Transcriptomic Profiling and Physiological Analysis of Haloxylon ammodendron in Response to Osmotic Stress
Abstract
:1. Introduction
2. Results
2.1. De Novo Transcriptome Assembly of Transcriptome
2.2. Functional Annotation of H. ammodendron Transcriptome
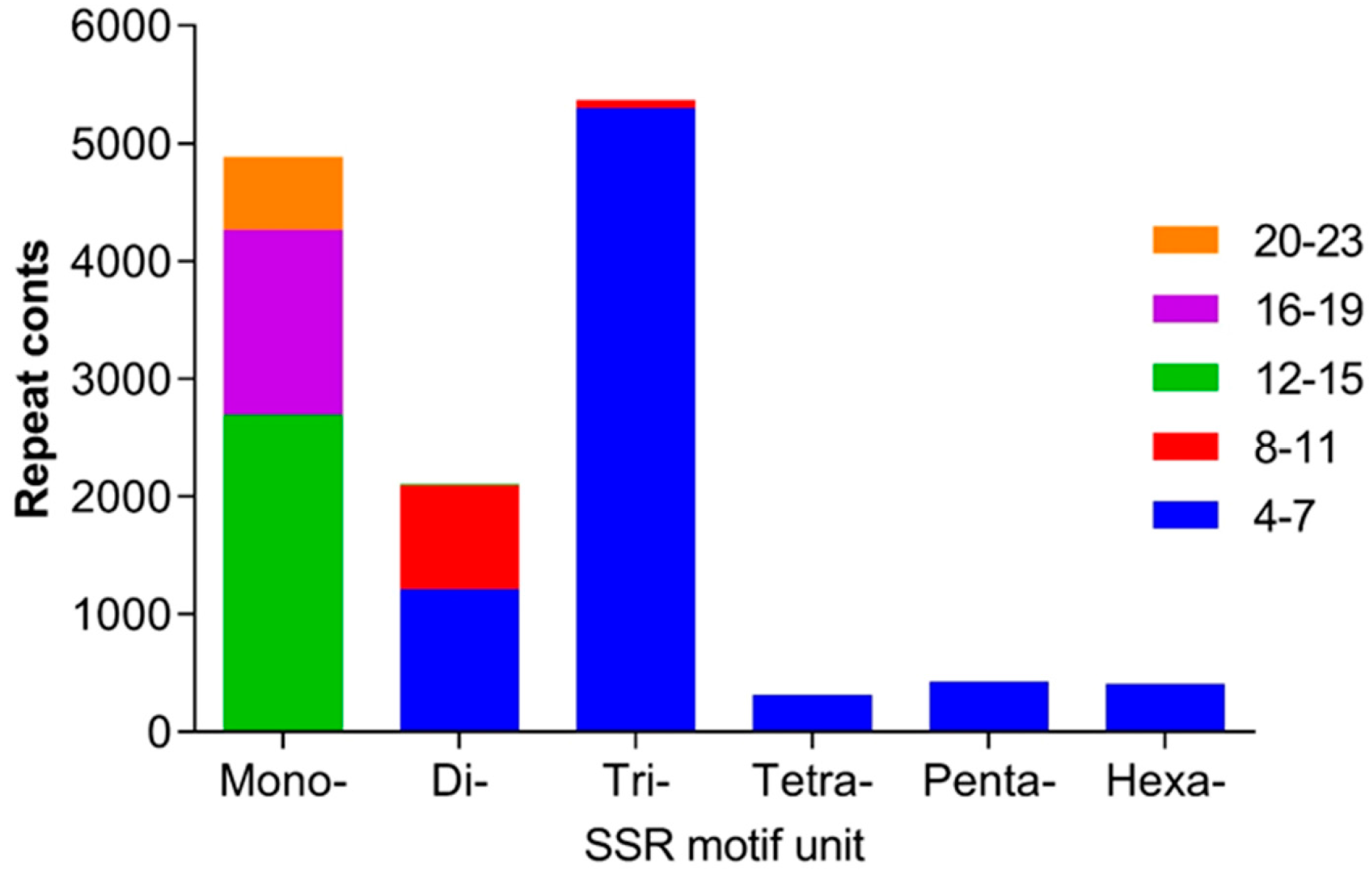
2.3. SSRs in the Transcriptome in H. ammodendron
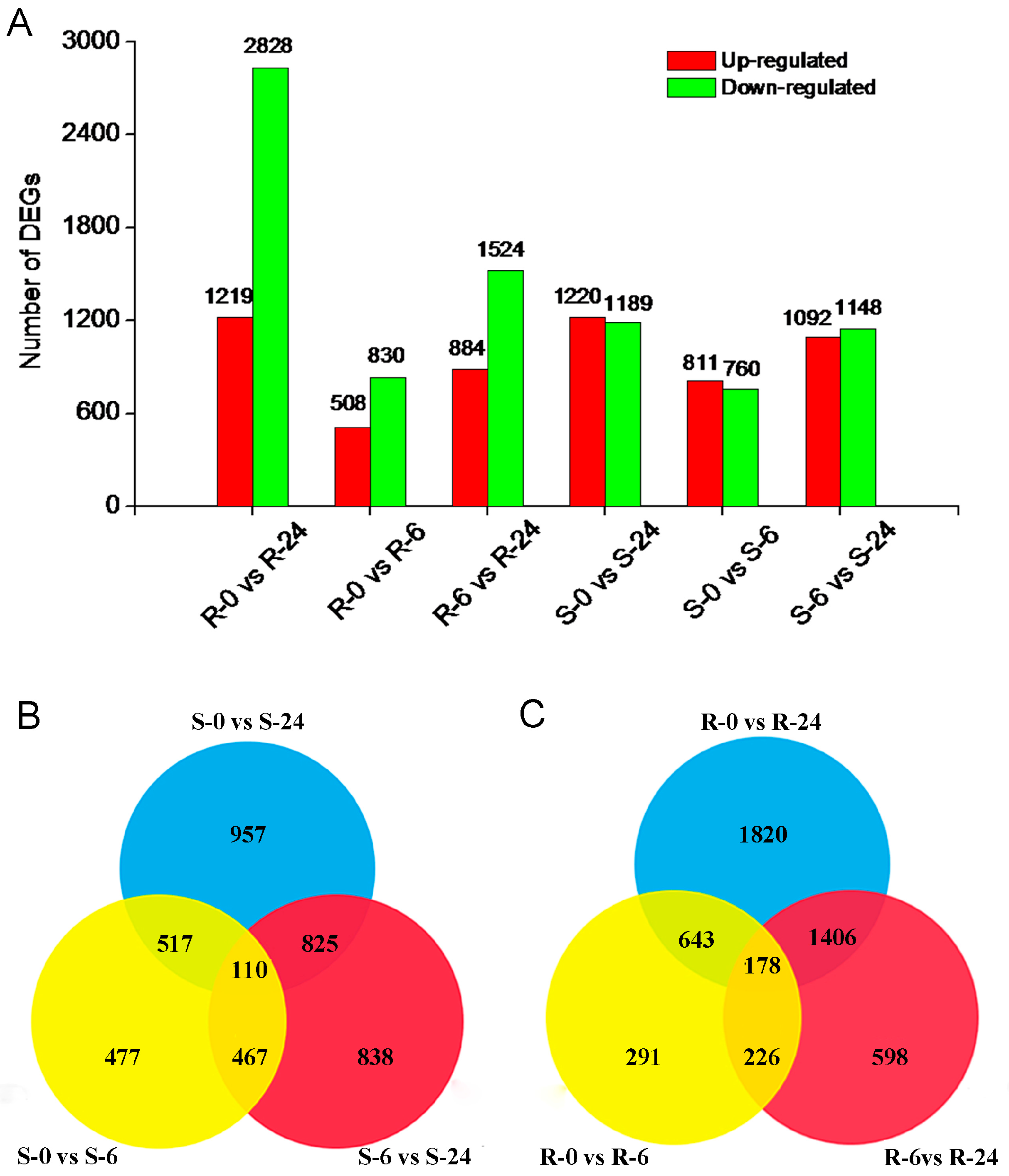
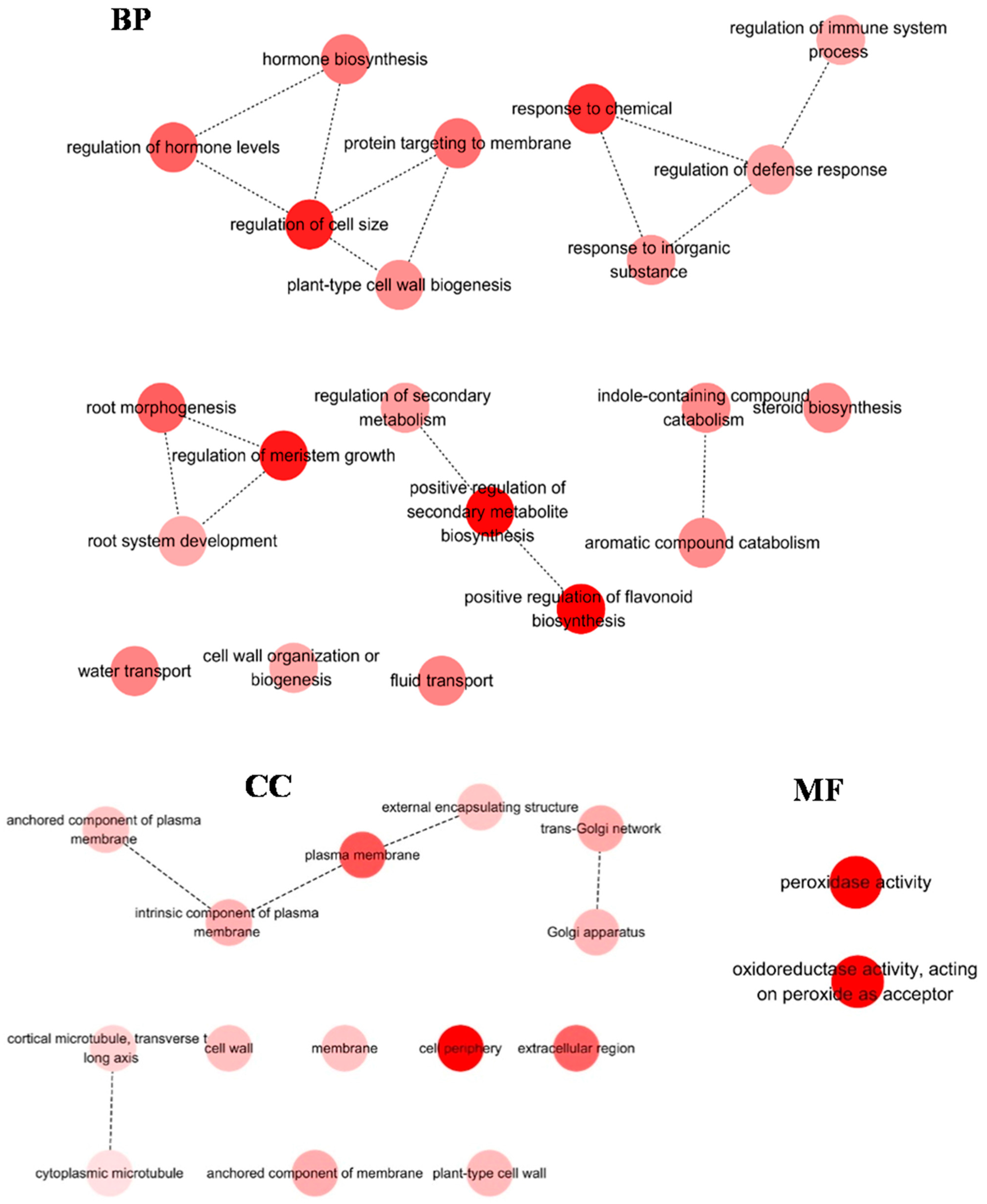
2.4. Identification of DEGs in H. ammodendron under Osmotic Stress
2.5. Screening and Characterization of the DEGs Related to Adaptation to Osmotic Stress in H. ammodendron
2.5.1. DEGs Related to Transporters
2.5.2. DEGs Related to Signal Transduction
2.5.3. DEGs Related to Reactive Oxygen Species (ROS)
2.5.4. DEGs Related to General Metabolism
2.5.5. General Stress Response Genes
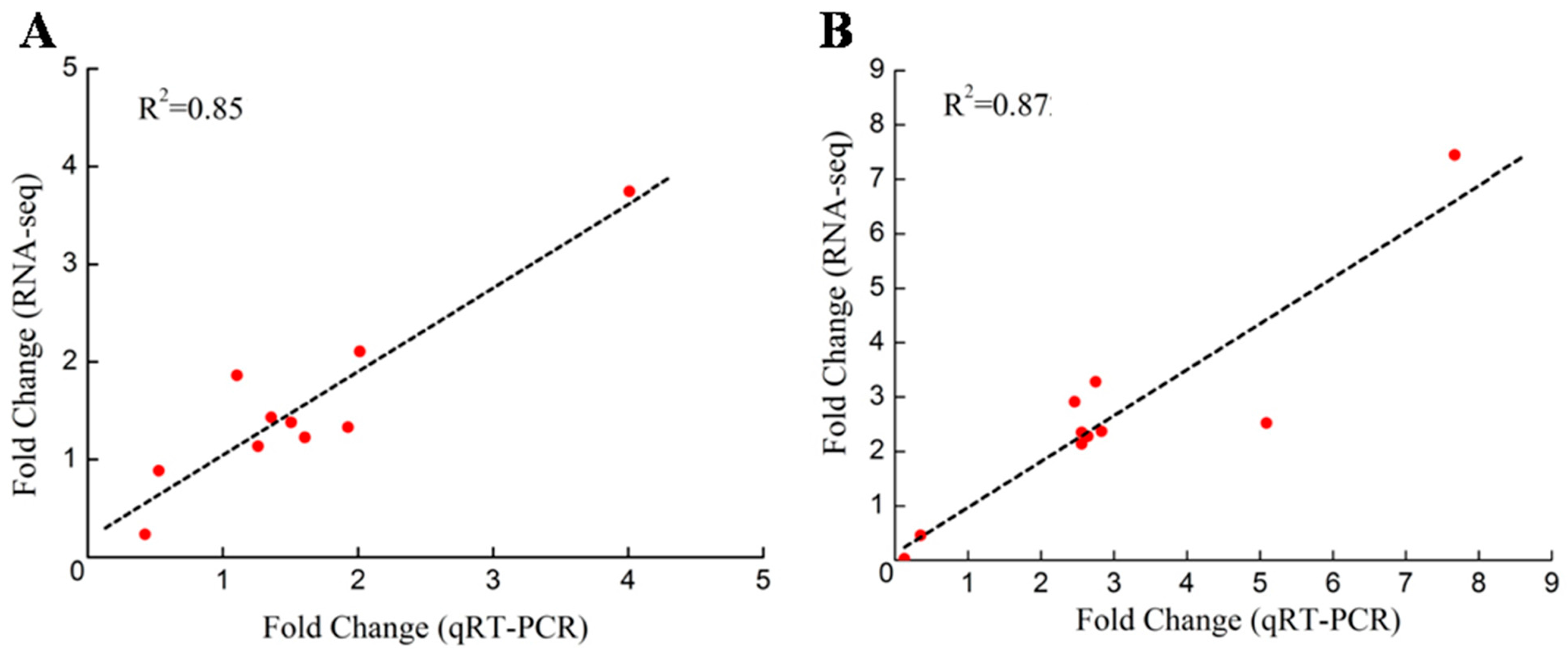
2.6. Validation of the DEGs through qRT-PCR
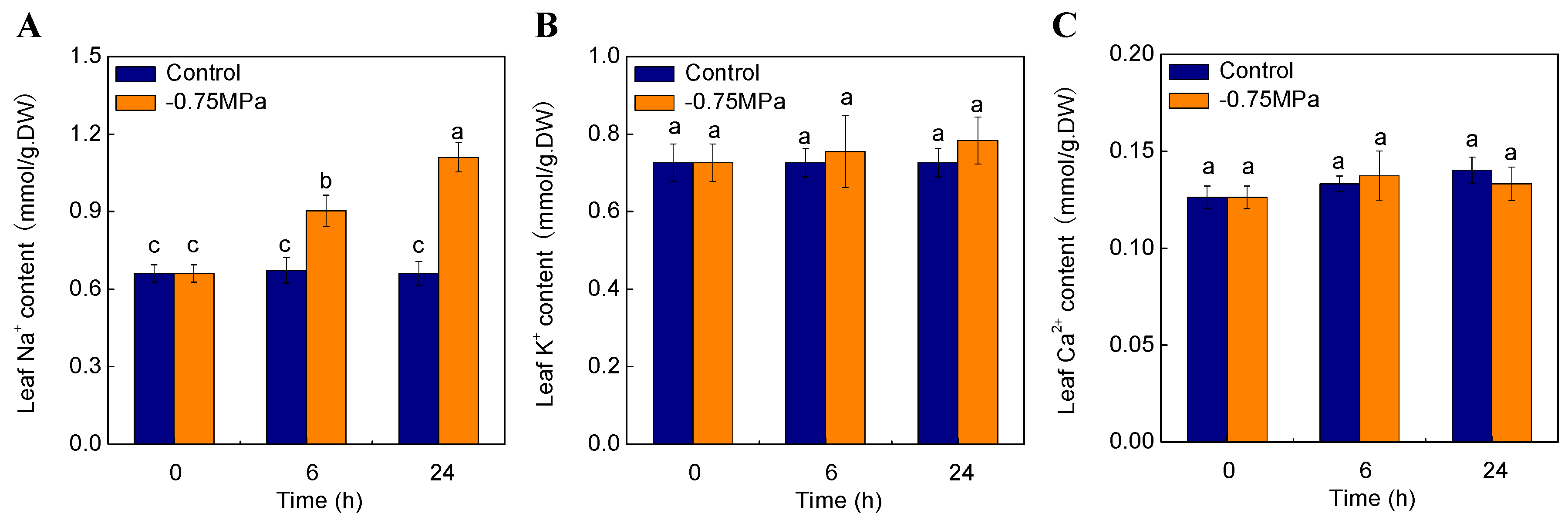
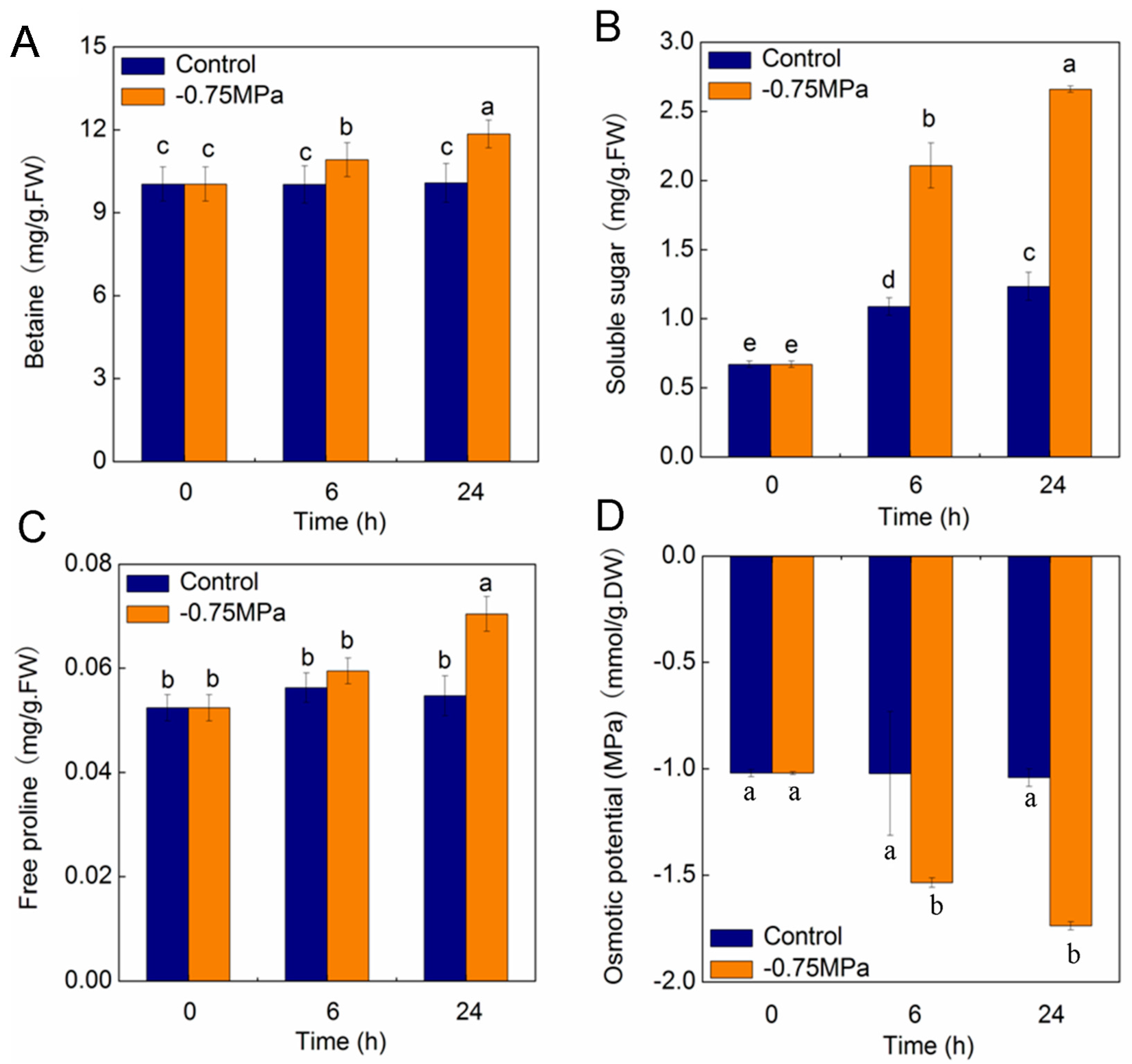
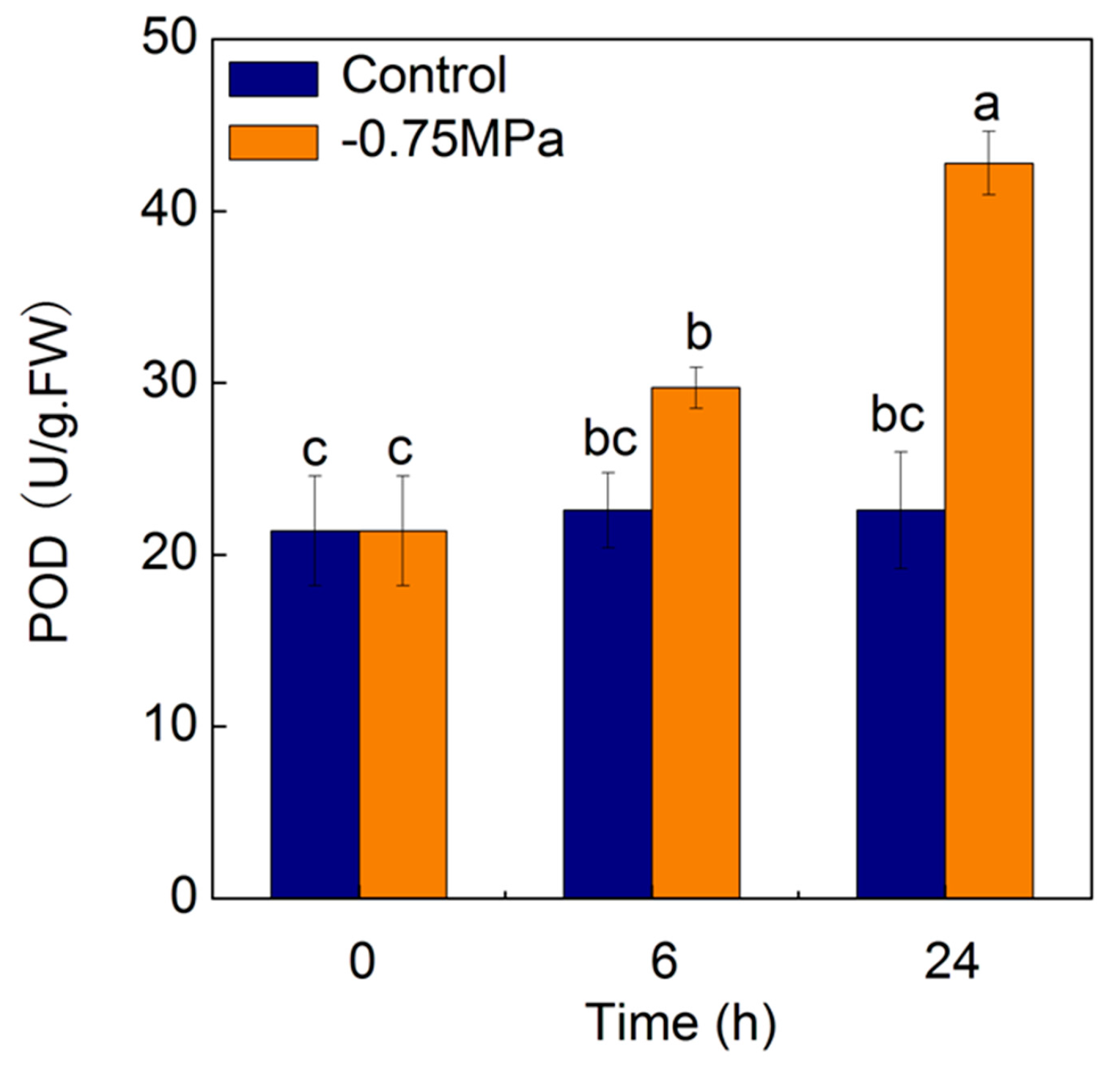
2.7. Changes of Physiological Parameters under Osmotic Stress
3. Discussion
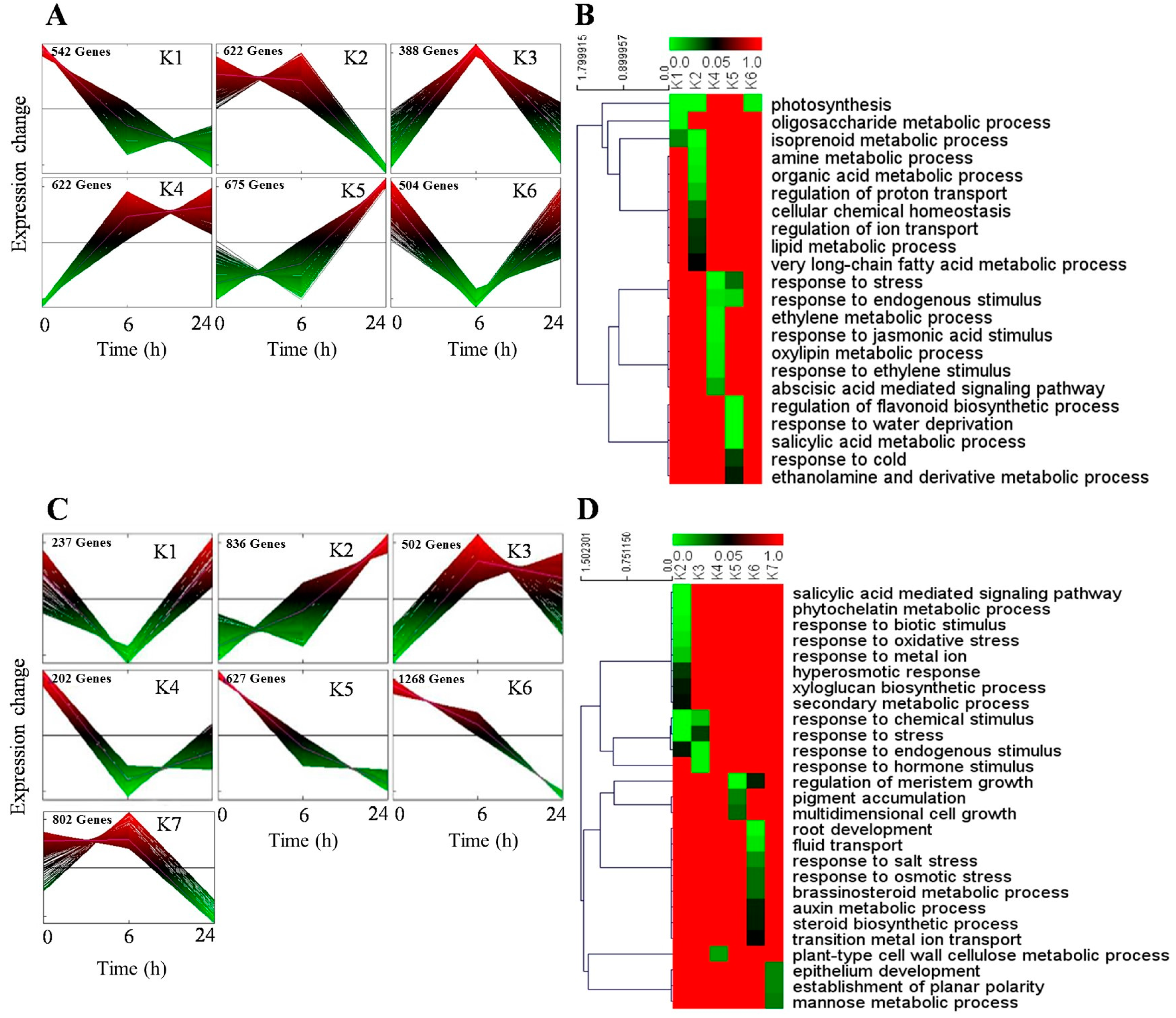
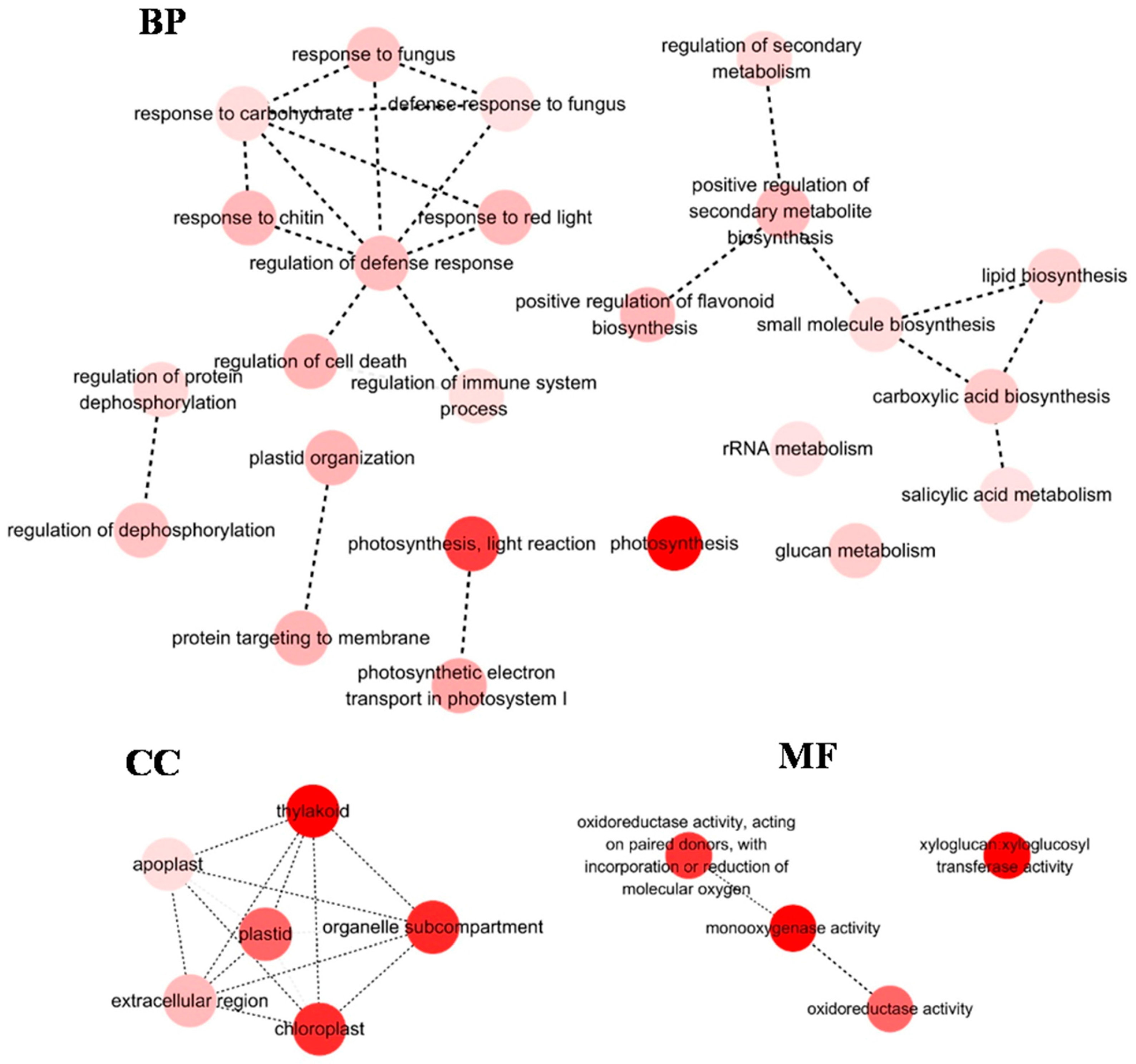
3.1. Analysis of Differentially Expressed Genes
3.2. Up-Regulation of the Genes Related to Transporters, Signal Transduction, ROS-Scavenging, Cell Wall and Membrane Stability, Secondary Metabolism Contributed to the Adaption Ability of H. ammodendron to Osmotic Stress
3.3. Altered Expression of Genes Related to Photosynthesis Also Contributed to the Adaption of H. ammodendron to Osmotic Stress
3.4. Physiological Mechanism for H. ammodendron to Adapt to Drought Stress
4. Materials and Methods
4.1. Plant Materials and Treatment Conditions
4.2. RNA Extraction, cDNA Library Creation, and Sequencing
4.3. De Novo Transcriptome Assembly and Functional Annotation
4.4. DGE Library Preparation and Sequencing
4.5. qRT-PCR Validation of Gene Expressions
4.6. Measurement of Physiological Parameters
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| ABA | Abscisic acid |
| AKT | Arabidopsis K+ transporter |
| BP | Biological process |
| C4H | Cinnamate 4-hydroxylase |
| CC | Cellular component |
| CNGC | Cyclic nucleotide-gated channel |
| COG | Clusters of orthologous groups of protein |
| CV | Coefficient of variation |
| CYP450 | Cytochrome P450 |
| DEGs | Differentially expressed genes |
| DGE | Digital gene expression |
| FBA | Fructose-1,6-bisphosphate aldolase |
| FDR | False discovery rate |
| GO | Gene ontology |
| JA | Jasmonic acid |
| KEGG | Kyoto encyclopedia of genes and genomes |
| LEA | Late embryogenesis abundant protein |
| MF | Molecular function |
| Nr | Non-redundant protein database |
| Nt | Non-redundant nucleotide sequences |
| POD | Peroxidase |
| qRT-PCR | Quantitative reverse transcription polymerase chain reaction |
| RLK | Receptor-like protein kinase |
| ROS | Reactive oxygen species |
| sHSP | small heat-shock protein |
| SSRs | Simple sequence repeats |
| TF | Transcription factor |
| TPM | Transcripts per million |
References
