Transcriptome-Wide Identification and Characterization of Potato Circular RNAs in Response to Pectobacterium carotovorum Subspecies brasiliense Infection
Abstract
:1. Introduction
2. Results and Discussion
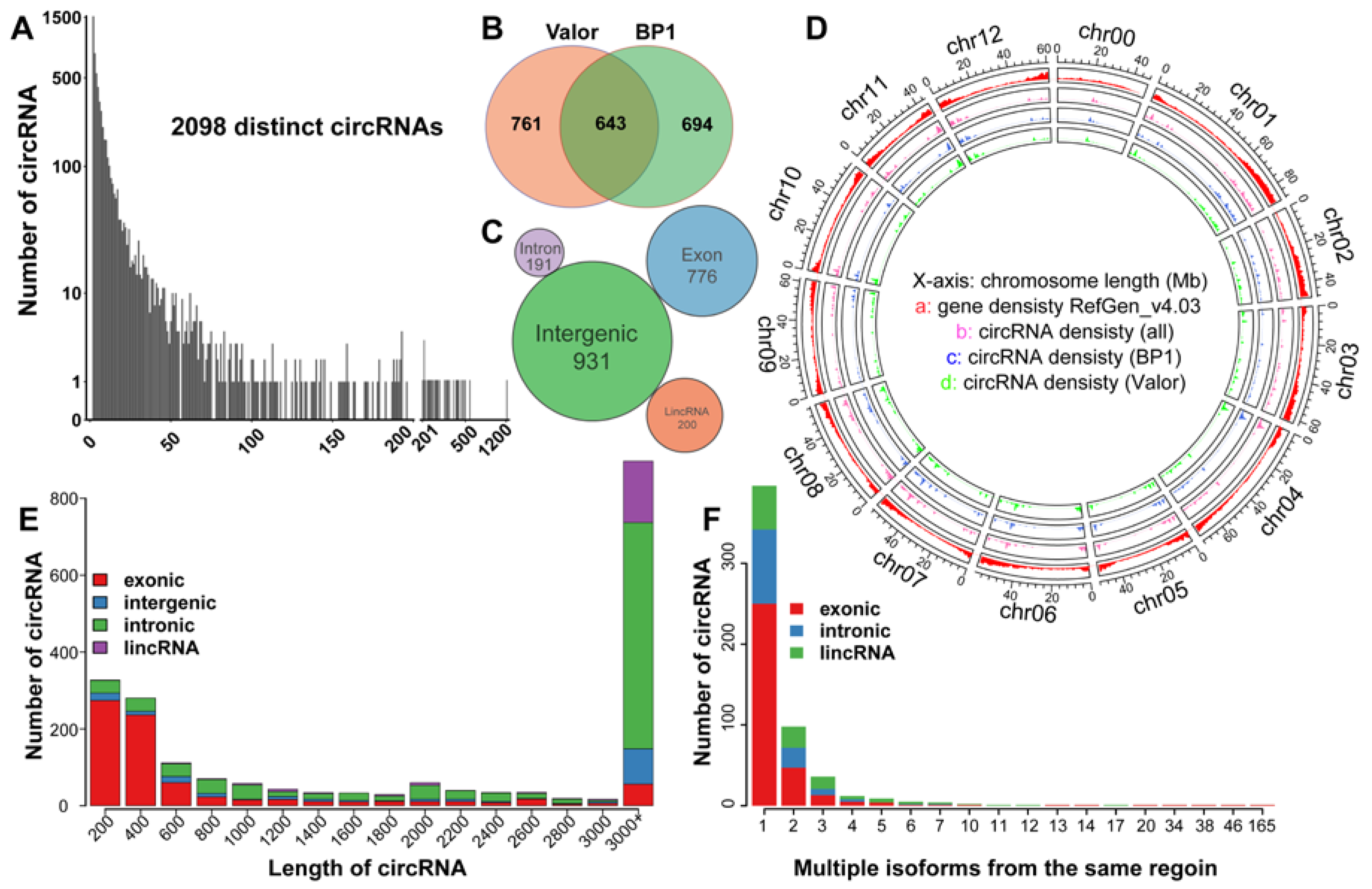
2.1. Genome-Wide Identification of circRNAs in Potato
2.2. The Characteristics of circRNA Abundance in Potato
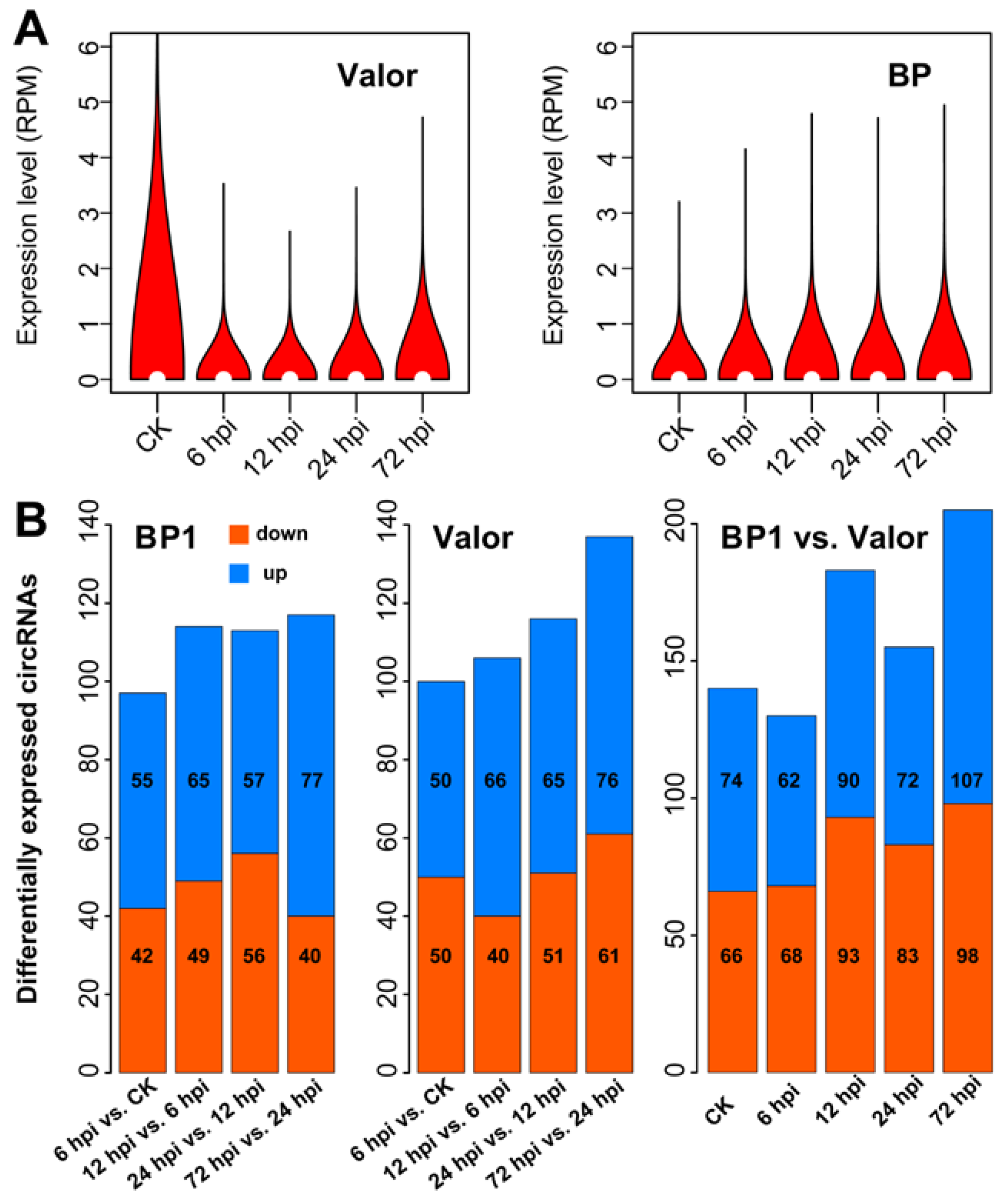
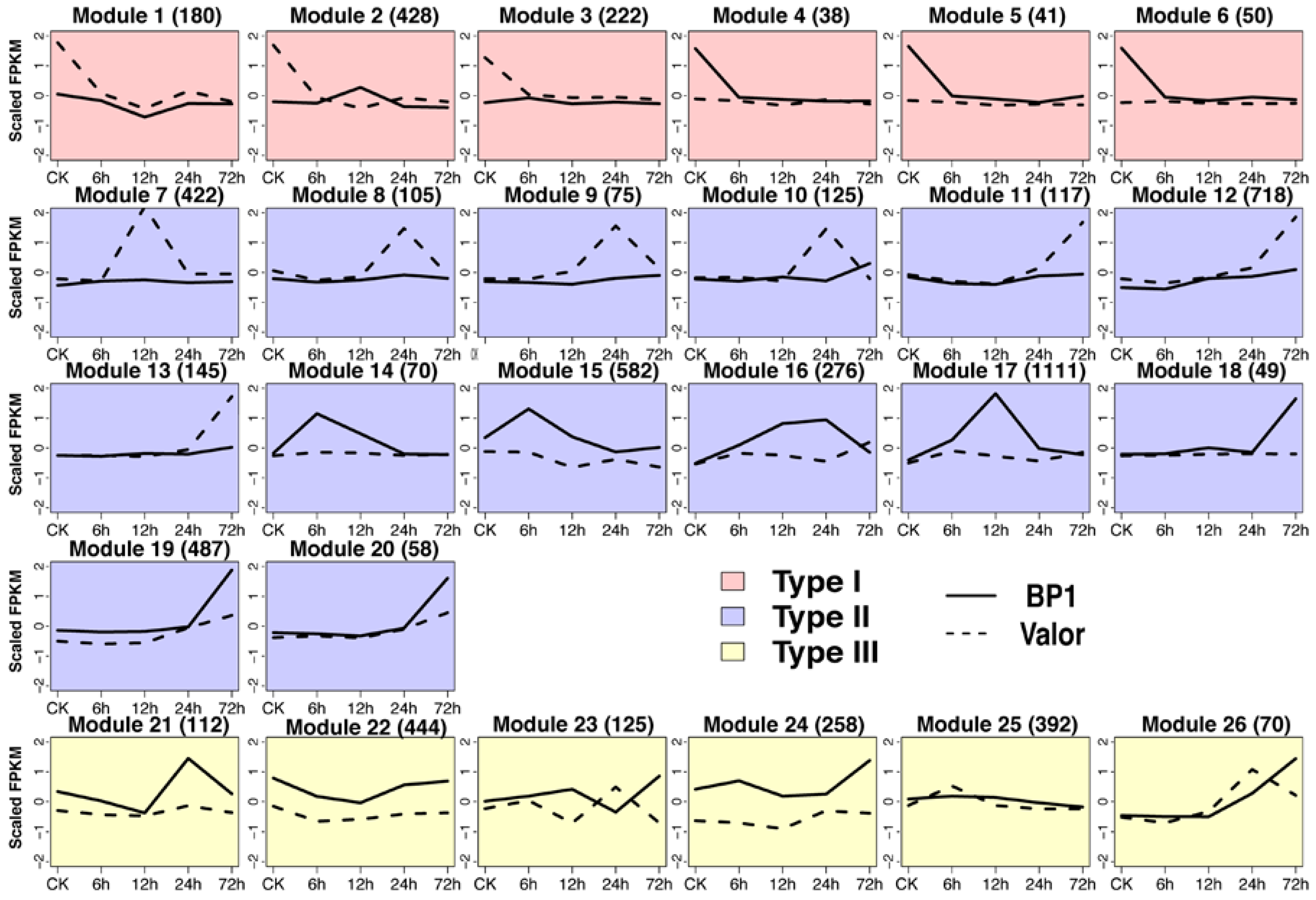
2.3. CircRNAs Are Differentially Expressed in Response to Pcb Infection
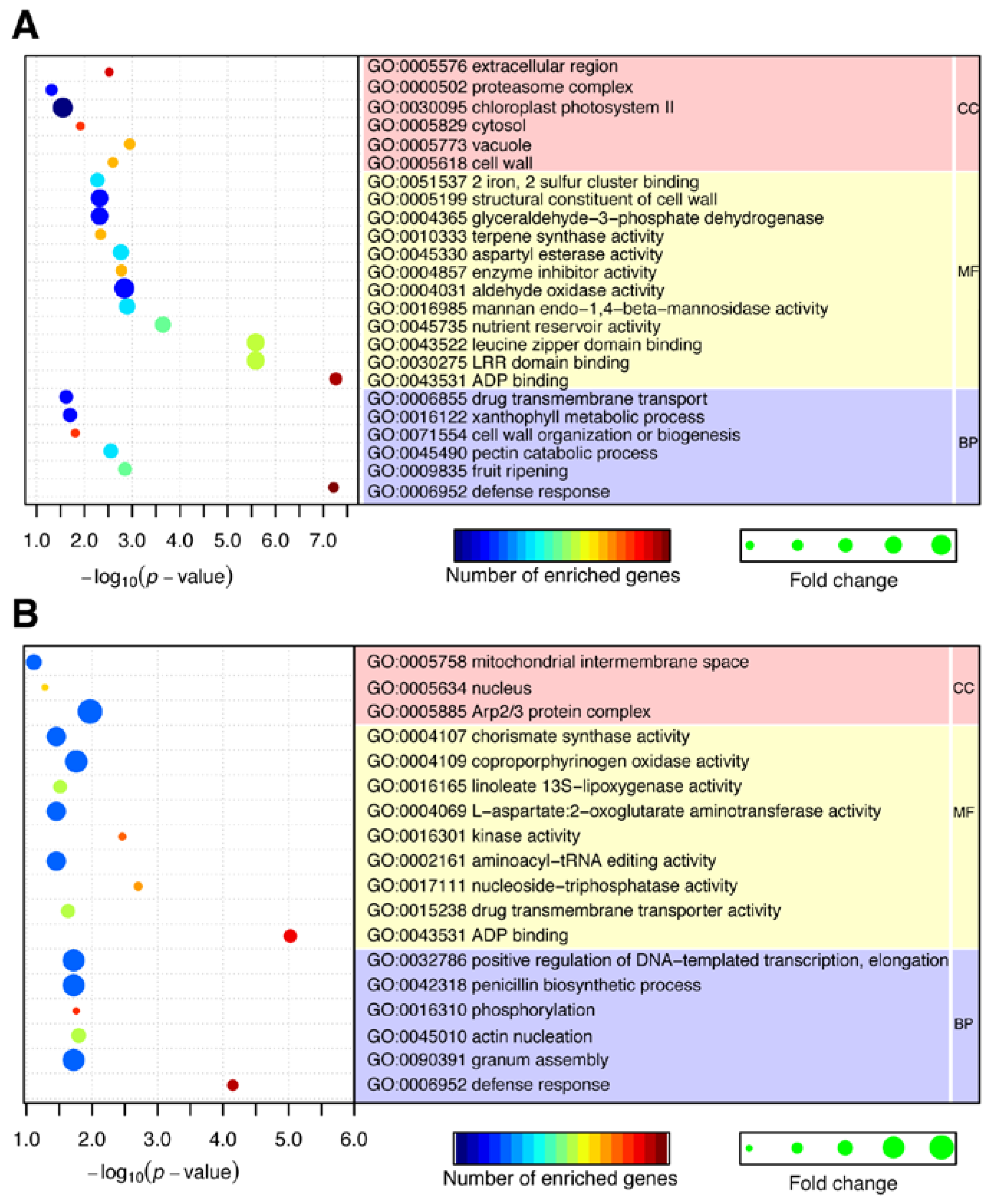
2.4. CircRNAs Act as Putative Regulators of Parental Genes
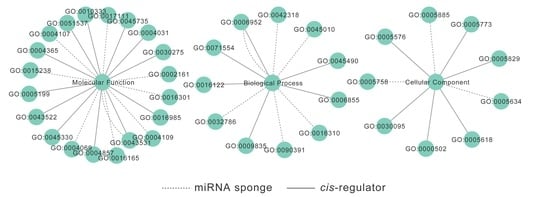
2.5. CircRNAs Act as Putative miRNA Sponges
2.6. mRNA-CircRNA-LincRNA Co-Expression Networks
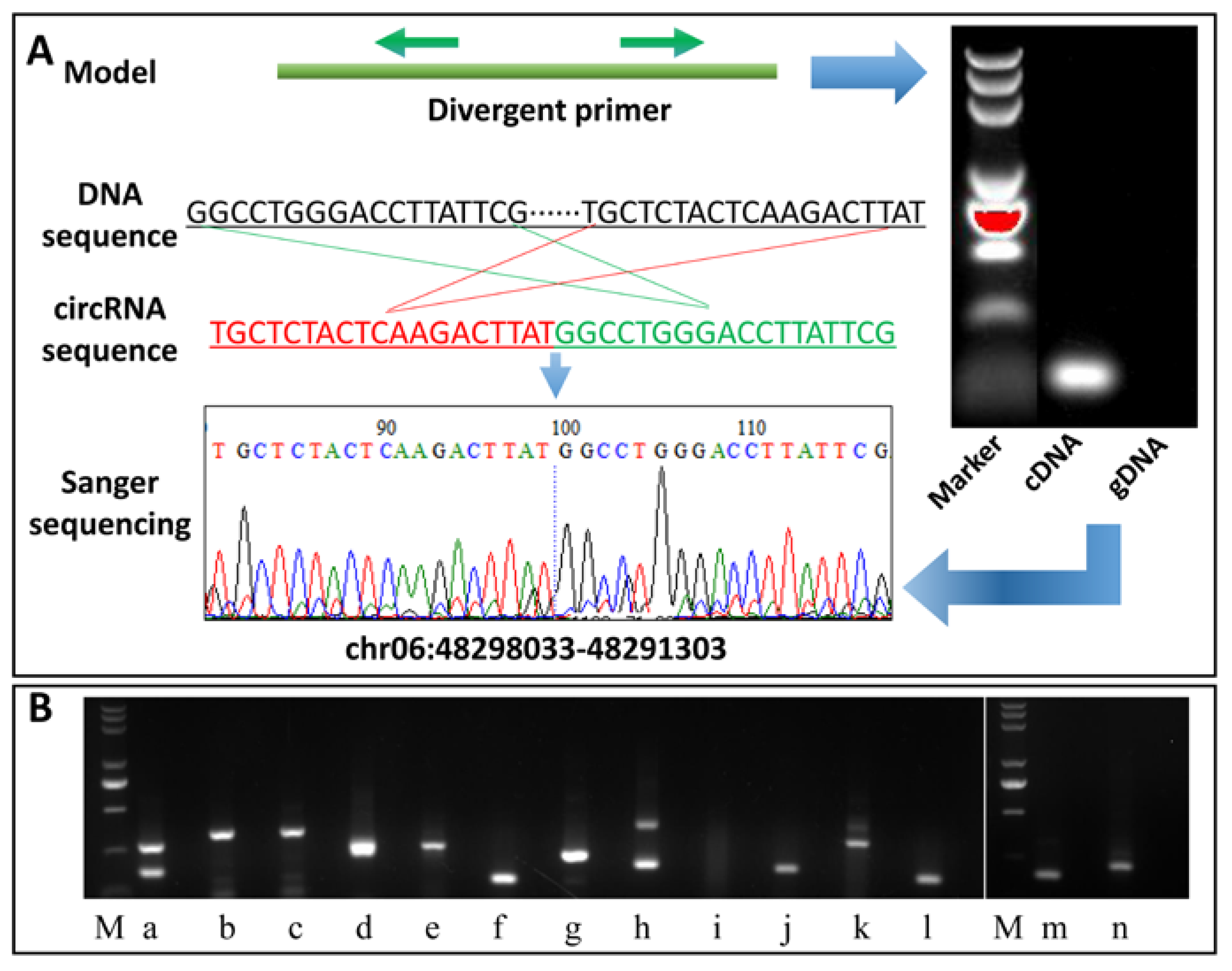
2.7. Validation of the Identified circRNAs
3. Materials and Methods
3.1. Potato RNA-seq Data Sets
3.2. Identification and Expression Analysis of Circular RNAs
3.3. Prediction of miRNA Target
3.4. Gene Co-Expression Network Analysis
3.5. Reverse Transcription (RT)-PCR
4. Conclusions and Perspectives
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| Pcb | Pectobacterium carotovorum subsp. brasiliense |
| circRNAs | Circular RNAs |
| miRNA | MicroRNA |
| hpi | Hours post-infection |
| WGCNA | Weighted gene co-expression network analysis |
| SRE | Soft rot Enterobacteriaceae |
| GO | Gene Ontology |
| WAK | Wall-associated kinases |
| BAK1 | Brassinosteroid insensitive 1-associated receptor kinase |
