Analysis of Transcriptional Responses of the Inflorescence Meristems in Jatropha curcas Following Gibberellin Treatment
Abstract
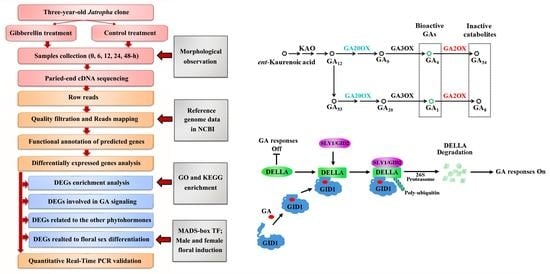
:1. Introduction
2. Results
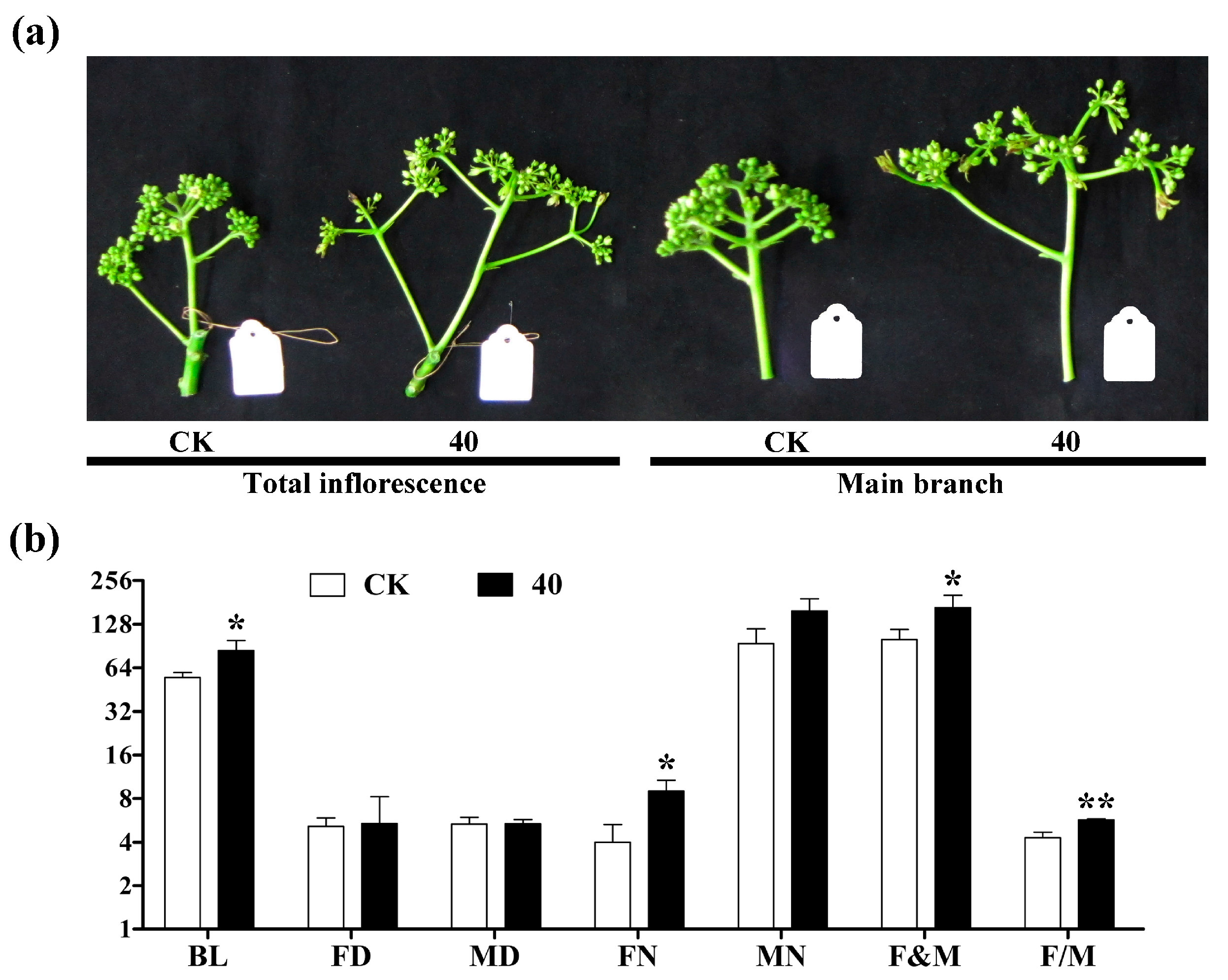
2.1. Morphological Observation after GA3 Treatment
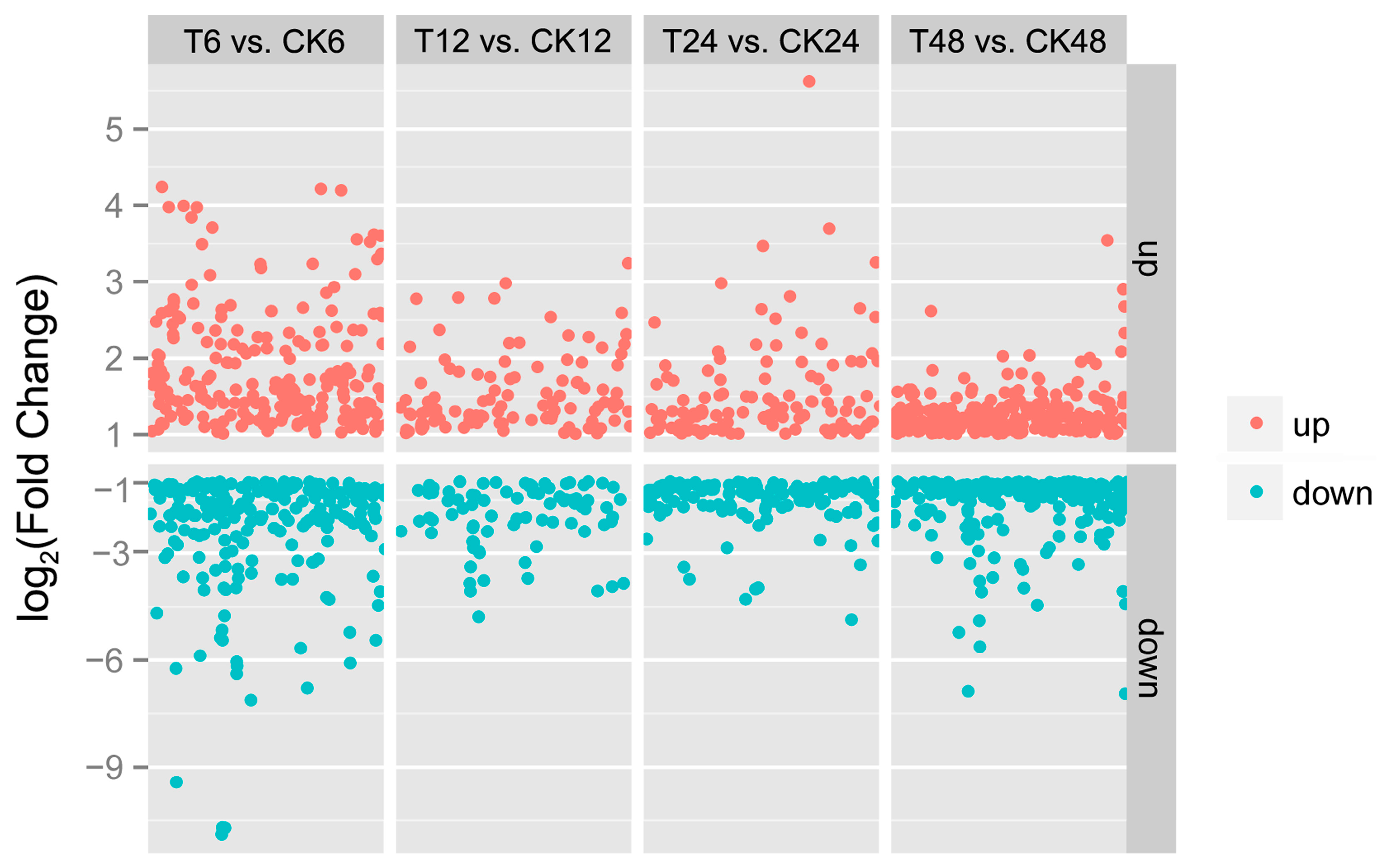
2.2. Illumina Sequencing of Different cDNA Libraries
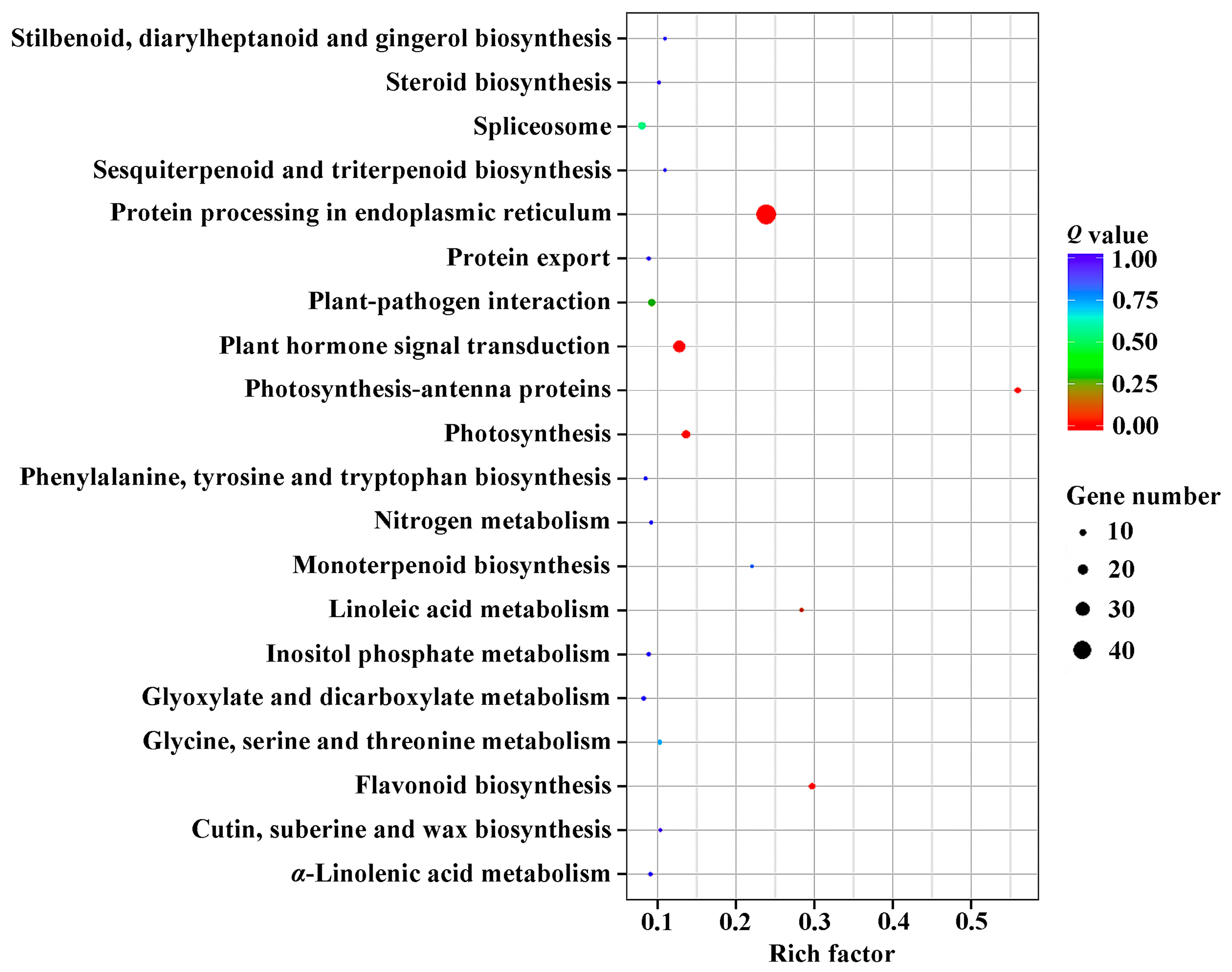
2.3. DEGs Annotation and Enrichment in Response to GA3 Treatment
2.4. DEGs Involved in Gibberellin Biosynthesis, Metabolism and Signaling
2.5. Gibberellin-Regulated DEGs Involved in the Signaling of Other Phytohormones
2.6. MADS-Box Transcription Factor in Response to GA3 Treatment
2.7. DEGs Involved in Male and Female Floral Differentiation
2.8. qRT-PCR Validation
3. Discussion
4. Materials and Methods
4.1. Plant Material and GA Treatment
4.2. Total RNA Extraction and Transcriptome Sequencing Analysis
4.3. Identification and Annotation of DEGs
4.4. Quantitative Real-Time PCR Validation
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Shabanimofrad, M.; Rafii, M.Y.; Wahab, P.E.M.; Biabani, A.R.; Latif, M.A. Phenotypic, genotypic and genetic divergence found in 48 newly collected Malaysian accessions of Jatropha curcas L. Ind. Crops Prod. 2013, 42, 543–551. [Google Scholar] [CrossRef] [Green Version]
- Fairless, D. Biofuel: The little shrub that could—maybe. Nature 2007, 449, 652–655. [Google Scholar] [CrossRef] [PubMed]
- Papalia, T.; Barreca, D.; Panuccio, M.R. Assessment of antioxidant and cytoprotective potential of Jatropha (Jatropha curcas) grown in Southern Italy. Int. J. Mol. Sci. 2017, 18, 660. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Chikara, J.; Chaudhary, D.R.; Prakash, A.R.; Boricha, G.; Zala, A. Paclobutrazol arrests vegetative growth and unveils unexpressed yield potential of Jatropha curcas. J. Plant Growth Regul. 2010, 29, 307–315. [Google Scholar] [CrossRef]
- Pan, B.; Xu, Z. Benzyladenine treatment significantly increases the seed yield of the biofuel plant Jatropha curcas. J. Plant Growth Regul. 2011, 30, 166–174. [Google Scholar] [CrossRef]
- Xu, G.; Luo, R.; Yao, Y. Paclobutrazol improved the reproductive growth and the quality of seed oil of Jatropha curcas. J. Plant Growth Regul. 2013, 32, 875–883. [Google Scholar] [CrossRef]
- Pan, B.; Luo, Y.; Song, L.; Chen, M.; Li, J.; Xu, Z. Thidiazuron increases fruit number in the biofuel plant Jatropha curcas by promoting pistil development. Ind. Crops Prod. 2016, 81, 202–210. [Google Scholar] [CrossRef]