- Zou, J.J.; Li, X.D.; Ratnasekera, D.; Wang, C.; Liu, W.X.; Song, L.F.; Zhang, W.Z.; Wu, W.H. Arabidopsis CALCIUM-DEPENDENT PROTEIN KINASE8 and CATALASE3 Function in Abscisic Acid-Mediated Signaling and H2O2 Homeostasis in Stomatal Guard Cells under Drought Stress. Plant Cell 2015, 27, 1445–1460. [Google Scholar] [CrossRef] [PubMed]
- Farooq, M.; Wahid, A.; Kobayashi, N.; Fujita, D.; Basra, S.M.A. Plant Drought Stress: Effects, mechanisms and management. Agron. Sustain. Dev. 2009, 29, 185–212. [Google Scholar] [CrossRef]
- Zhou, Y.J.; Gao, F.; Liu, R.; Feng, J.C.; Li, H.J. De novo sequencing and analysis of root transcriptome using 454 pyrosequencing to discover putative genes associated with drought tolerance in Ammopiptanthus mongolicus. BMC Genom. 2012, 13, 266. [Google Scholar] [CrossRef] [PubMed]
- Pangle, R.E.; Limousin, J.M.; Plaut, J.A.; Pockman, W.; McDowell, N.G. Prolonged experimental drought reduces plant hydraulic conductance and transpiration and increases mortality in a pinon–Atriplex woodland. Ecol. Evol. 2015, 5, 1618–1638. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, M. Inducing drought tolerance in plants: Recent advances. Biotechnol. Adv. 2010, 28, 169–183. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Xi, J.; Wang, Q.; Bao, A.; Ma, Q.; Zhang, J.; Wang, S.M. The ZxNHX gene encoding tonoplast Na+/H+ antiporter from the xerophyte Zygophyllum xanthoxylum plays important roles in response to salt and drought. J. Plant Physiol. 2011, 168, 758–767. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Zhang, X. Some observations of the adaptations of sandy shrubs to the arid environment in the Mu Us Sandland: Leaf water relations and anatomic features. J. Arid Environ. 2001, 48, 41–48. [Google Scholar] [CrossRef]
- Yang, W.B.; Feng, W.; Jia, Z.Q.; Zhu, Y.J.; Guo, J.Y. Soil water threshold for the growth of Haloxylon ammodendron in the Ulan Buh desert in arid northwest China. S. Afr. J. Bot. 2014, 92, 53–58. [Google Scholar] [CrossRef]
- Liu, J.L.; Wang, Y.G.; Yang, X.H.; Wang, B.F. Genetic variation in seed and seedling traits of six Haloxylon ammodendron shrub provenances in desert areas of China. Agroforest. Syst. 2010, 81, 135–146. [Google Scholar] [CrossRef]
- Kang, J.J.; Zhao, W.Z.; Su, P.X.; Yang, Z.H. Sodium (Na+) and silicon (Si) coexistence promotes growth and enhances drought resistance of the succulent xerophyte Haloxylon ammodendron. Soil Sci. Plant Nutr. 2014, 60, 659–669. [Google Scholar] [CrossRef]
- Guo, Q.; Tan, D.; Liu, Y.; Wang, C. Advance in studies of Haloxylon bunge’s mechanism of adapation and resistance to drought. Forest Res. 2004, 17, 796–803. [Google Scholar]
- Wang, S.; Wan, C.; Wang, Y.; Chen, H.; Zhou, Z.; Fu, H.; Sosebee, R.E. The characteristics of Na+, K+ and free proline distribution in several drought-resistant plants of the Alxa Desert, China. J. Arid Environ. 2004, 56, 525–539. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for Transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Q.; Ma, T.; Hu, Q.J.; Liu, B.B.; Wu, Y.X.; Zhou, H.H.; Wang, Q.; Wang, J.; Liu, J.Q. Genome-scale transcriptome analysis of the desert poplar, Populus euphratica. Tree Physiol. 2011, 31, 452–461. [Google Scholar] [CrossRef] [PubMed]
