Rings and Bricks: Expression of Cohesin Components is Dynamic during Development and Adult Life
Abstract
:1. Introduction
2. Results
2.1. Cohesin Genes Are Ubiquitously and Differentially Expressed in Human Tissues
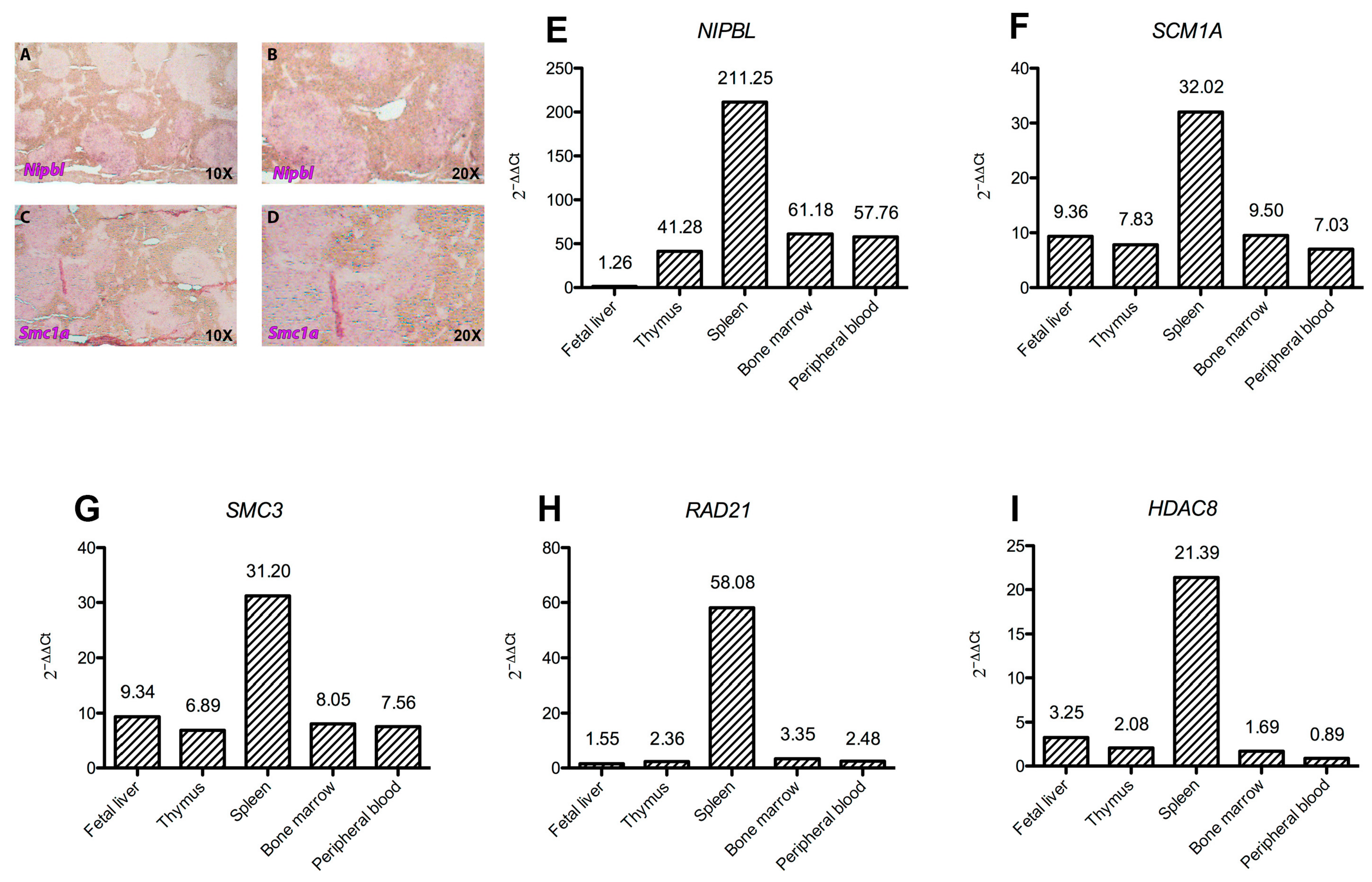
2.2. Cohesins Expression Is Intense in Hematopoietic Tissues