- Hui, W.K.; Yang, S.; Chen, H.; Li, S.; Peng, C.; Wu, G.; Chen, X. Female and male flower bud differentiation of Jatropha carcus L. by gibberellin. J. Nanjing For. Univ. 2016, 40, 174–180. [Google Scholar] [CrossRef]
- Boss, P.K.; Thomas, M.R. Association of dwarfism and floral induction with a grape ‘green revolution’ mutation. Nature 2002, 416, 847–850. [Google Scholar] [CrossRef] [PubMed]
- Daviere, J.; Achard, P. Gibberellin signaling in plants. Development 2013, 140, 1147–1151. [Google Scholar] [CrossRef] [PubMed]
- Olszewski, N.; Sun, T.P.; Gubler, F. Gibberellin signaling: Biosynthesis, catabolism, and response pathways. Plant Cell 2002, 14S, S61–S80. [Google Scholar] [CrossRef]
- Hu, Y.; Tao, Y.; Xu, Z. Overexpression of Jatropha Gibberellin 2-oxidase 6 (JcGA2ox6) induces dwarfism and smaller leaves, flowers and fruits in Arabidopsis and Jatropha. Front. Plant Sci. 2017, 8, 2103. [Google Scholar] [CrossRef] [PubMed]
- Sun, T. Gibberellin-GID1-DELLA: A pivotal regulatory module for plant growth and development. Plant Physiol. 2010, 154, 567–570. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.R.; Singh, R. Gibberellic acid influences the production of malformed and button berries, and fruit yield and quality in strawberry (Fragaria × ananassa Duch.). Sci. Hortic. 2009, 119, 430–433. [Google Scholar] [CrossRef]
- Canli, F.A.; Pektas, M. Improving fruit size and quality of low yielding and small fruited pear cultivars with benzyladenine and gibberellin applications. Eur. J. Hortic. Sci. 2015, 80, 103–108. [Google Scholar] [CrossRef]
- Goldberg-Moeller, R.; Shalom, L.; Shlizerman, L.; Samuels, S.; Zur, N.; Ophir, R.; Blumwald, E.; Sadka, A. Effects of gibberellin treatment during flowering induction period on global gene expression and the transcription of flowering-control genes in Citrus buds. Plant Sci. 2013, 198, 46–57. [Google Scholar] [CrossRef] [PubMed]
- David, A.K.; Tara, A.E.; Lucia, C.S. Auxin biosynthesis and storage forms. J. Exp. Bot. 2013, 64, 2541–2555. [Google Scholar] [CrossRef]
- Makwana, V.; Shukla, P.; Robin, P. GA application induces alteration in sex ratio and cell death in Jatropha curcas. Plant Growth Regul. 2010, 61, 121–125. [Google Scholar] [CrossRef]
- Pan, B.; Chen, M.; Ni, J.; Xu, Z.F. Transcriptome of the inflorescence meristems of the biofuel plant Jatropha curcas treated with cytokinin. BMC Genom. 2014, 15, 974. [Google Scholar] [CrossRef] [PubMed]
