Comparison of Human Dermal Fibroblasts and HaCat Cells Cultured in Medium with or without Serum via a Generic Tissue Engineering Research Platform
Abstract
:1. Introduction
2. Results
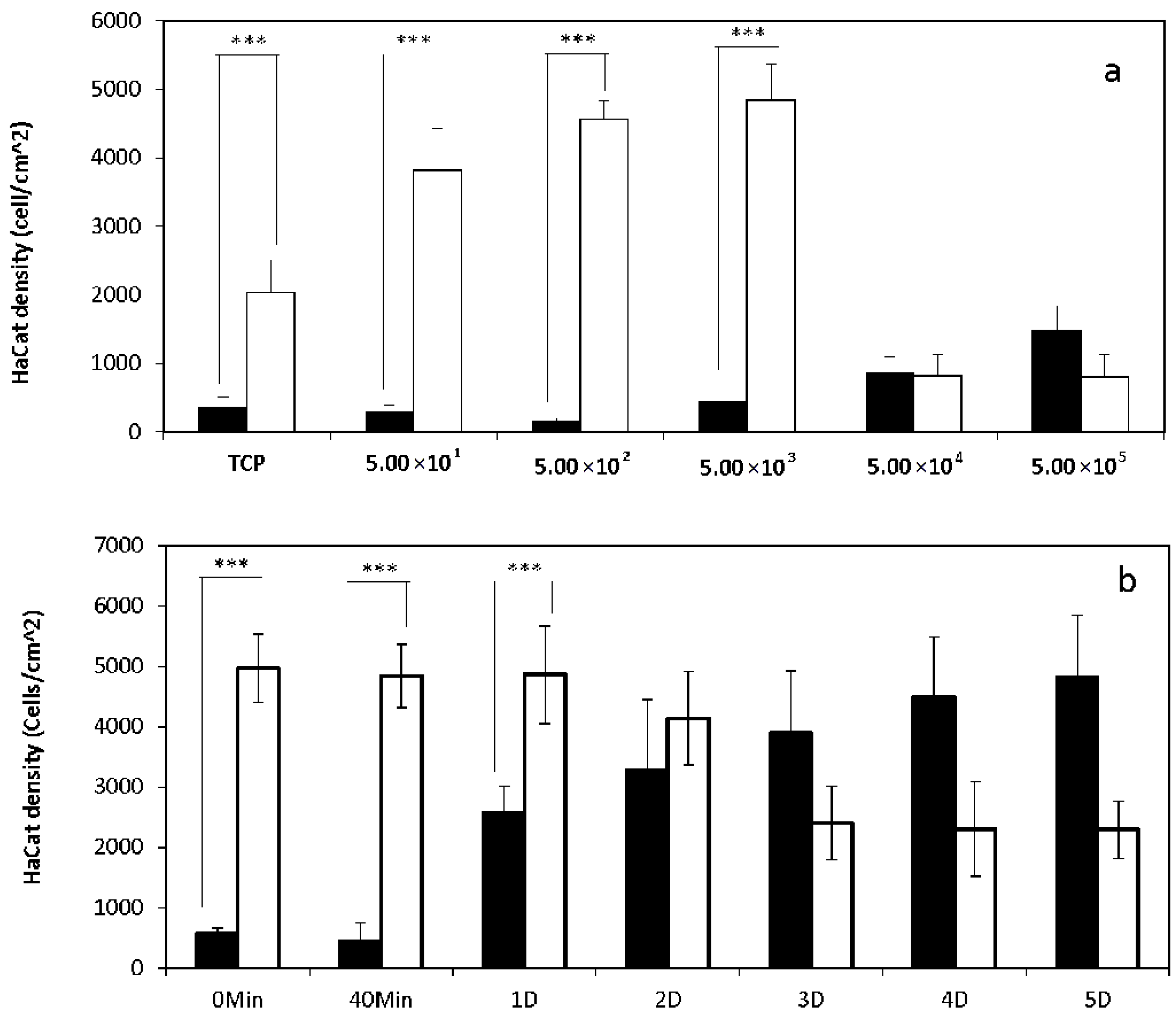
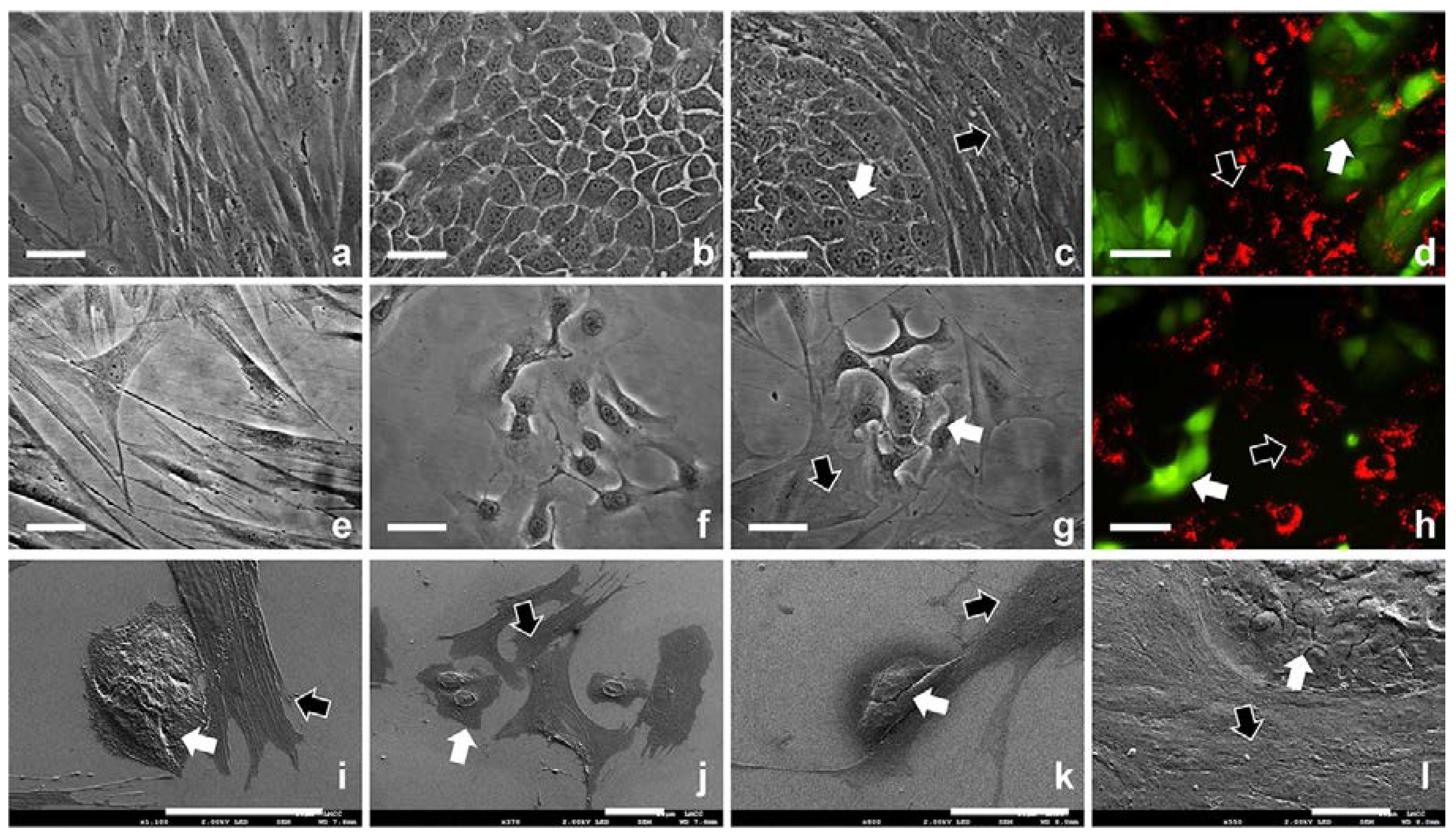
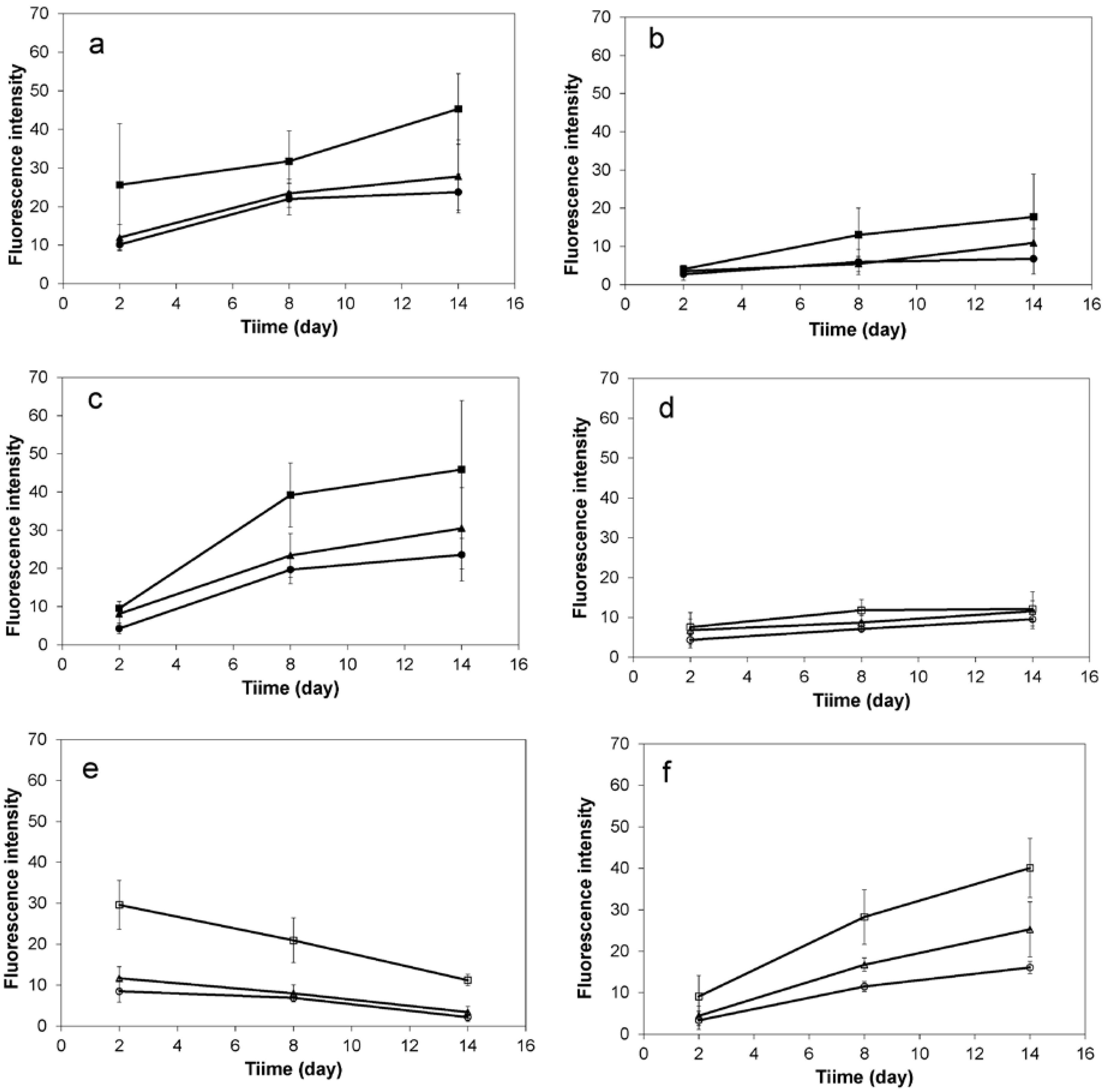
2.1. 2D Cell Culture in Medium with or without Serum
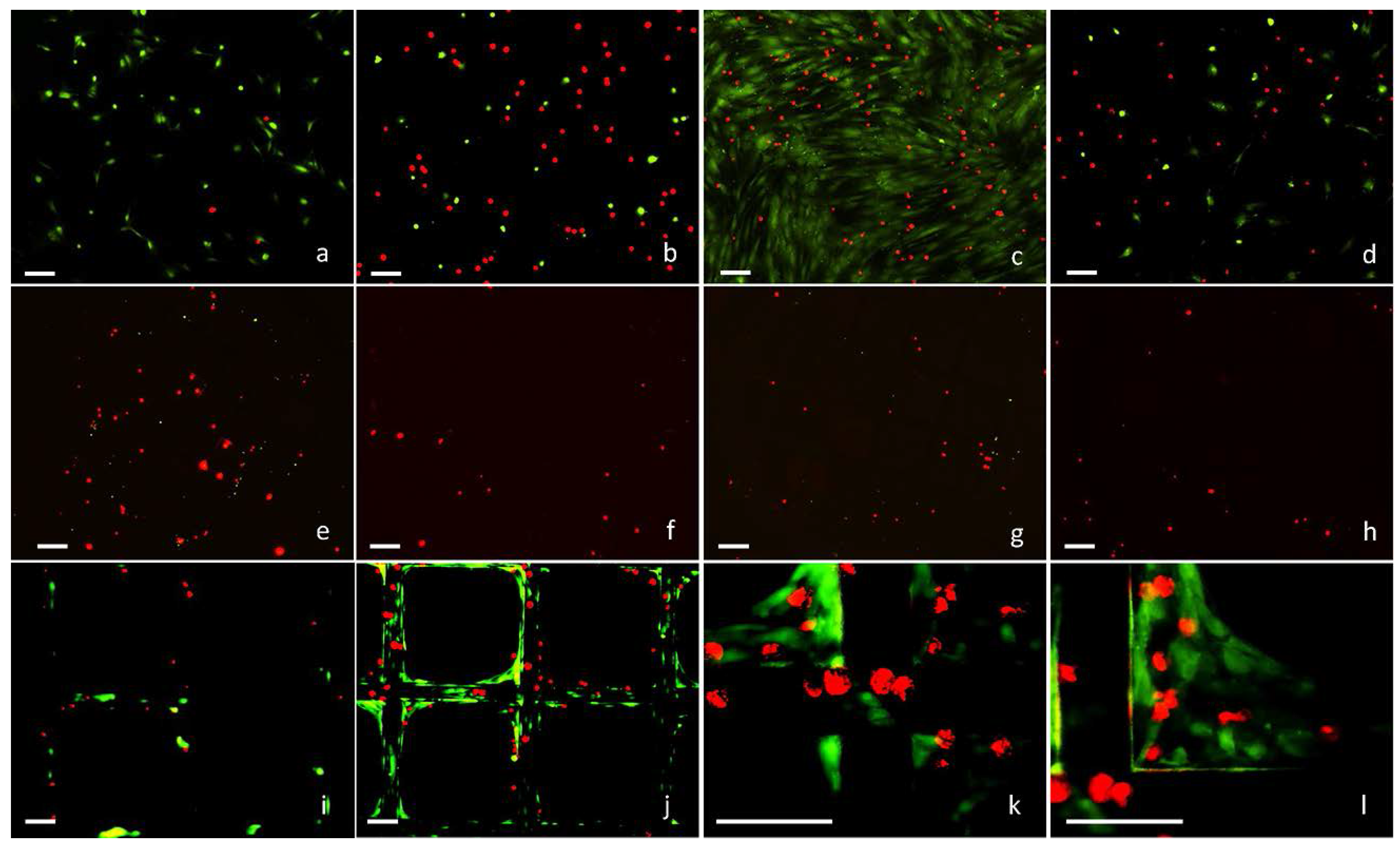
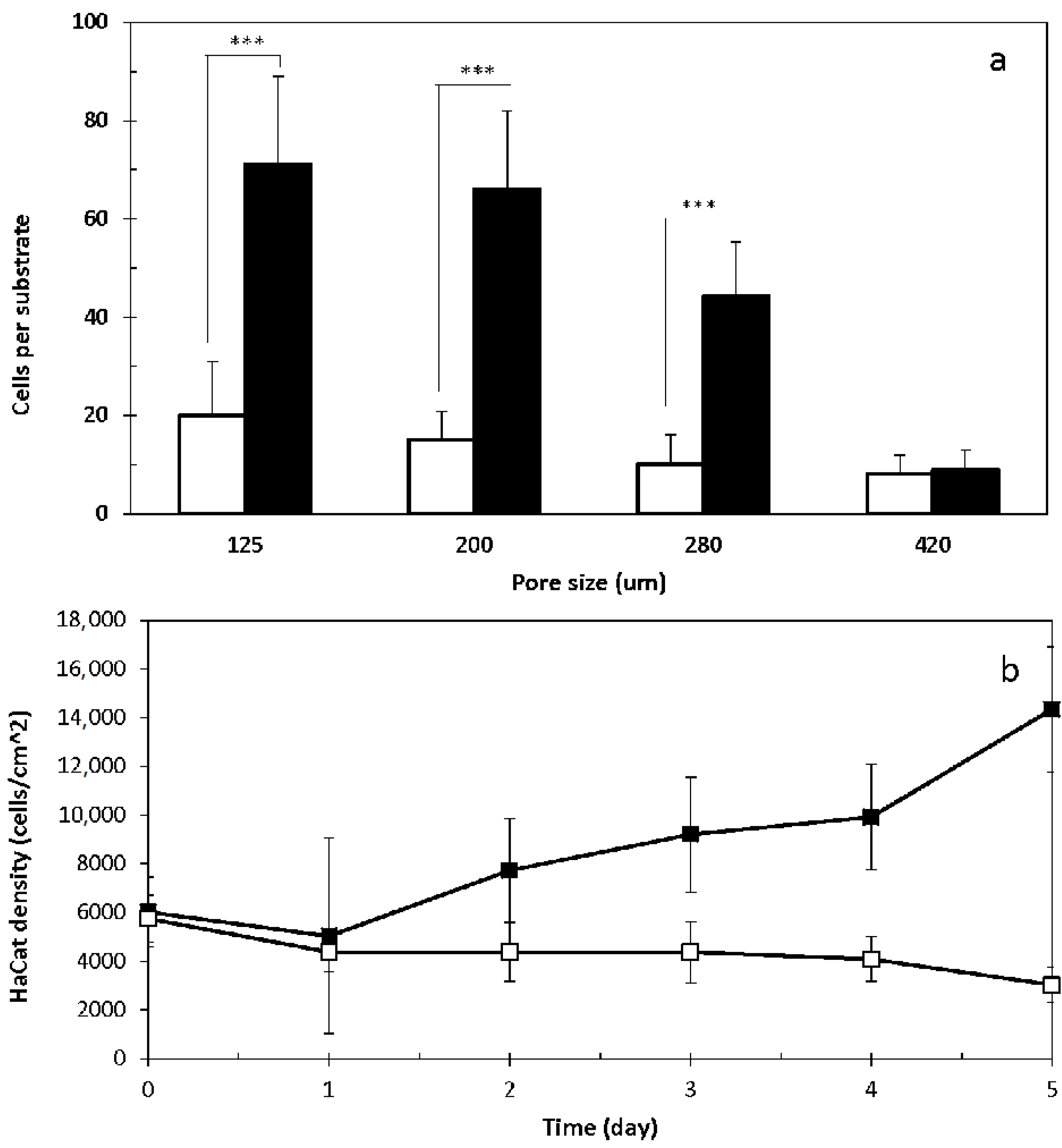
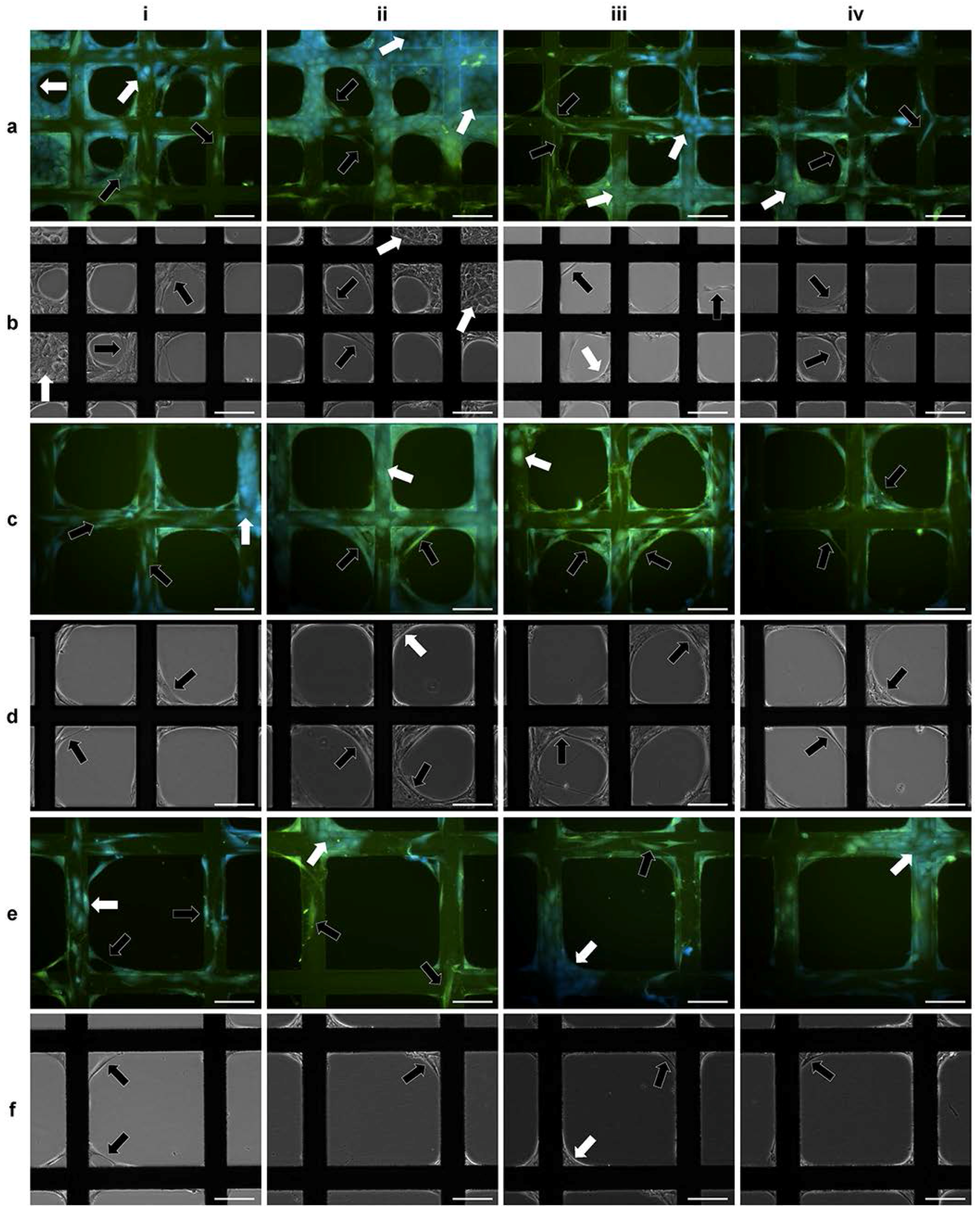
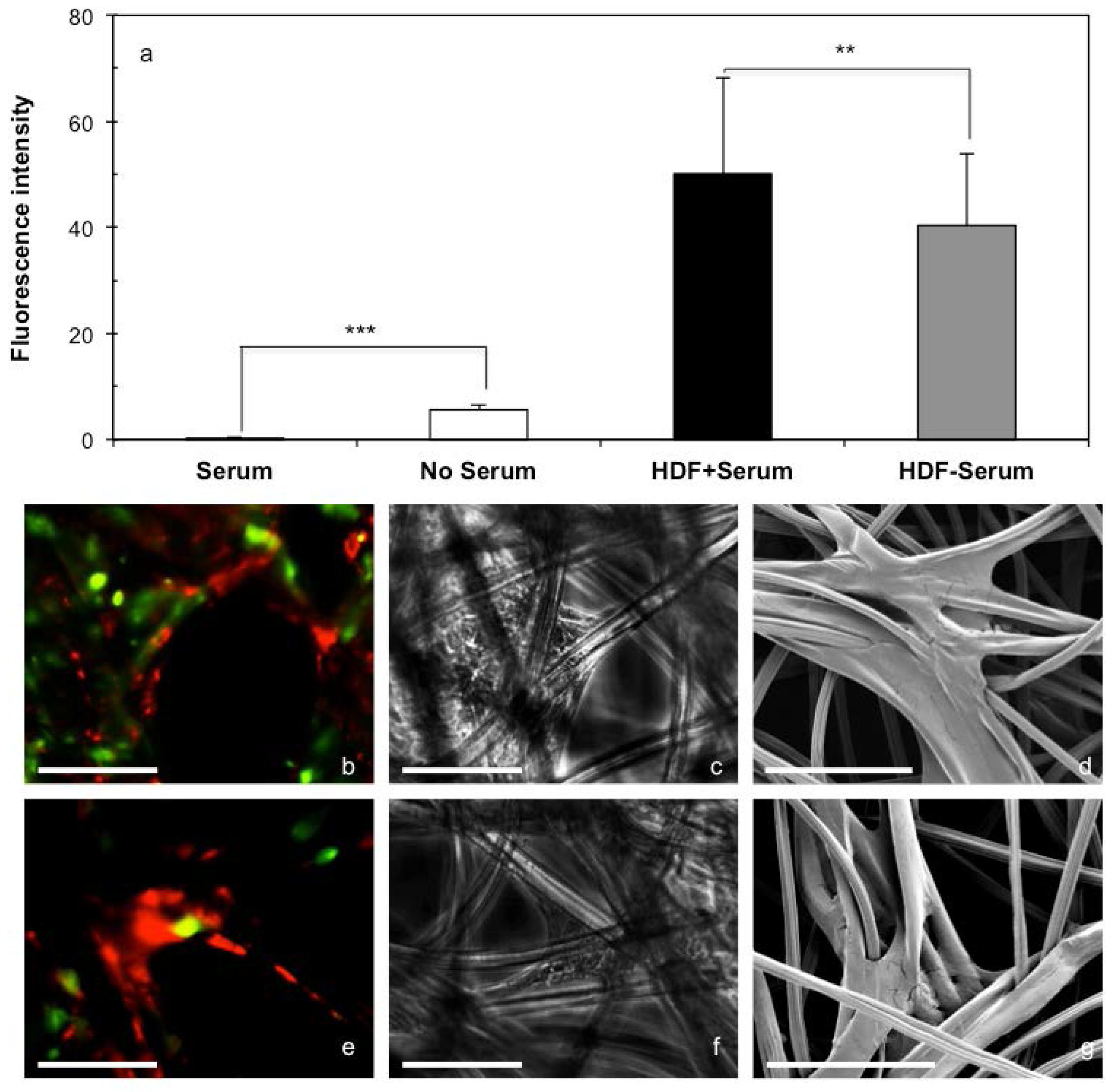
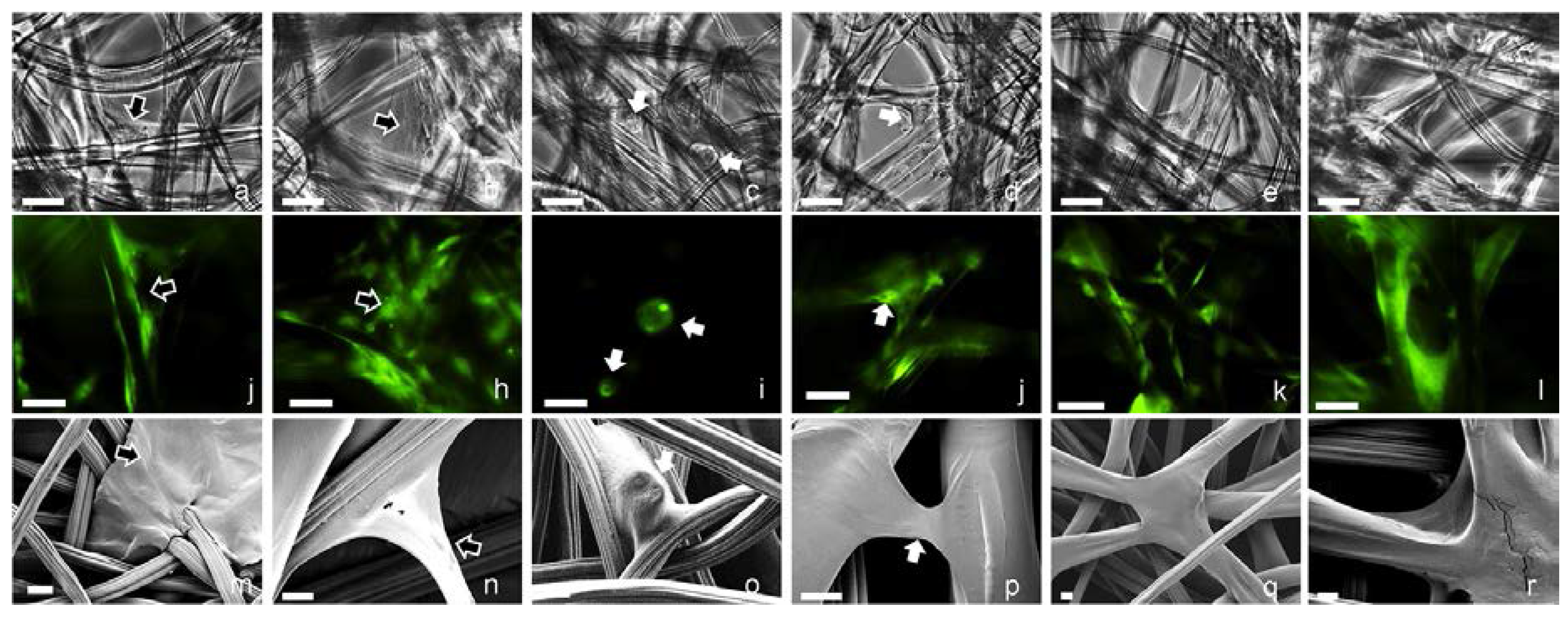
2.2. 3D Cell Culture on Porous Substrates in Medium with or without Serum
2.3. 3D Cell Culture on Cellulosic Scaffolds in Medium with or without Serum
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Cellulosic Scaffolds and Modular Substrates for 3D Cell Culture
4.3. Phase Contrast Microscopy
4.4. Live Cell Fluorescent Microscopy
4.5. Scanning Electron Microscopy
4.6. Statistics
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Zonari, A.; Novikoff, S.; Electo, N.R.P.; Breyner, N.M.; Gomes, D.A.; Martins, A. Endothelial differentiation of human stem cells seeded onto electrospun polyhydroxybutyrate/polyhydroxybutyrate-co-hydroxyvalerate fiber mesh. PLoS ONE 2012, 7, 335–353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gurtner, G.C.; Chapman, M.A. Regenerative Medicine: Charting a New Course in Wound Healing. Adv. Wound Care 2016, 5, 314–328. [Google Scholar] [CrossRef] [PubMed]
- Polak, D.J. Regenerative Medicine. Opportunities and Challenges: A brief overview. J. R. Soc. Interface 2010, 6, S777–S781. [Google Scholar]