| PTI | Pathogen-associated molecule patterns triggered immunity |
| ET | Ethylene |
| RLK | Receptor-like protein kinase |
| TFs | Transcription factors |
| MF | Molecular function |
| BP | Biological process |
| CC | Cellular component |
References
- Rybak-Wolf, A.; Stottmeister, C.; Glažar, P.; Jens, M.; Pino, N.; Giusti, S.; Hanan, M.; Behm, M.; Bartok, O.; Ashwal-Fluss, R.; et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell 2015, 58, 870–885. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, X.; Chen, T.; Xiang, J.; Yin, Q.; Xing, Y.; Zhu, S.; Yang, L.; Chen, L. Circular intronic long noncoding RNAs. Mol. Cell 2013, 51, 792–806. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Liu, T.; Wang, X.; He, A. Circles reshaping the RNA world: From waste to treasure. Mol. Cancer 2017, 16, 58. [Google Scholar] [CrossRef] [PubMed]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Du, W.W.; Yang, W.; Liu, E.; Yang, Z.; Dhaliwal, P.; Yang, B.B. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016, 44, 2846–2858. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, Z. Efficient backsplicing produces translatable circular mRNAs. RNA 2015, 21, 172–179. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Zhang, A.; Xiong, L.; Li, Y.; Huang, R.; Liao, L.; Zhu, Z.; Wang, Y. Deep circular RNA sequencing provides insights into the mechanism underlying grass carp reovirus infection. Int. J. Mol. Sci. 2017, 18, 1977. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Zhang, Y.; Chen, Y.; Yu, Y. Circular RNAs roll into the regulatory network of plants. Biochem. Biophys. Res. Commun. 