- Jewell, J.B.; Browse, J. Epidermal jasmonate perception is sufficient for all aspects of jasmonate-mediated male fertility in Arabidopsis. Plant J. 2016, 85, 634–647. [Google Scholar] [CrossRef] [PubMed]
- Tong, H.; Xiao, Y.; Liu, D.; Gao, S.; Liu, L.; Yin, Y.; Jin, Y.; Qian, Q.; Chu, C. Brassinosteroid regulates cell elongation by modulating gibberellin metabolism in rice. Plant Cell 2014, 26, 4376–4393. [Google Scholar] [CrossRef] [PubMed]
- Heijmans, K.; Morel, P.; Vandenbussche, M. MADS-box genes and floral development: The dark side. J. Exp. Bot. 2012, 63, 5397–5404. [Google Scholar] [CrossRef] [PubMed]
- Moon, J.; Suh, S.S.; Lee, H.; Choi, K.R.; Hong, C.B.; Paek, N.C.; Kim, S.G.; Lee, I. The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J. 2003, 35, 613–623. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, S.; Bohlenius, H.; Moritz, T.; Nilsson, O. GA4 is the active gibberellin in the regulation of LEAFY transcription and Arabidopsis floral initiation. Plant Cell 2006, 18, 2172–2181. [Google Scholar] [CrossRef] [PubMed]
- Galvao, V.C.; Horrer, D.; Kuettner, F.; Schmid, M. Spatial control of flowering by DELLA proteins in Arabidopsis thaliana. Development 2012, 139, 4072–4082. [Google Scholar] [CrossRef] [PubMed]
- Andrés, F.; Porri, A.; Torti, S.; Mateos, J.; Romera-Branchat, M.; García-Martínez, J.; Fornara, F.; Gregis, V.; Kater, M.; Coupland, G. SHORT VEGETATIVE PHASE reduces gibberellin biosynthesis at the Arabidopsis shoot apex to regulate the floral transition. Proc. Natl. Acad. Sci. USA 2014, 111, E2760–E2769. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, B.; Yang, S.; An, J.; Chen, C.; Zhang, X.; Ren, H. A cucumber DELLA homolog CsGAIP may inhibit staminate development through transcriptional repression of B class floral homeotic genes. PLoS ONE 2014, 9, e91804. [Google Scholar] [CrossRef] [PubMed]
- Dong, B.; Deng, Y.; Wang, H.; Gao, R.; Stephen, G.; Chen, S.; Jiang, J.; Chen, F. Gibberellic acid signaling is required to induce flowering of chrysanthemums grown under both short and long days. Int. J. Mol. Sci. 2017, 18, 1259. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Czech, B.; Weigel, D. miR156-Regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 2009, 138, 738–749. [Google Scholar] [CrossRef] [PubMed]