- Allickson, J.G. Emerging Translation of Regenerative Therapies. Clin. Pharmacol. Ther. 2017, 101, 28–30. [Google Scholar] [CrossRef] [PubMed]
- Hunsberger, J.; Neubert, J.; Wertheim, J.A.; Allickson, J.; Atala, A. Bioengineering Priorities on a Path to Ending Organ Shortage. Curr. Stem Cell Rep. 2016, 2, 118–127. [Google Scholar] [CrossRef]
- Saini, S.; Wick, T.M. Concentric cylinder bioreactor for production of tissue engineered cartilage: Effect of seeding density and hydrodynamic loading on construct development. Biotechnol. Prog. 2003, 19, 510–521. [Google Scholar] [CrossRef] [PubMed]
- Rayment, E.A.; Williams, D.J. Concise review: Mind the gap: Challenges in characterizing and quantifying cell- and tissue-based therapies for clinical translation. Stem Cells 2010, 28, 996–1004. [Google Scholar] [PubMed]
- Stassen, O.M.J.A.; Muylaert, D.E.P.; Bouten, C.V.C.; Hjortnaes, J. Current Challenges in Translating Tissue-Engineered Heart Valves. Curr. Treat. Options Cardiovasc. Med. 2017, 19, 71. [Google Scholar] [CrossRef] [PubMed]
- Vig, K.; Chaudhari, A.; Tripathi, S.; Dixit, S.; Sahu, R.; Pillai, S.; Dennis, V.A.; Singh, S.R. Advances in Skin Regeneration Using Tissue Engineering. Int. J. Mol. Sci. 2017, 18, 789. [Google Scholar] [CrossRef] [PubMed]
- Even, M.S.; Sandusky, C.B.; Barnard, N.D. Serum-free hybridoma culture: Eithical, scientific and safety considerations. Trends Biotechnol. 2006, 24, 105–108. [Google Scholar] [CrossRef] [PubMed]