- Ma, T.; Wang, J.Y.; Zhou, G.K.; Yue, Z.; Wang, J.; Liu, J.Q. Genomic insights into salt adaptation in a desert poplar. Nat. Commun. 2013, 4, 2797. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Yang, G.; Zhu, J.K.; Oliver, M.J.; He, Y. The resurrection genome of Boea hygrometrica: A blueprint for survival of dehydration. Proc. Natl. Acad. Sci. USA 2015, 112, 5833–5837. [Google Scholar] [CrossRef] [PubMed]
- Gugger, P.F.; Peñaloza-Ramírez, J.M.; Wright, J.W.; Sork, V.L. Whole-transcriptome response to water stress in a California endemic oak, Quercus lobata. Tree Physiol. 2016, 37, 632–644. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.B.; Zhou, J.; Shi, W.G.; Cao, X.; Luo, J.; Polle, A.; Luo, Z.B. Comparative transcriptomic analysis reveals the roles of overlapping heat-/drought responsive genes in poplars exposed to high temperature and drought. Sci. Rep. 2017, 7, 43215. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Bao, A.K.; Chai, W.W.; Wang, W.Y.; Zhang, J.L.; Li, Y.X.; Wang, S.W. Transcriptomic analysis of the succulent xerophyte Zygophyllum xanthoxylum in response to salt treatment and osmotic stress. Plant Soil 2016, 402, 343–361. [Google Scholar] [CrossRef]
- Jin, Y.; Jing, W.; Zhang, Q.; Zhang, W. Cyclic nucleotide gated channel 10 negatively regulates salt tolerance by mediating Na+ transport in Arabidopsis. J. Plant Res. 2015, 128, 211–220. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.M.; Zhang, J.L.; Flowers, T.J. Low-affinity Na+ uptake in the halophyte Suaeda maritima. Plant Physiol. 2007, 145, 559–571. [Google Scholar] [CrossRef] [PubMed]
- Qudeimat, E.; Faltusz, A.M.C.; Wheeler, G.; Lang, D.; Brownlee, C.; Reski, R.; Frank, W. A PIIB-type Ca2+ATPase is essential for stress adaptation in Physcomitrella patens. Proc. Natl. Acad. Sci. USA 2008, 105, 19555–19560. [Google Scholar] [CrossRef] [PubMed]
- Manohar, M.; Shigaki, T.; Hirschi, K.D. Plant cation/H+ exchangers (CAXs): Biological functions and genetic manipulations. Plant Biol. 2011, 13, 561–569. [Google Scholar] [CrossRef] [PubMed]
- Gaxiola, R.A.; Palmgren, M.G.; Schumacher, K. Plant proton pumps. FEBS Lett. 2007, 3, 330–344. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.C.; Zhang, F.L.; Yu, Y.J.; Zhang, D.S.; Zhao, X.Y. Transcriptome Profiling of Dehydration Stress in the Chinese Cabbage (Brassica rapa L. ssp. pekinensis) by Tag Sequencing. Plant Mol. Biol. Rep. 2012, 30, 17–28. [Google Scholar] [CrossRef]
- Bai, Z.Y.; Wang, T.; Wu, Y.H.; Wang, K.; Liang, Q.Y.; Pan, Y.Z. Whole-transcriptome sequence analysis of differentially expressed genes in Phormium tenaxunder drought stress. Sci. Rep. 2017, 7, 41700. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.; Fokar, M.; Abdelmageed, H.; Allen, R.D. Arabidopsis SAP5 functions as a positive regulator of stress responses and exhibits E3 ubiquitin ligase activity. Plant Mol. Biol. 2011, 75, 451–466. [Google Scholar] [CrossRef] [PubMed]