- Hui, W.K.; Yang, Y.; Wu, G.; Peng, C.; Chen, X.; Zayed, M. Transcriptome profile analysis reveals the regulation mechanism of floral sex differentiation in Jatropha curcas L. Sci. Rep. 2017, 7, 16421. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Pan, B.; Fu, Q.; Tao, Y.; Martínez-Herrera, J.; Niu, L.; Ni, J.; Dong, Y.; Zhao, M.; Xu, Z. Comparative transcriptome analysis between gynoecious and monoecious plants identifies regulatory networks controlling sex determination in Jatropha curcas. Front. Plant Sci. 2017, 7, 1953. [Google Scholar] [CrossRef] [PubMed]
- Nemhauser, J.L.; Hong, F.; Chory, J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 2006, 126, 467–475. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Jiao, C.; Singer, S.D.; Gao, M.; Xu, X.; Zhou, Y.; Li, Z.; Fei, Z.; Wang, Y.; Wang, X. Gibberellin-induced changes in the transcriptome of grapevine (Vitis labrusca × V. vinifera) cv. Kyoho flowers. BMC Genom. 2015, 16, 128. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Pan, B.; Wang, G.; Ni, J.; Niu, L.; Xu, Z. Analysis of the transcriptional responses in inflorescence buds of Jatropha curcas exposed to cytokinin treatment. BMC Plant Biol. 2014, 14, 318. [Google Scholar] [CrossRef] [PubMed]
- Rizza, A.; Walia, A.; Lanquar, V.; Frommer, W.B.; Jones, A.M. In vivo gibberellin gradients visualized in rapidly elongating tissues. Nat. Plants 2017, 3, 803–813. [Google Scholar] [CrossRef] [PubMed]
- Lantzouni, O.; Klermund, C.; Schwechheimer, C. Largely additive effects of gibberellin and strigolactone on gene expression in Arabidopsis thaliana seedlings. Plant J. 2017, 92, 924–938. [Google Scholar] [CrossRef] [PubMed]
- Hedden, P.; Phillips, A.L. Gibberellin metabolism: New insights revealed by the genes. Trends Plant Sci. 2000, 5, 523–530. [Google Scholar] [CrossRef]
- Ribeiro, D.; Araujo, W.; Fernie, A.; Schippers, J.; Mueller-Roeber, B. Translatome and metabolome effects triggered by gibberellins during rosette growth in Arabidopsis. J. Exp. Bot. 2012, 63, 2769–2786. [Google Scholar] [CrossRef] [PubMed]
- Hartweck, L.M. Gibberellin signaling. Planta 2008, 229, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.J.; Hur, Y.Y.; Yu, H.; Noh, J.; Park, K.; Lee, H.J. Gibberellin application at pre-bloom in grapevines down-regulates the expressions of VvIAA9 and VvARF7, negative regulators of fruit set initiation, during parthenocarpic fruit development. PLoS ONE 2014, 9, e95634. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Qi, T.; Huang, H.; Ren, Q.; Wu, D.; Chang, C.; Peng, W.; Liu, Y.; Peng, J.; Xie, D. The Jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect jasmonate-regulated stamen development in Arabidopsis. Plant Cell 2011, 23, 1000–1013. [Google Scholar] [CrossRef] [PubMed]
