification of ABA metabolism. J. Exp. Bot. 2015, 66, 1527–1542. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Gao, Z.; Du, P.; Xiao, W.; Tan, Q.; Chen, X.; Li, L.; Gao, D. Expression of ABA Metabolism-Related Genes Suggests Similarities and Differences Between Seed Dormancy and Bud Dormancy of Peach (Prunus persica). Front. Plant Sci. 2015, 6, 1248. [Google Scholar] [CrossRef] [PubMed]
- Tuan, P.A.; Bai, S.; Saito, T.; Ito, A.; Moriguchi, T. Dormancy-associated MADS-box (DAM) and Abscisic Acid Pathway Regulate Pear Endodormancy through a Feedback Mechanism. Plant Cell Physiol. 2017, 58, 1378–1390. [Google Scholar] [CrossRef] [PubMed]
- Nambara, E.; Marion-Poll, A. Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 2005, 56, 165–185. [Google Scholar] [CrossRef] [PubMed]
- Nambara, E.; Okamoto, M.; Tatematsu, K.; Yano, R.; Seo, M.; Kamiya, Y. Abscisic acid and the control of seed dormancy and germination. Seed Sci. Res. 2010, 20, 55–67. [Google Scholar] [CrossRef]
- Lefebvre, V.; North, H.; Frey, A.; Sotta, B.; Seo, M.; Okamoto, M.; Nambara, E.; Marionpoll, A. Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J. 2006, 45, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Cutler, A.J.; Krochko, J.E. Formation and breakdown of ABA. Trends Plant Sci. 1999, 4, 472. [Google Scholar] [CrossRef]
- Saito, S.; Hirai, N.; Matsumoto, C.; Ohigashi, H.; Ohta, D.; Sakata, K.; Mizutani, M. Arabidopsis CYP707As Encode (+)-Abscisic Acid 8′-Hydroxylase, a Key Enzyme in the Oxidative Catabolism of Abscisic Acid. Plant Physiol. 2004, 134, 1439. [Google Scholar] [CrossRef] [PubMed]
- Destefano-Beltrán, L.; Knauber, D.; Huckle, L.; Suttle, J.C. Effects of postharvest storage and dormancy status on ABA content, metabolism, and expression of genes involved in ABA biosynthesis and metabolism in potato tuber tissues. Plant Mol. Biol. 2006, 61, 687–697. [Google Scholar] [CrossRef] [PubMed]
- Horvath, D.P.; Anderson, J.V.; SotosuáRez, M.; Chao, W.S. Transcriptome analysis of leafy spurge (Euphorbia esula) crown buds during shifts in well-defined phases of dormancy. Weed Sci. 2006, 54, 821–827. [Google Scholar] [CrossRef]
- Bai, S.; Saito, T.; Sakamoto, D.; Ito, A.; Fujii, H.; Moriguchi, T. Transcriptome analysis of Japanese pear (Pyrus pyrifolia Nakai) flower buds transitioning through endodormancy. Plant Cell Physiol. 2013, 54, 1132–1151. [Google Scholar] [CrossRef] [PubMed]
- Hubbard, K.E.; Nishimura, N.; Hitomi, K.; Getzoff, E.D.; Schroeder, J.I. Early abscisic acid signal transduction mechanisms: Newly discovered components and newly emerging questions. Genes Dev. 2010, 24, 1695–1708. [Google Scholar] [CrossRef] [PubMed]
- Finkelstein, R.R. Mutations at two new Arabidopsis ABA response loci are similar to the abi3 mutations. Plant J. 1994, 5, 765–771. [Google Scholar] [CrossRef]
- Finkelstein, R. Abscisic Acid Synthesis and Response. Arabidopsis Book 2001, 11, e0058. [Google Scholar] [CrossRef] [PubMed]
- Niu, Q.; Li, J.; Cai, D.; Qian, M.; Jia, H.; Bai, S.; Hussain, S.; Liu, G.; Teng, Y.; Zheng, X. Dormancy-associated MADS-box genes and microRNAs jointly control dormancy transition in pear (Pyrus pyrifolia white pear group) flower bud. J. Exp. Bot. 2016, 67, 239–257. [Google Scholar] [CrossRef] [PubMed]
