Unfolded Protein Response-Dependent Communication and Contact among Endoplasmic Reticulum, Mitochondria, and Plasma Membrane
Abstract
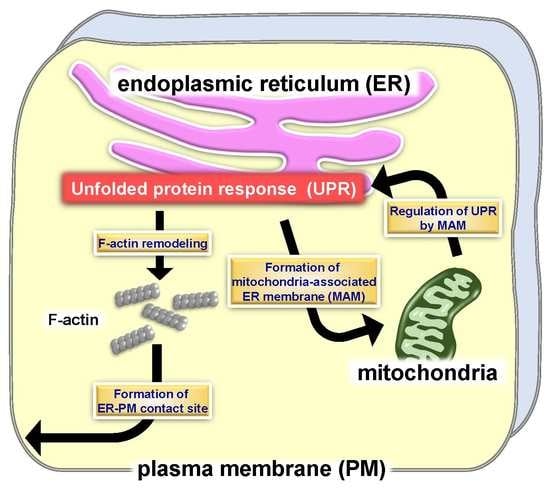
:1. Introduction
2. ER Stress Transducers and Key Molecules of the UPR
3. Morphological Changes to the ER by the UPR
4. Mitochondria-Associated ER Membrane (MAM) and UPR
5. ER-PM Contact Sites and the UPR
6. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kaufman, R.J. Orchestrating the unfolded protein response in health and disease. J. Clin. Investig. 2002, 110, 1389–1398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ron, D. Translational control in the endoplasmic reticulum stress response. J. Clin. Investig. 2002, 110, 1383–1388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rutkowski, D.T.; Kaufman, R.J. A trip to the er: Coping with stress. Trends Cell Biol. 2004, 14, 20–28. [Google Scholar] [CrossRef] [PubMed]
- Ron, D.; Walter, P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell. Biol. 2007, 8, 519–529. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Kaufman, R.J. From acute er stress to physiological roles of the unfolded protein response. Cell Death Differ. 2006, 13, 374–384. [Google Scholar] [CrossRef] [PubMed]
- Reimold, A.M.; Iwakoshi, N.N.; Manis, J.; Vallabhajosyula, P.; Szomolanyi-Tsuda, E.; Gravallese, E.M.; Friend, D.; Grusby, M.J.; Alt, F.; Glimcher, L.H. Plasma cell differentiation requires the transcription factor xbp-1. Nature 2001, 412, 300–307. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Shen, X.; Wu, J.; Sakaki, K.; Saunders, T.; Rutkowski, D.T.; Back, S.H.; Kaufman, R.J. Endoplasmic reticulum stress activates cleavage of crebh to induce a systemic inflammatory response. Cell 2006, 124, 587–599. [Google Scholar] [CrossRef] [PubMed]
- Vecchi, C.; Montosi, G.; Zhang, K.; Lamberti, I.; Duncan, S.A.; Kaufman, R.J.; Pietrangelo, A. ER stress controls iron metabolism through induction of hepcidin. Science 2009, 325, 877–880. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.H.; Scapa, E.F.; Cohen, D.E.; Glimcher, L.H. Regulation of hepatic lipogenesis by the transcription factor xbp1. Science 2008, 320, 1492–1496. [Google Scholar] [CrossRef] [PubMed]
- Mao, T.; Shao, M.; Qiu, Y.; Huang, J.; Zhang, Y.; Song, B.; Wang, Q.; Jiang, L.; Liu, Y.; Han, J.D.; et al. PKA phosphorylation couples hepatic inositol-requiring enzyme 1α to glucagon signaling in glucose metabolism. Proc. Natl. Acad. Sci. USA 2011, 108, 15852–15857. [Google Scholar] [CrossRef] [PubMed]
- Iwakoshi, N.N.; Lee, A.H.; Glimcher, L.H. The x-box binding protein-1 transcription factor is required for plasma cell differentiation and the unfolded protein response. Immunol. Rev. 2003, 194, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Gass, J.N.; Gifford, N.M.; Brewer, J.W. Activation of an unfolded protein response during differentiation of antibody-secreting B cells. J. Biol. Chem. 2002, 277, 49047–49054. [Google Scholar] [CrossRef] [PubMed]
- Saito, A.; Hino, S.; Murakami, T.; Kanemoto, S.; Kondo, S.; Saitoh, M.; Nishimura, R.; Yoneda, T.; Furuichi, T.; Ikegawa, S.; et al. Regulation of endoplasmic reticulum stress response by a bbf2h7-mediated sec23a pathway is essential for chondrogenesis. Nat. Cell Biol. 2009, 11, 1197–1204. [Google Scholar] [CrossRef] [PubMed]
- Murakami, T.; Saito, A.; Hino, S.; Kondo, S.; Kanemoto, S.; Chihara, K.; Sekiya, H.; Tsumagari, K.; Ochiai, K.; Yoshinaga, K.; et al. Signalling mediated by the endoplasmic reticulum stress transducer oasis is involved in bone formation. Nat. Cell Biol. 2009, 11, 1205–1211. [Google Scholar] [CrossRef] [PubMed]
- Sha, H.; He, Y.; Chen, H.; Wang, C.; Zenno, A.; Shi, H.; Yang, X.; Zhang, X.; Qi, L. The ire1alpha-xbp1 pathway of the unfolded protein response is required for adipogenesis. Cell Metab. 2009, 9, 556–564. [Google Scholar] [CrossRef] [PubMed]
- West, M.; Zurek, N.; Hoenger, A.; Voeltz, G.K. A 3d analysis of yeast er structure reveals how ER domains are organized by membrane curvature. J. Cell Biol. 2011, 193, 333–346. [Google Scholar] [CrossRef] [PubMed]
- Allison, R.; Edgar, J.R.; Pearson, G.; Rizo, T.; Newton, T.; Gunther, S.; Berner, F.; Hague, J.; Connell, J.W.; Winkler, J.; et al. Defects in ER-endosome contacts impact lysosome function in hereditary spastic paraplegia. J. Cell Biol. 2017, 216, 1337–1355. [Google Scholar] [CrossRef] [PubMed]
- Grimm, S. The ER-mitochondria interface: The social network of cell death. Biochim. Biophys. Acta 2012, 1823, 327–334. [Google Scholar] [CrossRef] [PubMed]