- Bonhomme, F.; Kurz, B.; Melzer, S.; Bernier, G.; Jacqmard, A. Cytokinin and gibberellin activate SaMADS A, a gene apparently involved in regulation of the floral transition in Sinapis alba. Plant J. 2000, 24, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Cerny, R.; Qi, Y.; Bhat, D.; Aydt, C.; Hanson, D.; Malloy, K.; Ness, L. Transgenic studies on the involvement of cytokinin and gibberellin in male development. Plant Physiol. 2003, 131, 1270–1282. [Google Scholar] [CrossRef] [PubMed]
- Jasinski, S.; Piazza, P.; Craft, J.; Hay, A.; Woolley, L.; Rieu, I.; Phillips, A.; Hedden, P.; Tsiantis, M. KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr. Biol. 2005, 15, 1560–1565. [Google Scholar] [CrossRef] [PubMed]
- Meer, I.M.; Stam, M.E.; Tunen, A.J.; Mol, J.N.; Stuitje, A.R. Antisense inhibition of flavonoid biosynthesis in petunia anthers results in male sterility. Plant Cell 1992, 4, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Burbulis, I.E.; Iacobucci, M.; Shirley, B.W. A null mutation in the first enzyme of flavonoid biosynthesis does not affect male fertility in Arabidopsis. Plant Cell 1996, 8, 1013–1025. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Liu, E.; Li, Y.; Li, X.; Ding, C. Transcriptome analysis reveals increases in visceral lipogenesis and storage and activation of the antigen processing and presentation pathway during the mouth-opening stage in zebrafish larvae. Int. J. Mol. Sci. 2017, 18, 1634. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Li, L.; Zhang, J.; Geng, Y.; Guo, F.; Wang, J.; Meng, J.; Sui, N.; Wan, S.; Li, X. Transcriptome and differential expression profiling analysis of the mechanism of Ca2+ regulation in Peanut (Arachis hypogaea) pod development. Front. Plant Sci. 2017, 8, 1609. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Zhao, L.; Qiu, L.; Xu, D.; Tong, Y.; Guo, W.; Yang, X.; Shen, C.; Yan, D.; Zheng, B. Transcriptome and hormonal analysis of grafting process by investigating the homeostasis of a series of metabolic pathways in Torreya grandis cv. Merrillii. Ind. Crops Prod. 2017, 108, 814–823. [Google Scholar] [CrossRef]