- Arora, R.; Rowland, L.J.; Tanino, K. Induction and Release of Bud Dormancy in Woody Perennials: A Science Comes of Age. Hortsci. A Publ. Am. Soc. Hortic. Sci. 2003, 38, 911–921. [Google Scholar]
- Horvath, D.P.; Anderson, J.V.; Chao, W.S.; Foley, M.E. Knowing when to grow: Signals regulating bud dormancy. Trends Plant Sci. 2003, 8, 534–540. [Google Scholar] [CrossRef] [PubMed]
- Rohde, A.; Bhalerao, R.P. Plant dormancy in the perennial context. Trends Plant Sci. 2007, 12, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Horvath, D.P.; Chao, W.S.; Suttle, J.C.; Thimmapuram, J.; Anderson, J.V. Transcriptome analysis identifies novel responses and potential regulatory genes involved in seasonal dormancy transitions of leafy spurge (Euphorbia esula L.). BMC Genom. 2008, 9, 536. [Google Scholar] [CrossRef] [PubMed]
- Guak, S.; Fuchigami, L.H. Effects of applied ABA on growth cessation, bud dormancy, cold acclimation, leaf senescence and N mobilization in apple nursery plants. J. Hortic. Sci. Biotechnol. 2001, 76, 459–464. [Google Scholar] [CrossRef]
- Palmer, J.P.; Jasrai, Y.T. Precocious Growth and Effect of ABA: Encapsulated Buds of Kalanchoe tubiflora. J. Plant Biochem. Biotechnol. 1996, 5, 103–104. [Google Scholar] [CrossRef]
- Bris, M.L.; Michaux-Ferrière, N.; Jacob, Y.; Poupet, A.; Barthe, P.; Guigonis, J.M. Regulation of bud dormancy by manipulation of ABA in isolated buds of Rosa hybrida cultured in vitro. Funct. Plant Biol. 2011, 26, 273–281. [Google Scholar] [CrossRef]
- Chono, M.; Honda, I.; Shinoda, S.; Kushiro, T.; Kamiya, Y.; Nambara, E.; Kawakami, N.; Kaneko, S.; Watanabe, Y. Field studies on the regulation of abscisic acid content and germinability during grain development of barley: Molecular and chemical analysis of pre-harvest sprouting. J. Exp. Bot. 2006, 57, 2421–2434. [Google Scholar] [CrossRef] [PubMed]
- Ruttink, T.; Arend, M.; Morreel, K.; Storme, V.; Rombauts, S.; Fromm, J.; Bhalerao, R.P.; Boerjan, W.; Rohde, A. A molecular timetable for apical bud formation and dormancy induction in poplar. Plant Cell 2007, 19, 2370–2390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, G.; Li, W.; Zheng, P.; Xu, T.; Chen, L.; Liu, D.; Hussain, S.; Teng, Y. Transcriptomic analysis of ‘Suli’ pear (Pyrus pyrifolia white pear group) buds during the dormancy by RNA-Seq. BMC Genom. 2012, 13, 700. [Google Scholar] [CrossRef] [PubMed]
- Destefano-Beltrán, L.; Knauber, D.; Huckle, L.; Suttle, J. Chemically forced dormancy termination mimics natural dormancy progression in potato tuber meristems by reducing ABA content and modifying expression of genes involved in regulating ABA synthesis and metabolism. J. Exp. Bot. 2006, 57, 2879. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Szostkiewicz, I.; Korte, A.; Moes, D.; Yang, Y.; Christmann, A.; Grill, E. Regulators of PP2C Phosphatase Activity Function as Abscisic Acid Sensors. Science 2009, 324, 1064. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Fung, P.; Nishimura, N.; Jensen, D.R.; Fujii, H.; Zhao, Y.; Lumba, S.; Santiago, J.; Rodrigues, A.; Chow, T.F. Abscisic Acid Inhibits Type 2C Protein Phosphatases via the PYR/PYL Family of START Proteins. Science 2009, 324, 1068–1071. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Hwang, H.; Hong, J.W.; Lee, Y.N.; Ahn, I.P.; Yoon, I.S.; Yoo, S.D.; Lee, S.; Lee, S.C.; Kim, B.G. A rice orthologue of the ABA receptor, OsPYL/RCAR5, is a positive regulator of the ABA signal transduction pathway in seed germination and early seedling growth. J. Exp. Bot. 2012, 63, 1013. [Google Scholar] [CrossRef] [PubMed]