- Reddy, A.S.; Ali, G.S.; Celesnik, H.; Day, I.S. Coping with stresses: Roles of calcium- and calcium/calmodulin-regulated gene expression. Plant Cell 2011, 23, 2010–2032. [Google Scholar] [CrossRef] [PubMed]
- Ichimura, K.; Shinozaki, K.; Tena, G.; Sheen, J.; Henry, Y.; Heberle-Bors, E. Mitogen-activated protein kinase cascades in plants: A new nomenclature. Trends Plant Sci. 2002, 7, 301–308. [Google Scholar]
- Vlad, F.; Rubio, S.; Rodrigues, A.; Sirichandra, C.; Belin, C.; Robert, N.; Leung, J.; Rodriguez, P.L.; Laurière, C.; Merlot, S. Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell 2009, 21, 3170–3184. [Google Scholar] [CrossRef] [PubMed]
- Nambara, E.; Marion-Poll, A. Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 2005, 56, 165–185. [Google Scholar] [CrossRef] [PubMed]
- Martin, R.C.; Mok, D.W.S.; Smets, R.; van Onckelen, H.A.; Mok, M.C. Development of transgenic tobacco harboring a zeatin O-glucosyl transferase gene from phaseolus. In Vitro Cell Dev. Plant 2001, 37, 354–360. [Google Scholar] [CrossRef]
- Martinelli, F.; Uratsu, S.L.; Albrecht, U.; Reagan, R.L.; Phu, M.L.; Britton, M.; Buffalo, V.; Fass, J.; Leicht, E.; Zhao, W.X.; et al. Transcriptome profiling of citrus fruit response to huanglongbing disease. PLoS ONE 2012, 7, 38039. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Yang, C.; Chen, S.; Zhang, X.; Wang, D.J. Transcriptomic basis for drought resistance in Brassica napus L. Sci. Rep. 2017, 7, 40532. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Liang, X.; Wan, Q.; Wang, X.; Bi, Y. Ethylene and nitric oxide are involved in maintaining ion homeostasis in Arabidopsis callus under salt stress. Planta 2009, 230, 293–307. [Google Scholar] [CrossRef] [PubMed]
- Cenzano, A.; Abdala, G.; Hause, B. Cytochemical immuno-localization of allene oxide cyclase, a jasmonic acid biosynthetic enzyme, in developing potato stolons. J. Plant Physiol. 2007, 164, 1449–1456. [Google Scholar] [CrossRef] [PubMed]
- Tani, T.; Sobajima, H.; Okada, K.; Chujo, T.; Arimura, S.I.; Tsutsumi, N.; Nishimura, M.; Seto, H.; Nojiri, H.; Yamane, H. Identification of the OsOPR7 gene encoding 12-oxophytodienoate reductase involved in the biosynthesis of jasmonic acid in rice. Planta 2007, 227, 517–526. [Google Scholar] [CrossRef] [PubMed]
- Shan, C.; Liang, Z. Jasmonic acid regulates ascorbate and glutathione metabolism in Agropyron cristatum leaves under water stress. Plant Sci. 2010, 178, 130–139. [Google Scholar] [CrossRef]
- Zalewski, W.; Galuszka, P.; Gasparis, S.; Orczyk, W.; Nadolska-Orczyk, A. Silencing of the HvCKX1 gene decreases the cytokinin oxidase/dehydrogenase level in barley and leads to higher plant productivity. J. Exp. Bot. 2010, 61, 1839–1851. [Google Scholar] [CrossRef] [PubMed]
- Reguera, M.; Peleg, Z.; Abdel-Tawab, Y.M.; Tumimbang, E.B.; Delatorre, C.A.; Blumwald, E. Stress-induced cytokinin synthesis increases drought tolerance through the coordinated regulation of carbon and nitrogen assimilation in rice. Plant Physiol. 2013, 163, 1609–1622. [Google Scholar] [CrossRef] [PubMed]