2017, 488, 382–386. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.L.; Bao, Y.; Yee, M.C.; Barrett, S.P.; Hogan, G.J.; Olsen, M.N.; Dinneny, J.R.; Brown, P.O.; Salzman, J. Circular RNA Is Expressed across the Eukaryotic Tree of Life. PLoS ONE 2014, 9, e90859. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.Y.; Chen, L.; Liu, C.; Zhu, Q.H.; Fan, L. Widespread noncoding circular RNAs in plants. New Phytol. 2015, 208, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yang, M.; Wei, S.; Qin, F.; Zhao, H.; Suo, B. Identification of circular RNAs and their targets in leaves of Triticum aestivum L. Under dehydration stress. Front. Plant Sci. 2016, 7, 2024. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Liu, Y.; Li, D.; Li, L.; Zhang, Q.; Wang, S.; Huang, H. Identification of circular RNAs in kiwifruit and their species-specific response to bacterial canker pathogen invasion. Front. Plant Sci. 2017, 8, 413. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.L.; Liu, M.Y.; Ma, D.F.; Wu, J.W.; Li, S.L.; Zhu, Y.X.; Han, B. Identification of circular RNAs and their targets during tomato fruit ripening. Postharvest Biol. Technol. 2018, 136, 90–98. [Google Scholar] [CrossRef]
- Zuo, J.; Wang, Q.; Zhu, B.; Luo, Y.; Gao, L. Deciphering the roles of circRNAs on chilling injury in tomato. Biochem. Biophys. Res. Commun. 2016, 479, 132–138. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, Q.; Gao, L.; Zhu, B.; Luo, Y.; Deng, Z.; Zuo, J. Integrative analysis of circRNAs acting as ceRNAs involved in ethylene pathway in tomato. Physiol. Plant. 2017, 161, 311–321. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Zhou, Z.; Niu, Y.; Sun, X.; Deng, Z. Identification and functional characterization of tomato circRNAs derived from genes involved in fruit pigment accumulation. Sci. Rep. 2017, 7, 8594. [Google Scholar] [CrossRef] [PubMed]
- Potato Genome Sequencing Constitute. Genome sequence and analysis of the tuber crop potato. Nature 2011, 475, 189–195. [Google Scholar]
- Van der Merwe, J.J.; Coutinho, T.A.; Korsten, L.; van der Waals, J.E. Pectobacterium carotovorum subsp. Brasiliensis causing blackleg on potatoes in south Africa. Eur. J. Plant Pathol. 2010, 126, 175–185. [Google Scholar] [CrossRef]
- Kwenda, S.; Birch, P.; Moleleki, L. Genome-wide identification of potato long intergenic noncoding RNAs responsive to Pectobacterium carotovorum subspecies brasiliense infection. BMC Genom. 2016, 17, 614. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.; Cui, L.; Zhou, Y.; Zhu, C.; Fan, D.; Gong, H.; Zhao, Q.; Zhou, C.; Zhao, Y.; Lu, D.; et al. Transcriptome-wide investigation of circular RNAs in rice. RNA 2015, 21, 2076–2087. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Meng, Y.; Zhang, M.; Tong, X.; Wang, Q.; Sun, Y.; Quan, J.; Govers, F.; Shan, W. Infection of Arabidopsis thaliana by Phytophthora parasitica and identification of variation in host specificity. Mol. Plant Pathol. 2011, 12, 187–201. [Google Scholar] [CrossRef] [PubMed]