- Guo, A.H.; Jahoda, C.A.B. An improved method of human keratinocyte culture from skin explants: Cell expansion is linked to markers of activated progenitor cells. Exp. Dermantol. 2009, 18, 720–726. [Google Scholar] [CrossRef] [PubMed]
- Van der Valk, J.; Mellor, D.; Brands, R.; Fischer, R.; Gruber, F.; Gstraunthaler, G.; Hellebrekers, L.; Hyllner, J.; Jonker, F.H.; Prieto, P.; et al. The humane collection of fetal bovine serum and possibilities for serum-free cell and tissue culture. Toxicol. In Vitro 2004, 18, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Hemeda, H.; Giebel, B.; Wagner, W. Evaluation of human platlet lysate versus fetal bovine serum for culture of mesenchymal stromal cells. Cytotherapy 2014, 16, 170–180. [Google Scholar] [CrossRef] [PubMed]
- Ang, L.P.K.; Tan, D.T.H.; Seah, C.J.Y.; Beuerman, R.W. The use of human serum in supporting the in vitro and in vivo proliferation of human conjunctival epithelial cells. Br. J. Ophthalmol. 2005, 89, 748–752. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.J. Human keratinocyte culture using porcine pituitary extract in serum-free medium. Burns 1995, 21, 503–506. [Google Scholar] [CrossRef]
- Sun, T.; Higham, M.; Layton, C.; Haycock, J.W.; Short, R.; MacNeil, S. Developments in xenobiotic free culture of human keratinocytes for clinical use. Wound Repair Regen. 2004, 12, 626–634. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Jackson, S.; Haycock, J.W.; MacNeil, S. Culture of skin cells in 3D rather than 2D improves their ability to survive exposure to cytotoxic agents. J. Biotechnol. 2006, 122, 372–381. [Google Scholar] [CrossRef] [PubMed]