- Sevilla, F.; Camejo, D.; Ortiz-Espin, A.; Calderon, A.; Lazaro, J.J.; Jimenez, A. The thioredoxin/peroxiredoxin/sulfiredoxin system: Current overview on its redox function in plants and regulation by reactive oxygen and nitrogen species. J. Exp. Bot. 2015, 66, 2945–2955. [Google Scholar] [CrossRef] [PubMed]
- Dahal, K.; Vanlerberghe, G.C. Alternative oxidase respiration maintains both mitochondrial and chloroplast function during drought. New Phytol. 2017, 213, 560–571. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.K.; Kim, J.E.; Park, J.A.; Eom, T.J.; Kim, W.T. Constitutive expression of abiotic stress-inducible hot pepper CaXTH3, which encodes a xyloglucan endotransglucosylase/hydrolase homolog, improves drought and salt tolerance in transgenic Arabidopsis plants. FEBS Lett. 2006, 580, 3136–3144. [Google Scholar] [CrossRef] [PubMed]
- Li, C.Y.; Deng, G.M.; Yang, J.; Viljoen, A.; Jin, Y.; Kuang, R.B.; Zuo, C.W.; Lv, Z.C.; Yang, Q.S.; Sheng, O.; et al. Transcriptome profiling of resistant and susceptible Cavendish banana roots following inoculation with Fusarium oxysporum f. sp. cubense tropical race 4. BMC Genom. 2012, 13, 1–11. [Google Scholar]
- Bray, E.A. Genes commonly regulated by water-deficit stress in Arabidopsis thaliana. J. Exp. Bot. 2004, 55, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Fu, J.; Gou, M.; Huai, J.; Liu, Y.; Jian, M.; Huang, Q.S.; Guo, X.Y.; Dong, Z.G.; Wang, H.Z.; et al. Genome-wide transcriptome analysis of two maize inbred lines under drought stress. Plant Mol. Biol. 2010, 72, 407–421. [Google Scholar] [CrossRef] [PubMed]
- Weng, J.; Chapple, C. The origin and evolution of lignin biosynthesis. New Phytol. 2010, 187, 273–285. [Google Scholar] [CrossRef] [PubMed]
- Liang, M.; Haroldsen, V.; Cai, X.; Wu, Y. Expression of a putative laccase gene, ZmLAC1, in maize primary roots under stress. Plant Cell Environ. 2006, 29, 746–753. [Google Scholar] [CrossRef] [PubMed]
- Bjurhager, I.; Olsson, A.; Zhang, B.; Gerber, L.R.; Kumar, M.S.; Berglund, L.A.; Burgert, I.; Sundberg, B.; Salme´n, L. Ultrastructure and mechanical properties of populus wood with reduced lignin content caused by transgenic down-regulation of cinnamate 4-hydroxylase. Biomacromolecules 2010, 11, 2359–2365. [Google Scholar] [CrossRef] [PubMed]
- Duan, J.; Zhang, M.; Zhang, H.; Xiong, H.; Liu, P.; Ali, J.; Li, J.J.; Li, Z.C. OsMIOX, a myo-inositol oxygenase gene, improves drought tolerance through scavenging of reactive oxygen species in rice (Oryza sativa L.). Plant Sci. 2012, 196, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Xu, C.; Libeisson, Y.; Philippar, K. Fatty acid and lipid transport in plant cells. Trends Plant Sci. 2016, 21, 145. [Google Scholar] [CrossRef] [PubMed]
- Dao, T.T.H.; Linthorst, H.J.M.; Verpoorte, R. Chalcone synthase and its functions in plant resistance. Phytochem. Rev. 2011, 10, 397–412. [Google Scholar] [CrossRef] [PubMed]
- Dehghan, S.; Sadeghi, M.; Poppel, A.; Fischer, R.; Lakes-Harlan, R.; Kavousi, H.R. Differential inductions of phenylalanine ammonia-lyase and chalcone synthase during wounding, salicylic acid treatment, and salinity stress in safflower, Carthamus tinctorius. Biosci. Rep. 2014, 34, 689–699. [Google Scholar] [CrossRef] [PubMed]