- Jupe, J.; Stam, R.; Howden, A.; Morris, J.; Zhang, R.; Hedley, P.; Huitema, E. Phytophthora capsici-tomato interaction features dramatic shifts in gene expression associated with a hemi-biotrophic lifestyle. Genome Biol. 2013, 14, R63. [Google Scholar] [CrossRef] [PubMed]
- Hückelhoven, R. Cell wall–associated mechanisms of disease resistance and susceptibility. Annu. Rev. Phytopathol. 2007, 45, 101–127. [Google Scholar] [CrossRef] [PubMed]
- Kishore, G.K.; Pande, S.; Podile, A.R. Pseudomonas aeruginosa GSE 18 inhibits the cell wall degrading enzymes of Aspergillus Niger and activates defence-related enzymes of groundnut in control of collar rot disease. Australas. Plant Pathol. 2006, 35, 259–263. [Google Scholar] [CrossRef]
- Ton, J.; Flors, V.; Mauch-Mani, B. The multifaceted role of ABA in disease resistance. Trends Plant Sci. 2009, 14, 310–317. [Google Scholar] [CrossRef] [PubMed]
- Luan, Y.; Cui, J.; Zhai, J.; Li, J.; Han, L.; Meng, J. High-throughput sequencing reveals differential expression of miRNAs in tomato inoculated with Phytophthora infestans. Planta 2015, 241, 1405–1416. [Google Scholar] [CrossRef] [PubMed]
- Aksoy, E.; Demirel, U.; ÖZTÜRK, Z.N.; ÇALIŞKAN, S.; ÇALIŞKAN, M.E. Recent advances in potato genomics, transcriptomics, and transgenicsunder drought and heat stresses: A review. Turk. J. Bot. 2015, 39, 920–940. [Google Scholar] [CrossRef]