- De la Puente, P.; Muz, B.; Gilson, R.C.; Azab, F.; Luderer, M.; King, J.; Achilefu, S.; Vij, R.; Azab, A.K. 3D tissue-engineered bone marrow as a novel model to study pathophysiology and drug resistance in multiple myeloma. Biomaterials 2015, 73, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Knight, E.; Przyborski, S. Advances in 3D cell culture technologies enabling tissue-like structures to be created in vitro. J. Anat. 2015, 227, 746–756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wrzesinski, K.; Fey, S.J. From 2D to 3D—A New Dimension for Modelling the Effect of Natural Products on Human Tissue. Curr. Pharm. Des. 2015, 21, 5605–5616. [Google Scholar] [CrossRef] [PubMed]
- Lowenthal, J.; Gerecht, S. Stem cell-derived vasculature: A potent and multidimensional technology for basic research, disease modeling, and tissue engineering Justin. Biochem. Biophys. Res. Commun. 2016, 473, 733–742. [Google Scholar] [CrossRef] [PubMed]
- Gabbott, C.M.; Zhou, Z.X.; Han, G.X.; Sun, T. A novel scale-down cell culture and imaging design for the mechanistic insight of cell colonization within porous substrate. J. Microsc. 2017, 267, 150–159. [Google Scholar] [CrossRef] [PubMed]
- Langer, R.; Vacanti, J.P. Tissue Engineering. Science 1993, 260, 920–926. [Google Scholar] [CrossRef] [PubMed]
- De Kemp, V.; de Graaf, P.; Fledderus, J.O.; Ruud Bosch, J.L.H.; de Kort, L.M.O. Tissue engineering for human urethral reconstruction: Systematic review of recent literature. PLoS ONE 2015, 10, e0118653. [Google Scholar] [CrossRef] [PubMed]