- Khalil-Ur-Rehman, M.; Long, S.; Li, C.X.; Faheem, M.; Wu, W.; Tao, J.M. Comparative RNA-seq based transcriptomic analysis of bud dormancy in grape. BMC Plant Biol. 2017, 17, 18. [Google Scholar] [CrossRef] [PubMed]
- Arend, M.; Schnitzler, J.P.; Ehlting, B.; Hänsch, R.; Lange, T.; Rennenberg, H.; Himmelbach, A.; Grill, E.; Fromm, J. Expression of the Arabidopsis Mutant abi1 Gene Alters Abscisic Acid Sensitivity, Stomatal Development, and Growth Morphology in Gray Poplars. Plant Physiol. 2009, 151, 2110–2119. [Google Scholar] [CrossRef] [PubMed]
- Umezawa, T.; Sugiyama, N.; Mizoguchi, M.; Hayashi, S.; Myouga, F.; Yamaguchi-Shinozaki, K.; Ishihama, Y.; Hirayama, T.; Shinozaki, K. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 17588–17593. [Google Scholar] [CrossRef] [PubMed]
- Finkelstein, R.R.; Lynch, T.J. The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 2000, 12, 599. [Google Scholar] [CrossRef] [PubMed]
- Furihata, T.; Maruyama, K.; Fujita, Y.; Umezawa, T.; Yoshida, R.; Shinozaki, K.; Yamaguchishinozaki, K. Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc. Natl. Acad. Sci. USA 2006, 103, 1988–1993. [Google Scholar] [CrossRef] [PubMed]
- Fujii, H.; Verslues, P.E.; Zhu, J.K. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 2007, 19, 485–494. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Fujita, Y.; Kanamori, N.; Katagiri, T.; Umezawa, T.; Kidokoro, S.; Maruyama, K.; Yoshida, T.; Ishiyama, K.; Kobayashi, M. Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 2009, 50, 1345. [Google Scholar] [CrossRef] [PubMed]
- Leida, C.; Conesa, A.; Llácer, G.; Badenes, M.L.; Ríos, G. Histone modifications and expression of DAM6 gene in peach are modulated during bud dormancy release in a cultivar-dependent manner. New Phytol. 2012, 193, 67. [Google Scholar] [CrossRef] [PubMed]
- Yooyongwech, S.; Sugaya, S.; Sekozawa, Y.; Gemma, H. Differential adaptation of high- and low-chill dormant peaches in winter through aquaporin gene expression and soluble sugar content. Plant Cell Rep. 2009, 28, 1709–1715. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wang, Z.; Shi, Z.; Zhang, S.; Ming, R.; Zhu, S.; Khan, M.A.; Tao, S.; Korban, S.S.; Wang, H.; et al. The genome of the pear (Pyrus bretschneideri Rehd.). Genome Res. 2013, 23, 396–408. [Google Scholar] [CrossRef] [PubMed]
- Niu, Q.; Zong, Y.; Qian, M.; Yang, F.; Teng, Y. Simultaneous quantitative determination of major plant hormones in pear flowers and fruit by UPLC/ESI-MS/MS. Anal. Methods 2014, 6, 1766. [Google Scholar] [CrossRef]
- Zhang, D.; Yu, B.; Bai, J.; Qian, M.; Shu, Q.; Su, J.; Teng, Y. Effects of high temperatures on UV-B/visible irradiation induced postharvest anthocyanin accumulation in ‘Yunhongli No. 1’ (Pyrus pyrifolia Nakai) pears. Sci. Hortic. 2012, 134, 53–59. [Google Scholar] [CrossRef]