- Csordas, G.; Renken, C.; Varnai, P.; Walter, L.; Weaver, D.; Buttle, K.F.; Balla, T.; Mannella, C.A.; Hajnoczky, G. Structural and functional features and significance of the physical linkage between ER and mitochondria. J. Cell Biol. 2006, 174, 915–921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tirasophon, W.; Welihinda, A.A.; Kaufman, R.J. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (ire1p) in mammalian cells. Genes Dev. 1998, 12, 1812–1824. [Google Scholar] [CrossRef] [PubMed]
- Harding, H.P.; Zhang, Y.; Ron, D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 1999, 397, 271–274. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, H.; Okada, T.; Haze, K.; Yanagi, H.; Yura, T.; Negishi, M.; Mori, K. Atf6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol. Cell. Biol. 2000, 20, 6755–6767. [Google Scholar] [CrossRef] [PubMed]
- Calfon, M.; Zeng, H.; Urano, F.; Till, J.H.; Hubbard, S.R.; Harding, H.P.; Clark, S.G.; Ron, D. Ire1 couples endoplasmic reticulum load to secretory capacity by processing the xbp-1 mRNA. Nature 2002, 415, 92–96. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, H.; Matsui, T.; Yamamoto, A.; Okada, T.; Mori, K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 2001, 107, 881–891. [Google Scholar] [CrossRef]
- Yoshida, H.; Matsui, T.; Hosokawa, N.; Kaufman, R.J.; Nagata, K.; Mori, K. A time-dependent phase shift in the mammalian unfolded protein response. Dev. Cell 2003, 4, 265–271. [Google Scholar] [CrossRef]
- Harding, H.P.; Novoa, I.; Zhang, Y.; Zeng, H.; Wek, R.; Schapira, M.; Ron, D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 2000, 6, 1099–1108. [Google Scholar] [CrossRef]
- Shi, Y.; Vattem, K.M.; Sood, R.; An, J.; Liang, J.; Stramm, L.; Wek, R.C. Identification and characterization of pancreatic eukaryotic initiation factor 2 α-subunit kinase, PEK, involved in translational control. Mol. Cell. Biol. 1998, 18, 7499–7509. [Google Scholar] [CrossRef] [PubMed]
- Vattem, K.M.; Wek, R.C. Reinitiation involving upstream orfs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. USA 2004, 101, 11269–11274. [Google Scholar] [CrossRef] [PubMed]
- Barbosa-Tessmann, I.P.; Chen, C.; Zhong, C.; Siu, F.; Schuster, S.M.; Nick, H.S.; Kilberg, M.S. Activation of the human asparagine synthetase gene by the amino acid response and the endoplasmic reticulum stress response pathways occurs by common genomic elements. J. Biol. Chem. 2000, 275, 26976–26985. [Google Scholar] [CrossRef] [PubMed]
- Roybal, C.N.; Hunsaker, L.A.; Barbash, O.; Vander Jagt, D.L.; Abcouwer, S.F. The oxidative stressor arsenite activates vascular endothelial growth factor mRNA transcription by an ATF4-dependent mechanism. J. Biol. Chem. 2005, 280, 20331–20339. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.Z.; Lawson, B.; Brewer, J.W.; Zinszner, H.; Sanjay, A.; Mi, L.J.; Boorstein, R.; Kreibich, G.; Hendershot, L.M.; Ron, D. Signals from the stressed endoplasmic reticulum induce C/EBP-homologous protein (CHOP/GADD153). Mol. Cell. Biol. 1996, 16, 4273–4280. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Brewer, J.W.; Diehl, J.A.; Hendershot, L.M. Two distinct stress signaling pathways converge upon the chop promoter during the mammalian unfolded protein response. J. Mol. Biol. 2002, 318, 1351–1365. [Google Scholar] [CrossRef]
- Harding, H.P.; Zhang, Y.; Zeng, H.; Novoa, I.; Lu, P.D.; Calfon, M.; Sadri, N.; Yun, C.; Popko, B.; Paules, R.; et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 2003, 11, 619–633. [Google Scholar] [CrossRef]
- Marciniak, S.J.; Yun, C.Y.; Oyadomari, S.; Novoa, I.; Zhang, Y.; Jungreis, R.; Nagata, K.; Harding, H.P.; Ron, D. Chop induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 2004, 18, 3066–3077. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Rawson, R.B.; Komuro, R.; Chen, X.; Dave, U.P.; Prywes, R.; Brown, M.S.; Goldstein, J.L. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process srebps. Mol. Cell 2000, 6, 1355–1364. [Google Scholar] [CrossRef]