- Steele, J.G.; Johnson, G.; Underwood, P.A. Role of serum vitronectin and fibronectin in adhesion of fibroblasts following seeding onto tissue culture polystyrene. J. Biomed. Mater. Res. 1992, 26, 861–884. [Google Scholar] [CrossRef] [PubMed]
- El-Ghalbzouri, A.; Gibbs, S.; Lamme, E.; Van Blitterswijk, C.A.; Ponec, M. Effect of fibroblasts on epidermal regeneration. Br. J. Dermatol. 2002, 147, 230–243. [Google Scholar] [CrossRef] [PubMed]
- Tracy, L.E.; Minasian, R.A.; Caterson, E.J. Extracellular Matrix and Dermal Fibroblast Function in the Healing Wound. Adv. Wound Care 2016, 5, 119–136. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, K.; Rochat, A.; Barrandon, Y. Segregation of keratinocyte colony-forming cells in the bulge of the rat vibrissae. Proc. Natl. Acad. Sci. USA 1993, 90, 7391–7395. [Google Scholar] [CrossRef] [PubMed]
- Savill, N.J.; Sherratt, J.A. Control of epidermal stem cell clusters by Notch-mediated lateral induction. Dev. Biol. 2003, 258, 141–153. [Google Scholar] [CrossRef]
- Popova, N.V.; Tryson, K.A.; Wu, K.Q.; Morris, R.J. Evidence that the keratinocyte colony number is genetically controlled. Exp. Dermatol. 2002, 11, 503–508. [Google Scholar] [CrossRef] [PubMed]
- Luchi, S.; Dabelsteen, S.; Easley, K.; Rheinwald, J.G.; Green, H. Immortalized keratinocyte lines derived from human embryonic stem cells. Proc. Natl. Acad. Sci. USA 2006, 103, 1792–1797. [Google Scholar]
- Roshan, A.; Murai, K.; Fowler, J.; Simons, B.D.; Nikolaidou-Neokosmidou, V.; Jones, P.H. Human keratinocytes have two interconvertible modes of proliferation. Nat. Cell Biol. 2016, 18, 145–156. [Google Scholar] [CrossRef] [PubMed]