| Gene Name | Genome Locus Tag | Chromosome Location | Start | End |
|---|---|---|---|---|
| PpPP2C-1 | Pbr013576.1 | Chr1 | 9763088 | 9766905 |
| PpPP2C-2 | Pbr041497.1 | Chr2 | 18358353 | 18360378 |
| PpPP2C-3 | Pbr013336.1 | Chr3 | 20846735 | 20850224 |
| PpPP2C-4 | Pbr028942.1 | Chr4 | 1769534 | 1772968 |
| PpPP2C-5 | Pbr009703.1 | Chr7 | 1815301 | 1820662 |
| PpPP2C-6 | Pbr025010.1 | Chr8 | 14605450 | 14607580 |
| PpPP2C-7 | Pbr015257.1 | Chr9 | 7829375 | 7832873 |
| PpPP2C-8 | Pbr041795.2 | Chr12 | 14167470 | 14172092 |
| PpPP2C-9 | Pbr028792.1 | Chr13 | 1840077 | 1843505 |
| PpPP2C-10 | Pbr026157.1 | Chr14 | 6552129 | 6556046 |
| PpPP2C-11 | Pbr019878.1 | Chr15 | 6270769 | 6272939 |
| PpPP2C-12 | Pbr015521.1 | Chr15 | 15615471 | 15619039 |
| PpPP2C-13 | Pbr019599.1 | Chr15 | 8273643 | 8277518 |
| PpPP2C-14 | Pbr022745.1 | Scaffold346.0.1 | 139061 | 141715 |
| PpABI4 | Pbr009544.1 | Chr1 | 7155273 | 7156489 |
| PpABI5-1 | Pbr017778.1 | Chr12 | 20360616 | 20363301 |
| PpABI5-2 | Pbr007589.1 | Chr14 | 344099 | 346885 |
| PpNCED-1 | Pbr039596.1 | Chr10 | 6419191 | 6421260 |
| PpNCED-2 | Pbr009089.1 | Chr10 | 10254167 | 10256724 |
| PpNCED-3 | Pbr006012.1 | Chr16 | 10335999 | 10337804 |
| PpSnRK2-1 | Pbr040276.1 | Chr6 | 16755401 | 16758728 |
| PpSnRK2-2 | Pbr026536.1 | Chr8 | 4160916 | 4164639 |
| PpSnRK2-3 | Pbr040625.1 | Chr15 | 36546195 | 36549943 |
| PpSnRK2-4 | Pbr003186.1 | Chr15 | 41233666 | 41237650 |
| PpSnRK2-5 | Pbr023607.1 | Scaffold364.0 | 289329 | 293241 |
| PpSnRK2-6 | Pbr042784.1 | Scaffold364.0 | 337528 | 342127 |
| PpCYP707A-1 | Pbr003860.1 | Chr3 | 26178912 | 26186900 |
| PpCYP707A-2 | Pbr006776.1 | Chr6 | 18879003 | 18881665 |
| PpCYP707A-3 | Pbr019636.1 | Chr15 | 7961660 | 7964906 |
| PpCYP707A-4 | Pbr029414.1 | Chr16 | 18720527 | 18723493 |
| PpCYP707A-5 | Pbr004630.1 | Scaffold1214.0 | 40622 | 43660 |
| PpPYL-1 | Pbr027457.1 | Chr1 | 3660265 | 3662621 |
| PpPYL-2 | Pbr013616.1 | Chr1 | 9469866 | 9472306 |
| PpPYL-3 | Pbr000497.1 | Chr5 | 25031100 | 25031660 |
| PpPYL-4 | Pbr036422.1 | Chr8 | 16927910 | 16928515 |
| PpPYL-5 | Pbr016128.1 | Chr10 | 3588258 | 3588818 |
| PpPYL-6 | Pbr010794.1 | Chr13 | 185820 | 188146 |
| PpPYL-7 | Pbr019827.1 | Chr15 | 6628678 | 6629283 |
| PpPYL-8 | Pbr042468.1 | Scaffold984.0 | 68401 | 69066 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Xu, Y.; Niu, Q.; He, L.; Teng, Y.; Bai, S. Abscisic Acid (ABA ) Promotes the Induction and Maintenance of Pear (Pyrus pyrifolia White Pear Group) Flower Bud Endodormancy. Int. J. Mol. Sci. 2018, 19, 310. https://doi.org/10.3390/ijms19010310
Li J, Xu Y, Niu Q, He L, Teng Y, Bai S. Abscisic Acid (ABA ) Promotes the Induction and Maintenance of Pear (Pyrus pyrifolia White Pear Group) Flower Bud Endodormancy. International Journal of Molecular Sciences. 2018; 19(1):310. https://doi.org/10.3390/ijms19010310
Chicago/Turabian StyleLi, Jianzhao, Ying Xu, Qingfeng Niu, Lufang He, Yuanwen Teng, and Songling Bai. 2018. "Abscisic Acid (ABA ) Promotes the Induction and Maintenance of Pear (Pyrus pyrifolia White Pear Group) Flower Bud Endodormancy" International Journal of Molecular Sciences 19, no. 1: 310. https://doi.org/10.3390/ijms19010310