- Liu, S.; Chen, J.; Li, S.; Zeng, X.; Meng, Z.; Guo, S. Comparative transcriptome analysis of genes involved in ga-gid1-della regulatory module in symbiotic and asymbiotic seed germination of Anoectochilus roxburghii (Wall.) Lind1. (Orchidaceae). Int. J. Mol. Sci. 2015, 16, 30190–30203. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef]
- Shen, S.; Park, J.; Lu, Z.; Lin, L.; Henry, M.; Wu, Y.; Zhou, Q.; Xing, Y. rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. USA 2014, 111, E5593–E5601. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Williams, B.; Pertea, G.; Mortazavi, A.; Kwan, G.; Baren, M.; Salzberg, S.; Wold, B.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.Z.; Cai, T.; Olyarchuk, J.G.; Wei, L.P. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef] [PubMed]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Jiao, C.; Sun, H.; Rosli, H.; Pombo, M.; Zhang, P.; Banf, M.; Dai, X.; Martin, G.; Giovannoni, J.; et al. iTAK: A program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Mol. Plant 2016, 9, 1667–1670. [Google Scholar] [CrossRef] [PubMed]
- Silva, T.S.; Richard, N. Visualization and differential analysis of protein expression data using R. In Methods in Molecular Biology; Jung, K., Ed.; Humana Press: New York, NY, UAS, 2016; Volume 1362, pp. 105–118. [Google Scholar]
- Zhang, L.; He, L.; Fu, Q.; Xu, Z. Selection of reliable reference genes for gene expression studies in the biofuel plant Jatropha curcas using real-time quantitative PCR. Int. J. Mol. Sci. 2013, 14, 24338–24354. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]

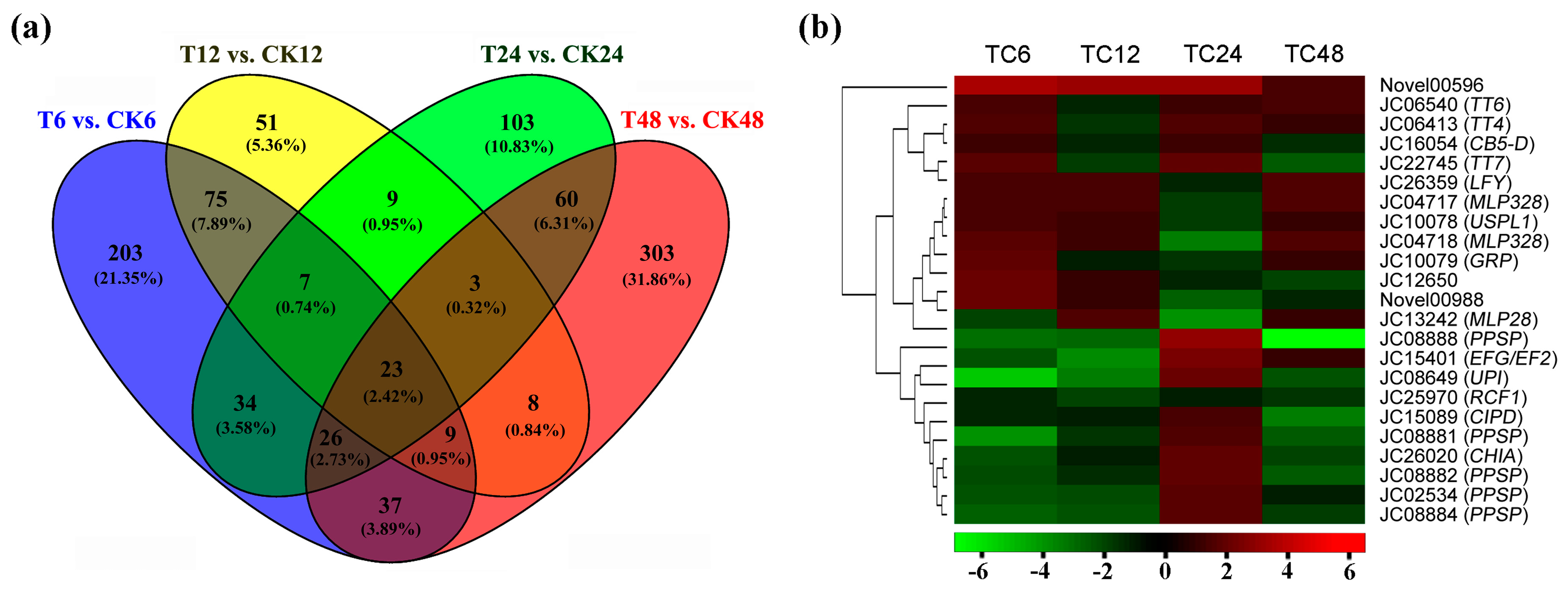
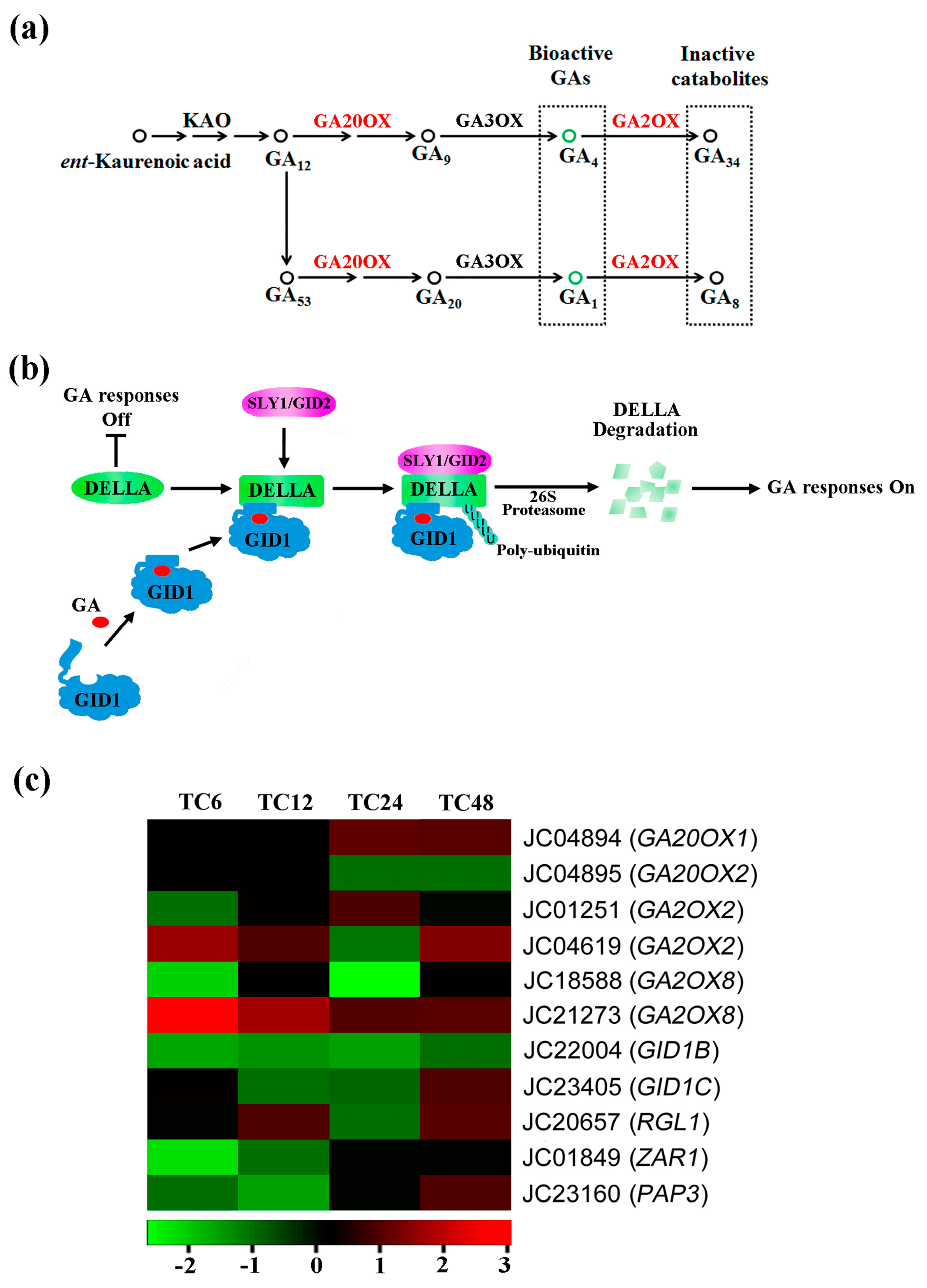
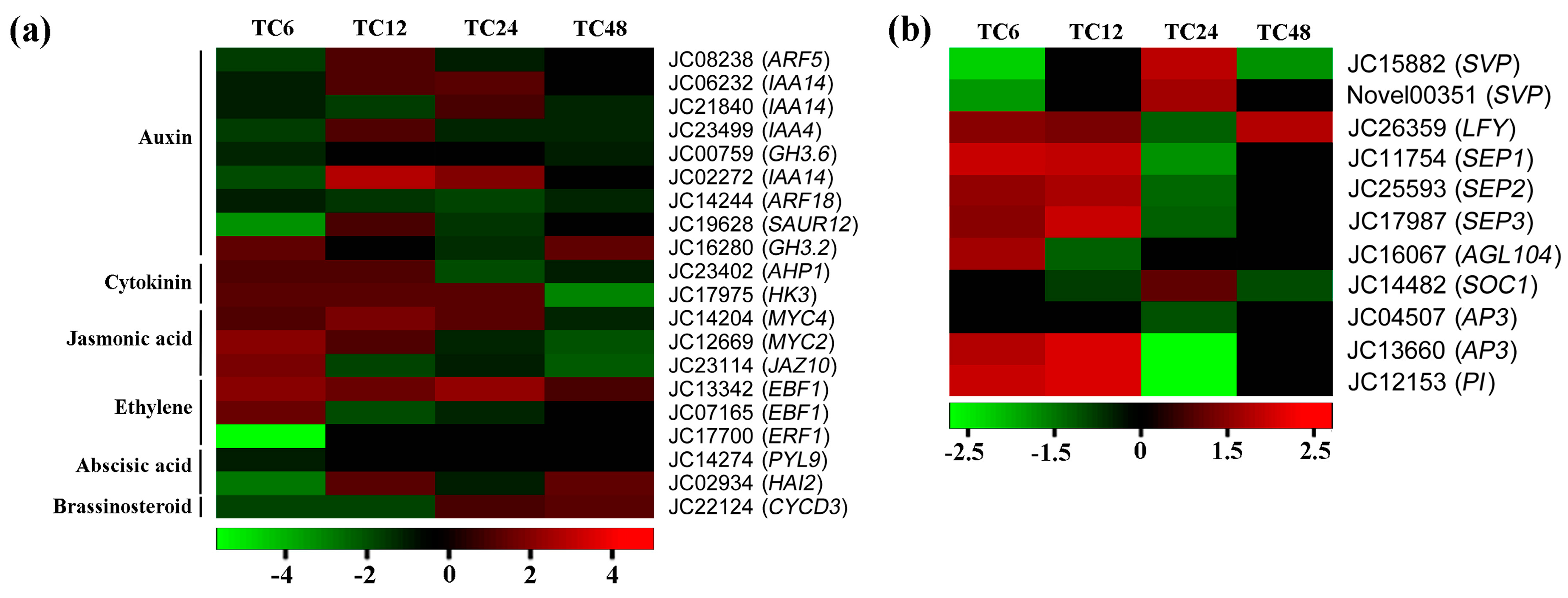

| Gene ID | At. locus | At. name | Blastx to TAIR10 Database |
|---|---|---|---|
| JC06413 | AT5G13930.1 | TT4 | Flavonoid biosynthetic process |
| JC06540 | AT3G51240.1 | TT6 | Flavonoid biosynthetic process |
| JC22745 | AT5G07990.1 | TT7 | Flavonoid biosynthetic process |
| JC26359 | AT5G61850.1 | LFY, LFY3 | Floral meristem determinacy |
| JC25970 | AT1G20920.1 | RCF1 | mRNA splicing via spliceosome |
| JC02534 | AT4G24340.1 | Phosphorylase superfamily protein | Nucleoside metabolic process |
| JC08881 | AT4G24340.1 | Phosphorylase superfamily protein | Nucleoside metabolic process |
| JC08882 | AT4G24340.2 | Phosphorylase superfamily protein | Nucleoside metabolic process |
| JC08884 | AT4G24340.3 | Phosphorylase superfamily protein | Nucleoside metabolic process |
| JC08888 | AT4G24340.4 | Phosphorylase superfamily protein | Nucleoside metabolic process |
| JC16054 | AT5G48810.1 | CB5-D | Oxidation-reduction process |