- Gailit, J.; Clark, R.A.F. Wound repair in the context of extracellular matrix. Curr. Opin. Cell Biol. 1994, 6, 717–725. [Google Scholar] [CrossRef]
- Angel, P.; Szabowski, A. Function of AP-1 target genes in mesenchymal-epithelial cross-talk in skin. Biochem. Pharmacol. 2002, 64, 949–956. [Google Scholar] [CrossRef]
- Maas-Szabowski, N.; Shimotoyodome, A.; Fusenig, N.E. Keratinocyte growth regulation in fibroblast cocultures via a double paracrine mechanism. J. Cell Sci. 1999, 112, 1843–1853. [Google Scholar] [PubMed]
- Steele, J.G.; McFarland, C.; Dalton, B.A.; Johnson, G.; Evans, M.D.; Howlett, C.R.; Underwood, P.A. Attachment of human bone cells to tissue culture polystyrene and to unmodified polystyrene: The effect of surface chemistry upon initial cell attachment. J. Biomater. Sci. Polym. Ed. 1993, 5, 245–257. [Google Scholar] [CrossRef] [PubMed]
- Wilson, C.J.; Clegg, R.E.; Leavesley, D.L.; Pearcy, M.J. Mediation of biomaterial-cell interactions by adsorbed proteins: A review. Tissue Eng. 2005, 11, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.S.; Chung, W.J.; Hsu, I.C.; Wu, C.M.; Chin, W.C. Force field measurements within the exclusion zone of water. J. Biol. Phys. 2012, 38, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Spriano, S.; Sarath, C.V.; Cochis, A.; Uberti, F.; Rimondini, L.; Bertone, E.; Vitale, A.; Scolaro, C.; Ferrari, M.; Cirisano, F.; et al. How do wettability, zeta potential and hydroxylation degree affect the biological response of biomaterials? Mater. Sci. Eng. C Mater. Biol. Appl. 2017, 74, 542–555. [Google Scholar] [CrossRef] [PubMed]