- Chen, X.; Shen, J.; Prywes, R. The luminal domain of atf6 senses endoplasmic reticulum (ER) stress and causes translocation of ATF6 from the ER to the golgi. J. Biol. Chem. 2002, 277, 13045–13052. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.; Sato, T.; Matsui, T.; Sato, M.; Okada, T.; Yoshida, H.; Harada, A.; Mori, K. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6α and XBP1. Dev. Cell 2007, 13, 365–376. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.C.; Dougan, S.K.; McGehee, A.M.; Love, J.C.; Ploegh, H.L. Xbp-1 regulates signal transduction, transcription factors and bone marrow colonization in b cells. EMBO J 2009, 28, 1624–1636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodnow, C.C.; Crosbie, J.; Adelstein, S.; Lavoie, T.B.; Smith-Gill, S.J.; Brink, R.A.; Pritchard-Briscoe, H.; Wotherspoon, J.S.; Loblay, R.H.; Raphael, K.; et al. Altered immunoglobulin expression and functional silencing of self-reactive b lymphocytes in transgenic mice. Nature 1988, 334, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Martinon, F.; Chen, X.; Lee, A.H.; Glimcher, L.H. Tlr activation of the transcription factor xbp1 regulates innate immune responses in macrophages. Nat. Immunol. 2010, 11, 411–418. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Mao, T.; Zhang, Y.; Shao, M.; You, J.; Ding, Q.; Chen, Y.; Wu, D.; Xie, D.; Lin, X.; et al. A crucial role for RACK1 in the regulation of glucose-stimulated IRE1α activation in pancreatic beta cells. Sci. Signal. 2010, 3, ra7. [Google Scholar] [CrossRef] [PubMed]
- Lipson, K.L.; Fonseca, S.G.; Ishigaki, S.; Nguyen, L.X.; Foss, E.; Bortell, R.; Rossini, A.A.; Urano, F. Regulation of insulin biosynthesis in pancreatic beta cells by an endoplasmic reticulum-resident protein kinase ire1. Cell Metab. 2006, 4, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Karali, E.; Bellou, S.; Stellas, D.; Klinakis, A.; Murphy, C.; Fotsis, T. VEGF signals through ATF6 and perk to promote endothelial cell survival and angiogenesis in the absence of ER stress. Mol. Cell 2014, 54, 559–572. [Google Scholar] [CrossRef] [PubMed]
- Hetz, C.; Chevet, E.; Oakes, S.A. Proteostasis control by the unfolded protein response. Nat. Cell Biol. 2015, 17, 829–838. [Google Scholar] [CrossRef] [PubMed]
- Schroder, M.; Kaufman, R.J. The mammalian unfolded protein response. Annu. Rev. Biochem. 2005, 74, 739–789. [Google Scholar] [CrossRef] [PubMed]
- Sriburi, R.; Jackowski, S.; Mori, K.; Brewer, J.W. Xbp1: A link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. J. Cell Biol. 2004, 167, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Shaffer, A.L.; Shapiro-Shelef, M.; Iwakoshi, N.N.; Lee, A.H.; Qian, S.B.; Zhao, H.; Yu, X.; Yang, L.; Tan, B.K.; Rosenwald, A.; et al. Xbp1, downstream of blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity 2004, 21, 81–93. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.H.; Iwakoshi, N.N.; Glimcher, L.H. Xbp-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol. Cell. Biol. 2003, 23, 7448–7459. [Google Scholar] [CrossRef] [PubMed]
- Calame, K.L.; Lin, K.I.; Tunyaplin, C. Regulatory mechanisms that determine the development and function of plasma cells. Annu. Rev. Immunol. 2003, 21, 205–230. [Google Scholar] [CrossRef] [PubMed]
- Wiest, D.L.; Burkhardt, J.K.; Hester, S.; Hortsch, M.; Meyer, D.I.; Argon, Y. Membrane biogenesis during B cell differentiation: Most endoplasmic reticulum proteins are expressed coordinately. J. Cell Biol. 1990, 110, 1501–1511. [Google Scholar] [CrossRef] [PubMed]
- McGehee, A.M.; Dougan, S.K.; Klemm, E.J.; Shui, G.; Park, B.; Kim, Y.M.; Watson, N.; Wenk, M.R.; Ploegh, H.L.; Hu, C.C. Xbp-1-deficient plasmablasts show normal protein folding but altered glycosylation and lipid synthesis. J. Immunol. 2009, 183, 3690–3699. [Google Scholar] [CrossRef] [PubMed]
- Lykidis, A.; Jackowski, S. Regulation of mammalian cell membrane biosynthesis. Prog. Nucleic Acid. Res. Mol. Biol. 2001, 65, 361–393. [Google Scholar] [PubMed]
- Kent, C. Ctp:Phosphocholine cytidylyltransferase. Biochim. Biophys. Acta 1997, 1348, 79–90. [Google Scholar] [CrossRef]
- Henneberry, A.L.; Wistow, G.; McMaster, C.R. Cloning, genomic organization, and characterization of a human cholinephosphotransferase. J. Biol. Chem. 2000, 275, 29808–29815. [Google Scholar] [CrossRef] [PubMed]
- Henneberry, A.L.; McMaster, C.R. Cloning and expression of a human choline/ethanolaminephosphotransferase: Synthesis of phosphatidylcholine and phosphatidylethanolamine. Biochem. J. 1999, 339, 291–298. [Google Scholar] [CrossRef] [PubMed]
- Sriburi, R.; Bommiasamy, H.; Buldak, G.L.; Robbins, G.R.; Frank, M.; Jackowski, S.; Brewer, J.W. Coordinate regulation of phospholipid biosynthesis and secretory pathway gene expression in xbp-1(s)-induced endoplasmic reticulum biogenesis. J. Biol. Chem. 2007, 282, 7024–7034. [Google Scholar] [CrossRef] [PubMed]
- Sprocati, T.; Ronchi, P.; Raimondi, A.; Francolini, M.; Borgese, N. Dynamic and reversible restructuring of the er induced by pdmp in cultured cells. J. Cell Sci. 2006, 119, 3249–3260. [Google Scholar] [CrossRef] [PubMed]