- Abrahams, S.; Lee, E.; Walker, A.R.; Tanner, G.J.; Larkin, P.J.; Ashton, A.R. The Arabidopsis TDS4 gene encodes leucoanthocyanidin dioxygenase (LDOX) and is essential for proanthocyanidin synthesis and vacuole development. Plant J. 2003, 35, 624–636. [Google Scholar] [CrossRef] [PubMed]
- Harish, M.C.; Dachinamoorthy, P.; Balamurugan, S.; Bala Murugan, S.; Sathishkumar, R. Overexpression of homogentisate phytyltransferase (HPT) and tocopherol cyclase (TC) enhances α-tocopherol content in transgenic tobacco. Biol. Plantarum 2013, 57, 395–400. [Google Scholar] [CrossRef]
- Nebert, D.W.; Dalton, T.P. The role of cytochrome P450 enzymes in endogenous signaling pathways and environmental carcinogenesis. Nat. Rev. Cancer 2006, 6, 947–960. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Michael, T.P.; Hudson, M.E.; Kay, S.A.; Chory, J.; Schuler, M.A. Cytochrome P450 monooxygenases as reporters for circadian-regulated pathways. Plant Physiol. 2009, 150, 858–878. [Google Scholar] [CrossRef] [PubMed]
- Kurepin, L.V.; Ivanov, A.G.; Zaman, M.; Pharis, R.P.; Allakhverdiev, S.I.; Hurry, V. Stress-related hormones and glycinebetaine interplay in protection of photosynthesis under abiotic stress conditions. Photosynth. Res. 2015, 126, 221–235. [Google Scholar] [CrossRef] [PubMed]
- Diray-Arce, J.; Clement, M.; Gul, B.; Khan, M.A.; Nielsen, B.L. Transcriptome assembly, profiling and differential gene expression analysis of the halophyte Suaeda fruticosa provides insights into salt tolerance. BMC Genom. 2015, 16, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, Z.; Wang, L.; Wu, R.; Phillips, J.; Deng, X. LEA 4 group genes from the resurrection plant Boea hygrometrica confer dehydration tolerance in transgenic tobacco. Plant Sci. 2009, 176, 90–98. [Google Scholar] [CrossRef]
- Kosova, K.; Vitamvas, P.; Prasil, I.T. Wheat and barley dehydrins under cold, drought, and salinity—What can LEA-II proteins tell us about plant stress response? Front. Plant Sci. 2014, 5, 343. [Google Scholar] [CrossRef] [PubMed]
- Stein, H.; Honig, A.; Miller, G.; Erster, O.; Eilenberg, H.; Csonka, L.N.; Szabados, L.; Koncz, C.; Zilberstein, A. Elevation of free proline and proline-rich protein levels by simultaneous manipulations of proline biosynthesis and degradation in plants. Plant Sci. 2011, 181, 140–150. [Google Scholar] [CrossRef] [PubMed]
- Malinovska, L.; Kroschwald, S.; Munder, M.C.; Richter, D.; Alberti, S. Molecular chaperones and stress-inducible protein-sorting factors coordinate the spatiotemporal distribution of protein aggregates. Mol. Biol. Cell 2012, 23, 3041–3056. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Yokoya, S. Enhanced tolerance to drought stress in transgenic rice plants overexpressing a small heat-shock protein, sHSP17.7. Plant Cell Rep. 2008, 27, 329–334. [Google Scholar] [CrossRef] [PubMed]