- Vogler, E.A. Structure and reactivity of water at biomaterial surfaces. Adv. Colloid Interface Sci. 1998, 74, 69–117. [Google Scholar] [CrossRef]
- Vogler, E.A. The Goldilocks surface. Biomaterials 2011, 32, 6670–6675. [Google Scholar] [CrossRef] [PubMed]
- Horner, C.B.; Ico, G.; Johnson, J.; Yi, Z.; Nam, J. Microstructure-dependent mechanical properties of electrospun core-shell scaffolds at multi-scale levels. J. Mech. Behav. Biomed. Mater. 2016, 59, 207–219. [Google Scholar] [CrossRef] [PubMed]
- Ramli, N.A.; Wong, T.W. Sodium carboxymethylcellulose scaffolds and their physicochemical effects on partial thickness wound healing. Int. J. Pharm. 2011, 403, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Jonsson, M.; Brackmann, C.; Puchades, M.; Brattås, K.; Ewing, A.; Gatenholm, P.; Enejder, A. Neuronal Networks on Nanocellulose Scaffolds. Tissue Eng. Part C Methods 2015, 21, 1162–1170. [Google Scholar] [CrossRef] [PubMed]
- Abraham, E.; Weber, D.E.; Sharon, S.; Lapidot, S.; Shoseyov, O. Multifunctional Cellulosic Scaffolds from Modified Cellulose Nanocrystals. ACS Appl. Mater. Interfaces 2017, 9, 2010–2015. [Google Scholar] [CrossRef] [PubMed]
| Cell | Density (Cells/mL) | With Serum | Without Serum | ||
|---|---|---|---|---|---|
| Maximum Confluence (%) | Time to Achieve Confluence (Day) | Maximum Confluence (%) | Time to Achieve Confluence (Day) | ||
| MONO-HDF | 5000 | 100.00 | 7 | 2.02 | 2 |
| 10,000 | 100.00 | 6 | 3.27 | 2 | |
| 20,000 | 100.00 | 5 | 25.07 | 8 | |
| 40,000 | 100.00 | 3 | 83.44 | 8 | |
| 80,000 | 100.00 | 2 | 100.00 | 6 | |
| 160,000 | 100.00 | 1 | 100.00 | 2 | |