- Maiuolo, J.; Bulotta, S.; Verderio, C.; Benfante, R.; Borgese, N. Selective activation of the transcription factor ATF6 mediates endoplasmic reticulum proliferation triggered by a membrane protein. Proc. Natl. Acad. Sci. USA 2011, 108, 7832–7837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bommiasamy, H.; Back, S.H.; Fagone, P.; Lee, K.; Meshinchi, S.; Vink, E.; Sriburi, R.; Frank, M.; Jackowski, S.; Kaufman, R.J.; et al. ATF6α induces XBP1-independent expansion of the endoplasmic reticulum. J. Cell Sci. 2009, 122, 1626–1636. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, M.; Hayashi, T.; Su, T.P. The role of cholesterol in the association of endoplasmic reticulum membranes with mitochondria. Biochem. Biophys. Res. Commun. 2012, 417, 635–639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayashi, T.; Rizzuto, R.; Hajnoczky, G.; Su, T.P. Mam: More than just a housekeeper. Trends Cell Biol. 2009, 19, 81–88. [Google Scholar] [CrossRef] [PubMed]
- de Brito, O.M.; Scorrano, L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature 2008, 456, 605–610. [Google Scholar] [CrossRef] [PubMed]
- De Vos, K.J.; Morotz, G.M.; Stoica, R.; Tudor, E.L.; Lau, K.F.; Ackerley, S.; Warley, A.; Shaw, C.E.; Miller, C.C. Vapb interacts with the mitochondrial protein ptpip51 to regulate calcium homeostasis. Hum. Mol. Genet. 2012, 21, 1299–1311. [Google Scholar] [CrossRef] [PubMed]
- Mendes, C.C.; Gomes, D.A.; Thompson, M.; Souto, N.C.; Goes, T.S.; Goes, A.M.; Rodrigues, M.A.; Gomez, M.V.; Nathanson, M.H.; Leite, M.F. The type III inositol 1,4,5-trisphosphate receptor preferentially transmits apoptotic Ca2+ signals into mitochondria. J. Biol. Chem. 2005, 280, 40892–40900. [Google Scholar] [CrossRef] [PubMed]
- Iwasawa, R.; Mahul-Mellier, A.L.; Datler, C.; Pazarentzos, E.; Grimm, S. Fis1 and bap31 bridge the mitochondria-ER interface to establish a platform for apoptosis induction. EMBO J. 2011, 30, 556–568. [Google Scholar] [CrossRef] [PubMed]
- Simmen, T.; Aslan, J.E.; Blagoveshchenskaya, A.D.; Thomas, L.; Wan, L.; Xiang, Y.; Feliciangeli, S.F.; Hung, C.H.; Crump, C.M.; Thomas, G. Pacs-2 controls endoplasmic reticulum-mitochondria communication and bid-mediated apoptosis. EMBO J. 2005, 24, 717–729. [Google Scholar] [CrossRef] [PubMed]
- van Vliet, A.R.; Verfaillie, T.; Agostinis, P. New functions of mitochondria associated membranes in cellular signaling. Biochim. Biophys. Acta 2014, 1843, 2253–2262. [Google Scholar] [CrossRef] [PubMed]
- Bononi, A.; Bonora, M.; Marchi, S.; Missiroli, S.; Poletti, F.; Giorgi, C.; Pandolfi, P.P.; Pinton, P. Identification of pten at the er and mams and its regulation of Ca2+ signaling and apoptosis in a protein phosphatase-dependent manner. Cell Death Differ. 2013, 20, 1631–1643. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Yazdi, A.S.; Menu, P.; Tschopp, J. A role for mitochondria in nlrp3 inflammasome activation. Nature 2011, 469, 221–225. [Google Scholar] [CrossRef] [PubMed]
- Vance, J.E. Mam (mitochondria-associated membranes) in mammalian cells: Lipids and beyond. Biochim. Biophys. Acta 2014, 1841, 595–609. [Google Scholar] [CrossRef] [PubMed]
- Tubbs, E.; Rieusset, J. Metabolic signaling functions of er-mitochondria contact sites: Role in metabolic diseases. J. Mol. Endocrinol. 2017, 58, R87–R106. [Google Scholar] [CrossRef] [PubMed]
- Hattori, N.; Arano, T.; Hatano, T.; Mori, A.; Imai, Y. Mitochondrial-associated membranes in Parkinson’s disease. Adv. Exp. Med. Biol. 2017, 997, 157–169. [Google Scholar] [PubMed]
- Theurey, P.; Tubbs, E.; Vial, G.; Jacquemetton, J.; Bendridi, N.; Chauvin, M.A.; Alam, M.R.; Le Romancer, M.; Vidal, H.; Rieusset, J. Mitochondria-associated endoplasmic reticulum membranes allow adaptation of mitochondrial metabolism to glucose availability in the liver. J. Mol. Cell. Biol. 2016, 8, 129–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tubbs, E.; Theurey, P.; Vial, G.; Bendridi, N.; Bravard, A.; Chauvin, M.A.; Ji-Cao, J.; Zoulim, F.; Bartosch, B.; Ovize, M.; et al. Mitochondria-associated endoplasmic reticulum membrane (MAM) integrity is required for insulin signaling and is implicated in hepatic insulin resistance. Diabetes 2014, 63, 3279–3294. [Google Scholar] [CrossRef] [PubMed]
- Betz, C.; Stracka, D.; Prescianotto-Baschong, C.; Frieden, M.; Demaurex, N.; Hall, M.N. Feature article: Mtor complex 2-akt signaling at mitochondria-associated endoplasmic reticulum membranes (MAM) regulates mitochondrial physiology. Proc. Natl. Acad. Sci. USA 2013, 110, 12526–12534. [Google Scholar] [CrossRef] [PubMed]