- Dana, M.D.L.M.; Pintor-Toro, J.A.; Cubero, B. Transgenic tobacco plants overexpressing chitinases of fungal origin show enhanced resistance to biotic and abiotic stress agents. Plant Physiol. 2006, 142, 722–730. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Yang, J.Y.; Yan, S.J.; Zhang, S.H.; Zhu, X.Y.; Liu, B. The germin-like protein OsGLP2-1 enhances resistance to fungal blast and bacterial blight in rice. Plant Mol. Biol. 2016, 92, 411–423. [Google Scholar] [CrossRef] [PubMed]
- Gokulakannan, G.G.; Niehaus, K. Characterization of the Medicago truncatula cell wall proteome in cell suspension culture upon elicitation and suppression of plant defense. J. Plant Physiol. 2010, 167, 1533–1541. [Google Scholar] [CrossRef] [PubMed]
- Chaves, M.M.; Flexas, J.; Pinheiro, C. Photosynthesis under drought and salt stress: Regulation mechanisms from whole plant to cell. Ann. Bot. 2009, 103, 551–560. [Google Scholar] [CrossRef] [PubMed]
- Li, C.L.; Wang, Y.; Liu, L.; Hu, Y.; Zhang, F.; Mergen, S.; Wang, G.D.; Schläppi, M.R.; Chu, C.C. A rice plastidial nucleotide sugar epimerase is involved in galactolipid biosynthesis and improves photosynthetic efficiency. PLoS Genet. 2011, 7, 545–547. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Wei, L.; Wang, Q.; Shi, D.; Chen, H. Increased activity of the non-regulated enzymes fructose-1,6-bisphosphate aldolase and triosephosphate isomerase in Anabaena sp. strain PCC 7120 increases photosynthetic yield. J. Appl. Phycol. 2006, 9, 207–213. [Google Scholar] [CrossRef]
- Woodson, J.D.; Perezruiz, J.M.; Chory, J. Heme synthesis by plastid ferrochelatase I regulates nuclear gene expression in Plants. Curr. Biol. 2011, 21, 897–903. [Google Scholar] [CrossRef] [PubMed]
- David, P.; des Francssmall, C.C.; Sevignac, M.; Thareau, V.; Macadre, C.; Langin, T.; Geffroy, V. Three highly similar formate dehydrogenase genes located in the vicinity of the B4 resistance gene cluster are differentially expressed under biotic and abiotic stresses in Phaseolus vulgaris. Theor. Appl. Genet. 2010, 121, 87–103. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Devaiah, S.P.; Narasimhan, R.; Pan, X.Q.; Zhang, Y.Y.; Zhang, W.H. Cytosolic glyceraldehyde-3-phosphate dehydrogenases interact with phospholipase D-δ to transduce hydrogen peroxide signals in the Arabidopsis response to stress. Plant Cell 2012, 24, 2200–2212. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.L.; Flowers, T.J.; Wang, S.M. Mechanisms of sodium uptake by roots of higher plants. Plant Soil 2010, 326, 45–60. [Google Scholar] [CrossRef]
- Anschütz, U.; Becker, D.; Shabala, S. Going beyond nutrition: Regulation of potassium homoeostasis as a common denominator of plant adaptive responses to environment. J. Plant Physiol. 2014, 171, 670–687. [Google Scholar] [CrossRef] [PubMed]
- Waditee, R.; Bhuiyan, N.H.; Hirata, E.; Hibino, T.; Tanaka, Y.; Shikata, M.; Takabe, T. Metabolic engineering for betaine accumulation in microbes and plants. J. Biol. Chem. 2007, 282, 34185–34193. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.H.H.; Murata, N. Glycinebetaine protects plants against abiotic stress: Mechanisms and biotechnological applications. Plant Cell Environ. 2011, 34, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.H.H.; Murata, N. Glycinebetaine: An effective protectant against abiotic stress in plants. Trends Plant Sci. 2008, 13, 499–505. [Google Scholar] [CrossRef] [PubMed]