| MONO-HaCat | 5000 | 66.99 | 16 | 0.42 | 1 |
| 10,000 | 91.50 | 16 | 0.61 | 1 | |
| 20,000 | 100.00 | 10 | 1.36 | 2 | |
| 40,000 | 100.00 | 8 | 7.11 | 6 | |
| 80,000 | 100.00 | 7 | 100.00 | 9 | |
| 160,000 | 100.00 | 3 | 100.00 | 3 | |
| CO-HDF | 2500 | 60.91 | 12 | 0.78 | 1 |
| 5000 | 60.10 | 10 | 1.35 | 2 | |
| 10,000 | 66.01 | 8 | 7.34 | 5 | |
| 20,000 | 66.36 | 6 | 26.80 | 7 | |
| 40,000 | 69.61 | 4 | 39.42 | 5 | |
| 80,000 | 59.87 | 2 | 44.87 | 3 | |
| CO-HaCat | 2500 | 67.42 | 16 | 0.11 | 1 |
| 5000 | 100.00 | 16 | 0.13 | 1 | |
| 10,000 | 100.00 | 14 | 0.47 | 1 | |
| 20,000 | 100.00 | 12 | 1.41 | 2 | |
| 40,000 | 100.00 | 10 | 28.59 | 8 | |
| 80,000 | 100.00 | 9 | 100.00 | 9 | |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gabbott, C.M.; Sun, T. Comparison of Human Dermal Fibroblasts and HaCat Cells Cultured in Medium with or without Serum via a Generic Tissue Engineering Research Platform. Int. J. Mol. Sci. 2018, 19, 388. https://doi.org/10.3390/ijms19020388
Gabbott CM, Sun T. Comparison of Human Dermal Fibroblasts and HaCat Cells Cultured in Medium with or without Serum via a Generic Tissue Engineering Research Platform. International Journal of Molecular Sciences. 2018; 19(2):388. https://doi.org/10.3390/ijms19020388
Chicago/Turabian StyleGabbott, Christopher Michael, and Tao Sun. 2018. "Comparison of Human Dermal Fibroblasts and HaCat Cells Cultured in Medium with or without Serum via a Generic Tissue Engineering Research Platform" International Journal of Molecular Sciences 19, no. 2: 388. https://doi.org/10.3390/ijms19020388