- Rieusset, J. Role of endoplasmic reticulum-mitochondria communication in type 2 diabetes. Adv. Exp. Med. Biol. 2017, 997, 171–186. [Google Scholar] [PubMed]
- Gomez-Suaga, P.; Bravo-San Pedro, J.M.; Gonzalez-Polo, R.A.; Fuentes, J.M.; Niso-Santano, M. ER-mitochondria signaling in Parkinson’s disease. Cell Death Dis. 2018, 9, 337. [Google Scholar] [CrossRef] [PubMed]
- Guardia-Laguarta, C.; Area-Gomez, E.; Rub, C.; Liu, Y.; Magrane, J.; Becker, D.; Voos, W.; Schon, E.A.; Przedborski, S. Alpha-synuclein is localized to mitochondria-associated er membranes. J. Neurosci. 2014, 34, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Pickrell, A.M.; Youle, R.J. The roles of pink1, parkin, and mitochondrial fidelity in Parkinson’s disease. Neuron 2015, 85, 257–273. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.Y.; Yang, W.Y. Bit-by-bit autophagic removal of parkin-labelled mitochondria. Nat. Commun. 2013, 4, 2428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Laar, V.S.; Roy, N.; Liu, A.; Rajprohat, S.; Arnold, B.; Dukes, A.A.; Holbein, C.D.; Berman, S.B. Glutamate excitotoxicity in neurons triggers mitochondrial and endoplasmic reticulum accumulation of parkin, and, in the presence of n-acetyl cysteine, mitophagy. Neurobiol. Dis. 2015, 74, 180–193. [Google Scholar] [CrossRef] [PubMed]
- Gelmetti, V.; De Rosa, P.; Torosantucci, L.; Marini, E.S.; Romagnoli, A.; Di Rienzo, M.; Arena, G.; Vignone, D.; Fimia, G.M.; Valente, E.M. PINK1 and BECN1 relocalize at mitochondria-associated membranes during mitophagy and promote ER-mitochondria tethering and autophagosome formation. Autophagy 2017, 13, 654–669. [Google Scholar] [CrossRef] [PubMed]
- Alto, N.M.; Soderling, J.; Scott, J.D. Rab32 is an a-kinase anchoring protein and participates in mitochondrial dynamics. J. Cell Biol. 2002, 158, 659–668. [Google Scholar] [CrossRef] [PubMed]
- Bui, M.; Gilady, S.Y.; Fitzsimmons, R.E.; Benson, M.D.; Lynes, E.M.; Gesson, K.; Alto, N.M.; Strack, S.; Scott, J.D.; Simmen, T. Rab32 modulates apoptosis onset and mitochondria-associated membrane (MAM) properties. J. Biol. Chem. 2010, 285, 31590–31602. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Sandoval, C.G.; Hughes, S.C.; Dacks, J.B.; Simmen, T. Interaction with the effector dynamin-related protein 1 (drp1) is an ancient function of rab32 subfamily proteins. Cell. Logist. 2014, 4, e986399. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Lin, S.; Zou, L.; Zhou, H.; Zhang, J.; Su, B.; Wan, Y. Expression profiling of rab gtpases reveals the involvement of rab20 and rab32 in acute brain inflammation in mice. Neurosci. Lett. 2012, 527, 110–114. [Google Scholar] [CrossRef] [PubMed]
- Haile, Y.; Deng, X.; Ortiz-Sandoval, C.; Tahbaz, N.; Janowicz, A.; Lu, J.Q.; Kerr, B.J.; Gutowski, N.J.; Holley, J.E.; Eggleton, P.; et al. Rab32 connects er stress to mitochondrial defects in multiple sclerosis. J. Neuroinflamm. 2017, 14, 19. [Google Scholar] [CrossRef] [PubMed]
- Mavlyutov, T.A.; Epstein, M.L.; Andersen, K.A.; Ziskind-Conhaim, L.; Ruoho, A.E. The sigma-1 receptor is enriched in postsynaptic sites of c-terminals in mouse motoneurons. An anatomical and behavioral study. Neuroscience 2010, 167, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, T.; Su, T.P. Sigma-1 receptor chaperones at the er-mitochondrion interface regulate Ca2+ signaling and cell survival. Cell 2007, 131, 596–610. [Google Scholar] [CrossRef] [PubMed]
- Bhuiyan, M.S.; Fukunaga, K. Targeting sigma-1 receptor signaling by endogenous ligands for cardioprotection. Expert Opin. Ther. Targets 2011, 15, 145–155. [Google Scholar] [CrossRef] [PubMed]
- Kourrich, S.; Hayashi, T.; Chuang, J.Y.; Tsai, S.Y.; Su, T.P.; Bonci, A. Dynamic interaction between sigma-1 receptor and kv1.2 shapes neuronal and behavioral responses to cocaine. Cell 2013, 152, 236–247. [Google Scholar] [CrossRef] [PubMed]
- Bernard-Marissal, N.; Medard, J.J.; Azzedine, H.; Chrast, R. Dysfunction in endoplasmic reticulum-mitochondria crosstalk underlies sigmar1 loss of function mediated motor neuron degeneration. Brain 2015, 138, 875–890. [Google Scholar] [CrossRef] [PubMed]
- Mitsuda, T.; Omi, T.; Tanimukai, H.; Sakagami, Y.; Tagami, S.; Okochi, M.; Kudo, T.; Takeda, M. Sigma-1Rs are upregulated via PERK/eIF2α/ATF4 pathway and execute protective function in ER stress. Biochem. Biophys. Res. Commun. 2011, 415, 519–525. [Google Scholar] [CrossRef] [PubMed]