- Din, M.; Barozai, M.Y.K.; Baloch, I.A. Identification and functional analysis of new conserved microRNAs and their targets in potato (Solanum tuberosum L.). Turk. J. Bot. 2015, 38, 1199–1213. [Google Scholar] [CrossRef]
- Gu, M.; Liu, W.; Meng, Q.; Zhang, W.; Chen, A.; Sun, S.; Xu, G. Identification of microRNAs in six Solanaceous plants and their potential link with phosphate and mycorrhizal signaling. J. Integr. Plant Biol. 2014, 56, 1164–1178. [Google Scholar] [CrossRef] [PubMed]
- Ficklin, S.P.; Feltus, F.A. Gene coexpression network alignment and conservation of gene modules between two grass species: Maize and rice. Plant Physiol. 2011, 156, 1244–1256. [Google Scholar] [CrossRef] [PubMed]
- Zipfel, C. Pattern-recognition receptors in plant innate immunity. Curr. Opin. Immunol. 2008, 20, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Meng, X.; Shan, L.; He, P. Transcriptional regulation of pattern-triggered immunity in plants. Cell Host Microbe 2016, 19, 641–650. [Google Scholar] [CrossRef] [PubMed]
- Cass, C.L.; Peraldi, A.; Dowd, P.F.; Mottiar, Y.; Santoro, N.; Karlen, S.D.; Bukhman, Y.V.; Foster, C.E.; Thrower, N.; Bruno, L.C. Effects of phenylalanine ammonia lyase (PAL) knockdown on cell wall composition, biomass digestibility, and biotic and abiotic stress responses in Brachypodium. J. Exp. Bot. 2015, 66, 4317–4335. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Li, F.; Tian, L.; Huang, M.; Deng, R.; Li, X.; Chen, W.; Wu, P.; Li, M.; Jiang, H. The phenylalanine ammonia lyase gene LJPAL1 is involved in plant defense responses to pathogens and plays diverse roles in Lotus japonicus-rhizobium symbioses. Mol. Plant Microbe Interact. 2017, 30, 739–753. [Google Scholar] [CrossRef] [PubMed]