| JC15089 | AT3G01680.1 | Contains interpro domain/s | Phloem development |
| JC04718 | AT1G70830.1 | MLP-like protein 28 | Response to biotic stimulus |
| JC15401 | AT2G45030.1 | EFG/EF2 | Translation elongation factor |
| JC26020 | AT5G24090.1 | CHIA | Response to light intensity |
| JC10078 | AT1G49320.1 | USPL1 | Seed development |
| JC08649 | AT5G43580.1 | UPI | Serine protease inhibitor |
| JC04717 | AT2G01520.1 | MLP-like protein 328 | Vegetative to reproductive phase transition |
| JC13242 | AT1G70830.1 | MLP-like protein 28 | Defense response |
| JC10079 | AT3G29075.1 | Glycine-rich protein | - |
| JC12650 | - | - | - |
| Novel00596 | - | - | - |
| Novel00988 | - | - | - |
| Gene ID | At. locus | At. name | Blastx to TAIR10 Database |
|---|---|---|---|
| JC04894 | AT4G25420.1 | GA20OX1 | Gibberellin biosynthetic process |
| JC04895 | AT4G25420.2 | GA20OX2 | Gibberellin biosynthetic process |
| JC01251 | AT1G30040.1 | GA2OX2 | Gibberellin oxidation-reduction process |
| JC04619 | AT1G30040.1 | GA2OX2 | Gibberellin oxidation-reduction process |
| JC18588 | AT4G21200.1 | GA2OX8 | Gibberellin oxidation-reduction process |
| JC21273 | AT4G21200.1 | GA2OX8 | Gibberellin oxidation-reduction process |
| JC22004 | AT3G63010.1 | GID1B | Positive regulation of gibberellin mediated signaling pathway |
| JC23405 | AT5G27320.1 | GID1C | Positive regulation of gibberellin mediated signaling pathway |
| JC20657 | AT1G66350.1 | RGL1 | Gibberellic acid mediated signaling pathway |
| JC01849 | AT3G50950.2 | ZAR1 | Gibberellic acid signal transduction |
| JC23160 | AT1G09530.1 | PAP3 | Gibberellic acid mediated signaling pathway |
| Gene ID | At. locus | At. name | Blastx to TAIR10 Database |
|---|---|---|---|
| JC08238 | AT1G19850.1 | ARF5 | Auxin-activated signaling pathway |
| JC06232 | AT4G14550.1 | IAA14 | Auxin-activated signaling pathway |
| JC21840 | AT4G14550.1 | IAA14 | Auxin-activated signaling pathway |
| JC23499 | AT5G43700.1 | IAA4 | Auxin-activated signaling pathway |
| JC00759 | AT5G54510.1 | GH3.6 | Auxin-activated signaling pathway |
| JC02272 | AT4G14550.1 | IAA14 | Auxin-activated signaling pathway |
| JC14244 | AT3G61830.1 | ARF18 | Auxin-activated signaling pathway |
| JC16280 | AT2G14960.1 | GH3.2 | Response to auxin |
| JC19628 | AT2G21220.1 | SAUR12 | Response to auxin |
| JC23402 | AT3G21510.1 | AHP1 | Cytokinin-activated signaling pathway |
| JC17975 | AT1G27320.1 | HK3 | Cytokinin-activated signaling pathway |