- Mori, T.; Hayashi, T.; Hayashi, E.; Su, T.P. Sigma-1 receptor chaperone at the er-mitochondrion interface mediates the mitochondrion-er-nucleus signaling for cellular survival. PLoS ONE 2013, 8, e76941. [Google Scholar] [CrossRef] [PubMed]
- Selkoe, D.J. Translating cell biology into therapeutic advances in Alzheimer’s disease. Nature 1999, 399, A23–A31. [Google Scholar] [CrossRef] [PubMed]
- Price, D.L.; Sisodia, S.S. Mutant genes in familial Alzheimer’s disease and transgenic models. Annu. Rev. Neurosci. 1998, 21, 479–505. [Google Scholar] [CrossRef] [PubMed]
- Hedskog, L.; Pinho, C.M.; Filadi, R.; Ronnback, A.; Hertwig, L.; Wiehager, B.; Larssen, P.; Gellhaar, S.; Sandebring, A.; Westerlund, M.; et al. Modulation of the endoplasmic reticulum-mitochondria interface in Alzheimer’s disease and related models. Proc. Natl. Acad. Sci. USA 2013, 110, 7916–7921. [Google Scholar] [CrossRef] [PubMed]
- Hutter-Paier, B.; Huttunen, H.J.; Puglielli, L.; Eckman, C.B.; Kim, D.Y.; Hofmeister, A.; Moir, R.D.; Domnitz, S.B.; Frosch, M.P.; Windisch, M.; et al. The acat inhibitor cp-113,818 markedly reduces amyloid pathology in a mouse model of Alzheimer’s disease. Neuron 2004, 44, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Toyohara, J.; Sakata, M.; Ishiwata, K. Imaging of sigma1 receptors in the human brain using pet and [11c]sa4503. Cent. Nerv. Syst. Agents Med. Chem. 2009, 9, 190–196. [Google Scholar] [CrossRef] [PubMed]
- Prause, J.; Goswami, A.; Katona, I.; Roos, A.; Schnizler, M.; Bushuven, E.; Dreier, A.; Buchkremer, S.; Johann, S.; Beyer, C.; et al. Altered localization, abnormal modification and loss of function of sigma receptor-1 in amyotrophic lateral sclerosis. Hum. Mol. Genet. 2013, 22, 1581–1600. [Google Scholar] [CrossRef] [PubMed]
- Mori, T.; Hayashi, T.; Su, T.P. Compromising sigma-1 receptors at the endoplasmic reticulum render cytotoxicity to physiologically relevant concentrations of dopamine in a nuclear factor-κB/bcl-2-dependent mechanism: Potential relevance to Parkinson’s disease. J. Pharmacol. Exp. Ther. 2012, 341, 663–671. [Google Scholar] [CrossRef] [PubMed]
- Lindholm, D.; Wootz, H.; Korhonen, L. Er stress and neurodegenerative diseases. Cell Death Differ. 2006, 13, 385–392. [Google Scholar] [CrossRef] [PubMed]
- Munoz, J.P.; Ivanova, S.; Sanchez-Wandelmer, J.; Martinez-Cristobal, P.; Noguera, E.; Sancho, A.; Diaz-Ramos, A.; Hernandez-Alvarez, M.I.; Sebastian, D.; Mauvezin, C.; et al. Mfn2 modulates the UPR and mitochondrial function via repression of perk. EMBO J 2013, 32, 2348–2361. [Google Scholar] [CrossRef] [PubMed]
- Verfaillie, T.; Rubio, N.; Garg, A.D.; Bultynck, G.; Rizzuto, R.; Decuypere, J.P.; Piette, J.; Linehan, C.; Gupta, S.; Samali, A.; et al. Perk is required at the er-mitochondrial contact sites to convey apoptosis after ros-based er stress. Cell Death Differ. 2012, 19, 1880–1891. [Google Scholar] [CrossRef] [PubMed]
- Porter, K.R.; Palade, G.E. Studies on the endoplasmic reticulum. III. Its form and distribution in striated muscle cells. J. Biophys. Biochem. Cytol. 1957, 3, 269–300. [Google Scholar] [CrossRef] [PubMed]
- Saheki, Y.; De Camilli, P. Endoplasmic reticulum-plasma membrane contact sites. Annu. Rev. Biochem. 2017, 86, 659–684. [Google Scholar] [CrossRef] [PubMed]
- Henne, W.M.; Liou, J.; Emr, S.D. Molecular mechanisms of inter-organelle er-pm contact sites. Curr. Opin. Cell Biol. 2015, 35, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Dickson, E.J.; Jensen, J.B.; Hille, B. Regulation of calcium and phosphoinositides at endoplasmic reticulum-membrane junctions. Biochem. Soc. Trans. 2016, 44, 467–473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stefan, C.J.; Manford, A.G.; Emr, S.D. Er-pm connections: Sites of information transfer and inter-organelle communication. Curr. Opin. Cell Biol. 2013, 25, 434–442. [Google Scholar] [CrossRef] [PubMed]
- Carrasco, S.; Meyer, T. Stim proteins and the endoplasmic reticulum-plasma membrane junctions. Annu. Rev. Biochem. 2011, 80, 973–1000. [Google Scholar] [CrossRef] [PubMed]
- Lewis, R.S. Store-operated calcium channels: New perspectives on mechanism and function. Cold Spring Harb. Perspect. Biol. 2011, 3. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.L.; Yu, Y.; Roos, J.; Kozak, J.A.; Deerinck, T.J.; Ellisman, M.H.; Stauderman, K.A.; Cahalan, M.D. Stim1 is a Ca2+ sensor that activates crac channels and migrates from the Ca2+ store to the plasma membrane. Nature 2005, 437, 902–905. [Google Scholar] [CrossRef] [PubMed]