- Macho, A.P.; Zipfel, C. Plant PRRs and the activation of innate immune signaling. Mol. Cell 2014, 54, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Mersmann, S.; Bourdais, G.; Rietz, S.; Robatzek, S. Ethylene signaling regulates accumulation of the FLS2 receptor and is required for the oxidative burst contributing to plant immunity. Plant Physiol. 2010, 154, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Serrano, M.; Coluccia, F.; Torres, M.; L’Haridon, F.; Métraux, J. The cuticle and plant defense to pathogens. Front. Plant Sci. 2014, 5, 274. [Google Scholar] [CrossRef] [PubMed]
- Valdivia, E.R.; Herrera, M.T.; Gianzo, C.; Fidalgo, J.; Revilla, G.; Zarra, I.; Sampedro, J. Regulation of secondary wall synthesis and cell death by NAC transcription factors in the monocot Brachypodium distachyon. J. Exp. Bot. 2013, 64, 1333–1343. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DEseq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Alexa, A.; Rahnenführer, J.; Lengauer, T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 2006, 22, 1600–1607. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2go: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Köster, J.; Rahmann, S. Snakemake—A scalable bioinformatics workflow engine. Bioinformatics 2012, 28, 2520–2522. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with burrows—Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Zhang, J.; Zhao, F. Circular RNA identification based on multiple seed matching. Brief. Bioinform. 