- Loreti, E.; Bellis, L.D.; Alpi, A.; Perata, P. Why and how do plant cells sense sugars? Ann. Bot. 2001, 88, 803–812. [Google Scholar] [CrossRef]
- Hu, H.H.; Dai, M.Q.; Yao, J.L.; Xiao, B.Z.; Li, X.G.; Zhang, Q.F.; Xiong, L.Z. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc. Natl. Acad. Sci. USA 2006, 103, 12987–12992. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Liang, D.; Li, C.; Hao, Y.; Ma, F.; Shu, H. Influence of drought stress on the cellular ultrastructure and antioxidant system in leaves of drought-tolerant and drought-sensitive apple rootstocks. Plant Physiol. Biochem. 2012, 51, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Pertea, G.; Huang, X.; Liang, F.; Antonescu, V.; Sultana, R.; Karamycheva, S.; Lee, Y.; White, J.; Cheung, F.; Quackenbush, J. TIGR Gene Indices clustering tools (TGICL): A software system for fast clustering of large EST datasets. Bioinformatics 2003, 19, 651–652. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Fang, L.; Zheng, H.; Zhang, Y.; Chen, J.; Zhang, Z.; Wang, J.; Li, S.; Li, R.; Bolund, L.; et al. WEGO: A web tool for plotting GO annotations. Nucleic Acids Res. 2006, 34, 293–297. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.; Bao, Y.Y.; Li, B.-L.; Cheng, Y.B.; Peng, Z.Y.; Liu, H.; Xu, H.J.; Zhu, Z.R.; Lou, Y.G.; Cheng, J.A.; Zhang, C.X. Transcriptome analysis of the brown planthopper Nilaparvata lugens. PLoS ONE 2010, 5, e14233. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Yu, C.Y.; Li, H.X.; Ouyang, B.; Wang, T.T.; Zhang, J.H. Overexpression of ShDHN, a dehydrin gene from Solanum habrochaites enhances tolerance to multiple abiotic stresses in tomato. Plant Sci. 2015, 231, 198–211. [Google Scholar] [CrossRef] [PubMed]
| Unigenes | Total Number | Total Length (bp) | Mean Length (bp) | N50 (bp) |
|---|---|---|---|---|
| Shoots | 82,736 | 46,038,066 | 556 | 867 |
| Roots | 99,624 | 53,273,063 | 535 | 828 |
| All | 87,109 | 59,244,982 | 680 | 1064 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, H.-J.; Lü, X.-P.; Zhang, L.; Qiao, Y.; Zhao, Q.; Wang, Y.-P.; Li, M.-F.; Zhang, J.-L. Transcriptomic Profiling and Physiological Analysis of Haloxylon ammodendron in Response to Osmotic Stress. Int. J. Mol. Sci. 2018, 19, 84. https://doi.org/10.3390/ijms19010084
Gao H-J, Lü X-P, Zhang L, Qiao Y, Zhao Q, Wang Y-P, Li M-F, Zhang J-L. Transcriptomic Profiling and Physiological Analysis of Haloxylon ammodendron in Response to Osmotic Stress. International Journal of Molecular Sciences. 2018; 19(1):84. https://doi.org/10.3390/ijms19010084
Chicago/Turabian StyleGao, Hui-Juan, Xin-Pei Lü, Ling Zhang, Yan Qiao, Qi Zhao, Yong-Ping Wang, Meng-Fei Li, and Jin-Lin Zhang. 2018. "Transcriptomic Profiling and Physiological Analysis of Haloxylon ammodendron in Response to Osmotic Stress" International Journal of Molecular Sciences 19, no. 1: 84. https://doi.org/10.3390/ijms19010084