- Varnai, P.; Toth, B.; Toth, D.J.; Hunyady, L.; Balla, T. Visualization and manipulation of plasma membrane-endoplasmic reticulum contact sites indicates the presence of additional molecular components within the stim1-orai1 complex. J. Biol. Chem. 2007, 282, 29678–29690. [Google Scholar] [CrossRef] [PubMed]
- Maleth, J.; Choi, S.; Muallem, S.; Ahuja, M. Translocation between pi(4,5)p2-poor and pi(4,5)p2-rich microdomains during store depletion determines stim1 conformation and orai1 gating. Nat. Commun. 2014, 5, 5843. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Quintana, A.; Findlay, G.M.; Mettlen, M.; Baust, B.; Jain, M.; Nilsson, R.; Rao, A.; Hogan, P.G. An siRNA screen for nfat activation identifies septins as coordinators of store-operated Ca2+ entry. Nature 2013, 499, 238–242. [Google Scholar] [CrossRef] [PubMed]
- Manford, A.G.; Stefan, C.J.; Yuan, H.L.; Macgurn, J.A.; Emr, S.D. Er-to-plasma membrane tethering proteins regulate cell signaling and er morphology. Dev. Cell 2012, 23, 1129–1140. [Google Scholar] [CrossRef] [PubMed]
- Giordano, F.; Saheki, Y.; Idevall-Hagren, O.; Colombo, S.F.; Pirruccello, M.; Milosevic, I.; Gracheva, E.O.; Bagriantsev, S.N.; Borgese, N.; De Camilli, P. Pi(4,5)p(2)-dependent and Ca2+-regulated er-pm interactions mediated by the extended synaptotagmins. Cell 2013, 153, 1494–1509. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.L.; Hsieh, T.S.; Yang, T.T.; Rothberg, K.G.; Azizoglu, D.B.; Volk, E.; Liao, J.C.; Liou, J. Feedback regulation of receptor-induced Ca2+ signaling mediated by e-syt1 and nir2 at endoplasmic reticulum-plasma membrane junctions. Cell Rep. 2013, 5, 813–825. [Google Scholar] [CrossRef] [PubMed]
- Takeshima, H.; Komazaki, S.; Nishi, M.; Iino, M.; Kangawa, K. Junctophilins: A novel family of junctional membrane complex proteins. Mol. Cell 2000, 6, 11–22. [Google Scholar] [PubMed]
- Nishi, M.; Sakagami, H.; Komazaki, S.; Kondo, H.; Takeshima, H. Coexpression of junctophilin type 3 and type 4 in brain. Brain Res. Mol. Brain Res. 2003, 118, 102–110. [Google Scholar] [CrossRef]
- Bennett, H.J.; Davenport, J.B.; Collins, R.F.; Trafford, A.W.; Pinali, C.; Kitmitto, A. Human junctophilin-2 undergoes a structural rearrangement upon binding ptdins(3,4,5)p3 and the s101r mutation identified in hypertrophic cardiomyopathy obviates this response. Biochem. J. 2013, 456, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Hirata, Y.; Brotto, M.; Weisleder, N.; Chu, Y.; Lin, P.; Zhao, X.; Thornton, A.; Komazaki, S.; Takeshima, H.; Ma, J.; et al. Uncoupling store-operated Ca2+ entry and altered Ca2+ release from sarcoplasmic reticulum through silencing of junctophilin genes. Biophys. J. 2006, 90, 4418–4427. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Ding, X.; Lopez, J.R.; Takeshima, H.; Ma, J.; Allen, P.D.; Eltit, J.M. Impaired orai1-mediated resting Ca2+ entry reduces the cytosolic [Ca2+] and sarcoplasmic reticulum Ca2+ loading in quiescent junctophilin 1 knock-out myotubes. J. Biol. Chem. 2010, 285, 39171–39179. [Google Scholar] [CrossRef] [PubMed]
- Varga-Szabo, D.; Braun, A.; Kleinschnitz, C.; Bender, M.; Pleines, I.; Pham, M.; Renne, T.; Stoll, G.; Nieswandt, B. The calcium sensor stim1 is an essential mediator of arterial thrombosis and ischemic brain infarction. J. Exp. Med. 2008, 205, 1583–1591. [Google Scholar] [CrossRef] [PubMed]
- Stiber, J.; Hawkins, A.; Zhang, Z.S.; Wang, S.; Burch, J.; Graham, V.; Ward, C.C.; Seth, M.; Finch, E.; Malouf, N.; et al. Stim1 signalling controls store-operated calcium entry required for development and contractile function in skeletal muscle. Nat. Cell Biol. 2008, 10, 688–697. [Google Scholar] [CrossRef] [PubMed]
- Feske, S.; Picard, C.; Fischer, A. Immunodeficiency due to mutations in orai1 and stim1. Clin. Immunol. 2010, 135, 169–182. [Google Scholar] [CrossRef] [PubMed]
- Klein, M.; Klein-Hessling, S.; Palmetshofer, A.; Serfling, E.; Tertilt, C.; Bopp, T.; Heib, V.; Becker, M.; Taube, C.; Schild, H.; et al. Specific and redundant roles for nfat transcription factors in the expression of mast cell-derived cytokines. J. Immunol. 2006, 177, 6667–6674. [Google Scholar] [CrossRef] [PubMed]