2017, 1, 8. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server. Nucleic Acids Res. 2011, 39, W155–W159. [Google Scholar] [CrossRef] [PubMed]
- Fahlgren, N.; Carrington, J.C. miRNA target prediction in plants. In Plant Micrornas: Methods and Protocols; Meyers, B.C., Green, P.J., Eds.; Humana Press: Totowa, NJ, USA, 2010; Volume 592, pp. 51–57. [Google Scholar]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, R.; Zhu, Y.; Zhao, J.; Fang, Z.; Wang, S.; Yin, J.; Chu, Z.; Ma, D. Transcriptome-Wide Identification and Characterization of Potato Circular RNAs in Response to Pectobacterium carotovorum Subspecies brasiliense Infection. Int. J. Mol. Sci. 2018, 19, 71. https://doi.org/10.3390/ijms19010071
Zhou R, Zhu Y, Zhao J, Fang Z, Wang S, Yin J, Chu Z, Ma D. Transcriptome-Wide Identification and Characterization of Potato Circular RNAs in Response to Pectobacterium carotovorum Subspecies brasiliense Infection. International Journal of Molecular Sciences. 2018; 19(1):71. https://doi.org/10.3390/ijms19010071
Chicago/Turabian StyleZhou, Ran, Yongxing Zhu, Jiao Zhao, Zhengwu Fang, Shuping Wang, Junliang Yin, Zhaohui Chu, and Dongfang Ma. 2018. "Transcriptome-Wide Identification and Characterization of Potato Circular RNAs in Response to Pectobacterium carotovorum Subspecies brasiliense Infection" International Journal of Molecular Sciences 19, no. 1: 71. https://doi.org/10.3390/ijms19010071