| JC14204 | AT4G17880.1 | MYC4 | Jasmonic acid-activated signaling pathway |
| JC12669 | AT1G32640.1 | MYC2 | Jasmonic acid-activated signaling pathway |
| JC23114 | AT5G13220.1 | JAZ10 | Negative regulation of JA signaling pathway |
| JC13342 | AT2G25490.1 | EBF1 | Negative regulation of ethylene-activated signaling pathway |
| JC07165 | AT2G25490.1 | EBF1, | Negative regulation of ethylene-activated signaling pathway |
| JC17700 | AT3G23240.1 | ERF1, | Ethylene-activated signaling pathway |
| JC14274 | AT1G01360.1 | PYL9 | Abscisic acid-activated signaling pathway |
| JC02934 | AT1G07430.1 | HAI2 | Negative regulation of ABA signaling pathway |
| JC22124 | AT4G34160.1 | CYCD3 | Response to brassinosteroid |
| Gene ID | At. locus | At. name | Blastx to TAIR10 Database |
|---|---|---|---|
| JC15882 | AT2G22540.1 | SVP, AGL22 | Floral meristem determinacy |
| Novel00351 | AT2G22540.1 | SVP, AGL22 | Floral meristem determinacy |
| JC26359 | AT5G61850.1 | LFY, LFY3 | Floral meristem determinacy |
| JC11754 | AT5G15800.1 | SEP1, AGL2 | Floral meristem differentiation |
| JC25593 | AT5G15800.1 | SEP2, AGL2 | Floral meristem differentiation |
| JC17987 | AT1G24260.1 | SEP3, AGL9 | Floral meristem differentiation |
| JC16067 | AT1G22130.1 | AGL104 | Pollen maturation |
| JC14482 | AT2G45660.1 | SOC1, AGL20 | Positive regulation of flower development |
| JC04507 | AT3G54340.1 | AP3 | Petal and stamen development |
| JC13660 | AT3G54340.1 | AP3 | Petal and stamen development |
| JC12153 | AT5G20240.1 | PI | Petal identity |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hui, W.-K.; Wang, Y.; Chen, X.-Y.; Zayed, M.Z.; Wu, G.-J. Analysis of Transcriptional Responses of the Inflorescence Meristems in Jatropha curcas Following Gibberellin Treatment. Int. J. Mol. Sci. 2018, 19, 432. https://doi.org/10.3390/ijms19020432
Hui W-K, Wang Y, Chen X-Y, Zayed MZ, Wu G-J. Analysis of Transcriptional Responses of the Inflorescence Meristems in Jatropha curcas Following Gibberellin Treatment. International Journal of Molecular Sciences. 2018; 19(2):432. https://doi.org/10.3390/ijms19020432
Chicago/Turabian StyleHui, Wen-Kai, Yi Wang, Xiao-Yang Chen, Mohamed Zaky Zayed, and Guo-Jiang Wu. 2018. "Analysis of Transcriptional Responses of the Inflorescence Meristems in Jatropha curcas Following Gibberellin Treatment" International Journal of Molecular Sciences 19, no. 2: 432. https://doi.org/10.3390/ijms19020432