- Baba, Y.; Nishida, K.; Fujii, Y.; Hirano, T.; Hikida, M.; Kurosaki, T. Essential function for the calcium sensor stim1 in mast cell activation and anaphylactic responses. Nat. Immunol. 2008, 9, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Vig, M.; DeHaven, W.I.; Bird, G.S.; Billingsley, J.M.; Wang, H.; Rao, P.E.; Hutchings, A.B.; Jouvin, M.H.; Putney, J.W.; Kinet, J.P. Defective mast cell effector functions in mice lacking the cracm1 pore subunit of store-operated calcium release-activated calcium channels. Nat. Immunol. 2008, 9, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Oh-Hora, M.; Yamashita, M.; Hogan, P.G.; Sharma, S.; Lamperti, E.; Chung, W.; Prakriya, M.; Feske, S.; Rao, A. Dual functions for the endoplasmic reticulum calcium sensors stim1 and stim2 in T cell activation and tolerance. Nat. Immunol. 2008, 9, 432–443. [Google Scholar] [CrossRef] [PubMed]
- Gwack, Y.; Srikanth, S.; Oh-Hora, M.; Hogan, P.G.; Lamperti, E.D.; Yamashita, M.; Gelinas, C.; Neems, D.S.; Sasaki, Y.; Feske, S.; et al. Hair loss and defective T- and B-cell function in mice lacking orai1. Mol. Cell. Biol. 2008, 28, 5209–5222. [Google Scholar] [CrossRef] [PubMed]
- Lynch, C.D.; Gauthier, N.C.; Biais, N.; Lazar, A.M.; Roca-Cusachs, P.; Yu, C.H.; Sheetz, M.P. Filamin depletion blocks endoplasmic spreading and destabilizes force-bearing adhesions. Mol. Biol. Cell 2011, 22, 1263–1273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.H.; Zhu, P.P.; Parker, R.L.; Blackstone, C. Hereditary spastic paraplegia proteins reep1, spastin, and atlastin-1 coordinate microtubule interactions with the tubular er network. J. Clin. Investig. 2010, 120, 1097–1110. [Google Scholar] [CrossRef] [PubMed]
- van Vliet, A.R.; Giordano, F.; Gerlo, S.; Segura, I.; Van Eygen, S.; Molenberghs, G.; Rocha, S.; Houcine, A.; Derua, R.; Verfaillie, T.; et al. The ER stress sensor perk coordinates er-plasma membrane contact site formation through interaction with filamin-a and f-actin remodeling. Mol. Cell 2017, 65, 885–899. [Google Scholar] [CrossRef] [PubMed]
- Urra, H.; Henriquez, D.R.; Canovas, J.; Villarroel-Campos, D.; Carreras-Sureda, A.; Pulgar, E.; Molina, E.; Hazari, Y.M.; Limia, C.M.; Alvarez-Rojas, S.; et al. IRE1α governs cytoskeleton remodelling and cell migration through a direct interaction with filamin a. Nat. Cell Biol. 2018, 20, 942–953. [Google Scholar] [CrossRef] [PubMed]
- Druelle, C.; Drullion, C.; Desle, J.; Martin, N.; Saas, L.; Cormenier, J.; Malaquin, N.; Huot, L.; Slomianny, C.; Bouali, F.; et al. ATF6α regulates morphological changes associated with senescence in human fibroblasts. Oncotarget 2016, 7, 67699–67715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cormenier, J.; Martin, N.; Desle, J.; Salazar-Cardozo, C.; Pourtier, A.; Abbadie, C.; Pluquet, O. The ATF6α arm of the unfolded protein response mediates replicative senescence in human fibroblasts through a cox2/prostaglandin e2 intracrine pathway. Mech. Ageing Dev. 2018, 170, 82–91. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.; Gundersen, D. Increased organization of cytoskeleton accompanying the aging of human fibroblasts in vitro. Exp. Cell Res. 1984, 154, 191–202. [Google Scholar] [CrossRef]
- Wang, E. Are cross-bridging structures involved in the bundle formation of intermediate filaments and the decrease in locomotion that accompany cell aging? J. Cell Biol. 1985, 100, 1466–1473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishio, K.; Inoue, A.; Qiao, S.; Kondo, H.; Mimura, A. Senescence and cytoskeleton: Overproduction of vimentin induces senescent-like morphology in human fibroblasts. Histochem. Cell Biol. 2001, 116, 321–327. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Liang, X.; Li, M.; Guo, T.; Duan, N.; Wang, Y.; Rong, G.; Yang, L.; Zhang, S.; Zhang, J. ATF6 pathway of unfolded protein response mediates advanced oxidation protein product-induced hypertrophy and epithelial-to-mesenchymal transition in HK-2 cells. Mol. Cell Biochem. 2015, 407, 197–207. [Google Scholar] [CrossRef] [PubMed]


© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saito, A.; Imaizumi, K. Unfolded Protein Response-Dependent Communication and Contact among Endoplasmic Reticulum, Mitochondria, and Plasma Membrane. Int. J. Mol. Sci. 2018, 19, 3215. https://doi.org/10.3390/ijms19103215
Saito A, Imaizumi K. Unfolded Protein Response-Dependent Communication and Contact among Endoplasmic Reticulum, Mitochondria, and Plasma Membrane. International Journal of Molecular Sciences. 2018; 19(10):3215. https://doi.org/10.3390/ijms19103215
Chicago/Turabian StyleSaito, Atsushi, and Kazunori Imaizumi. 2018. "Unfolded Protein Response-Dependent Communication and Contact among Endoplasmic Reticulum, Mitochondria, and Plasma Membrane" International Journal of Molecular Sciences 19, no. 10: 3215. https://doi.org/10.3390/ijms19103215