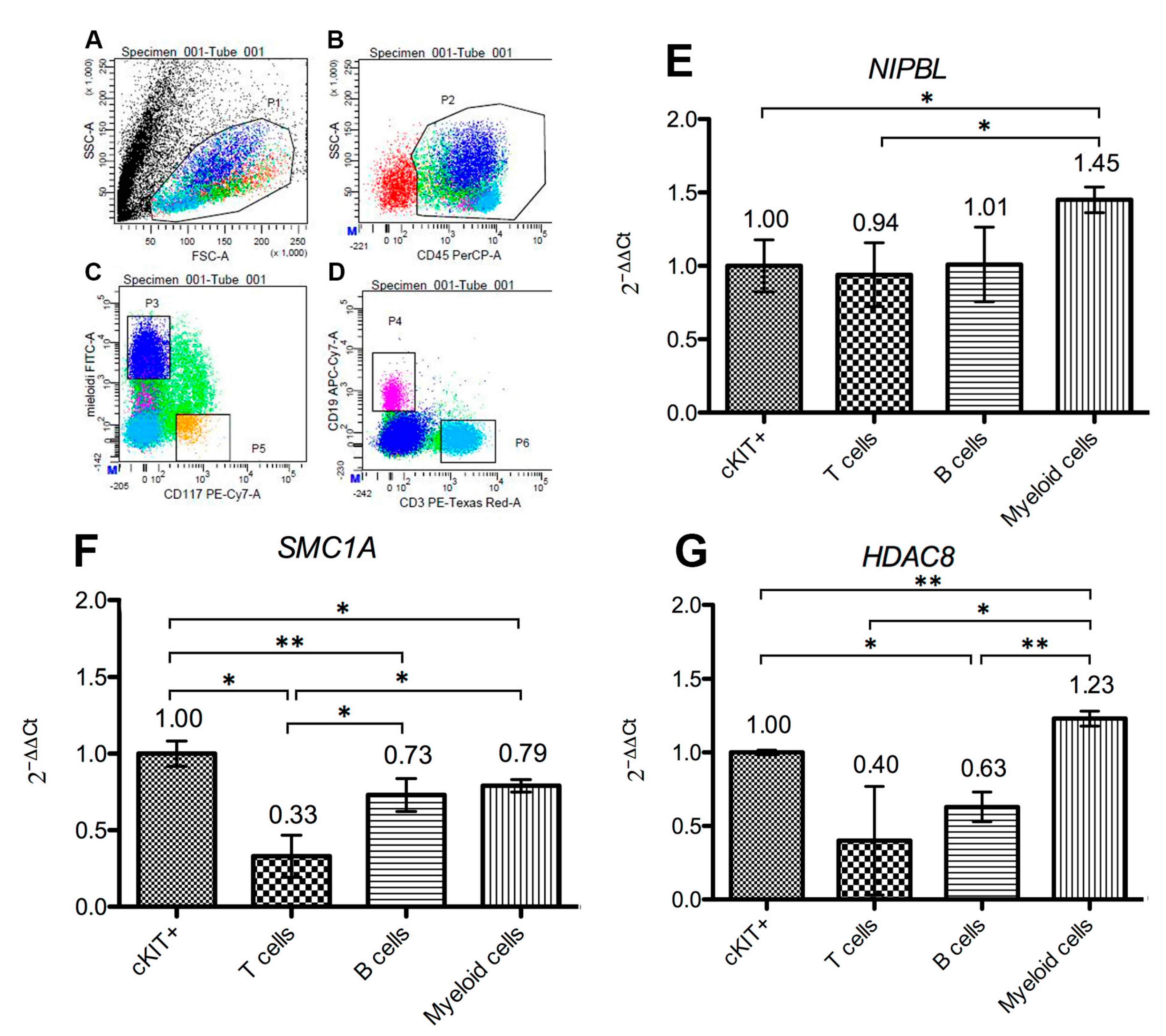
2.3. Cohesins Are Differentially Expressed in Bone Marrow Cells
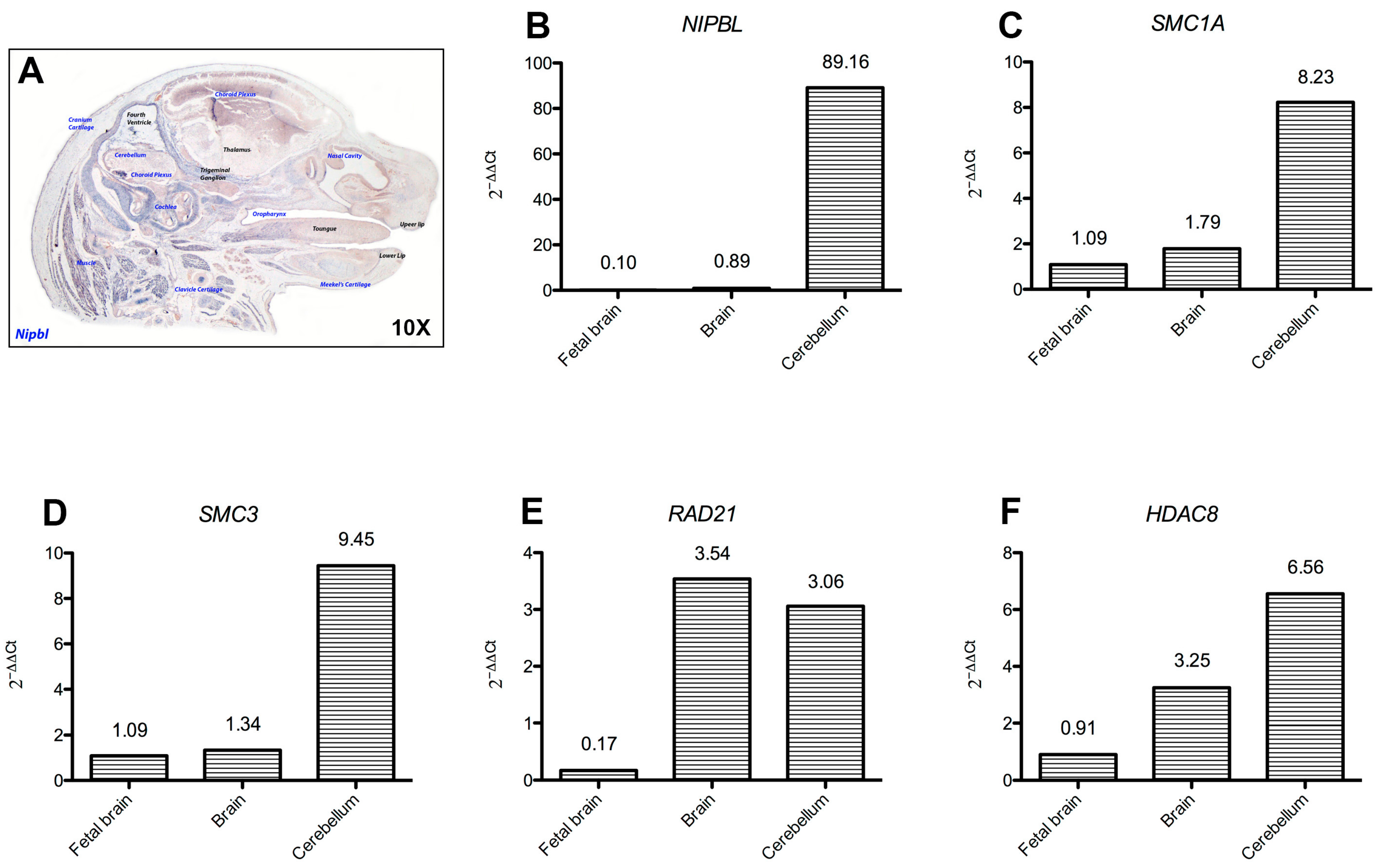
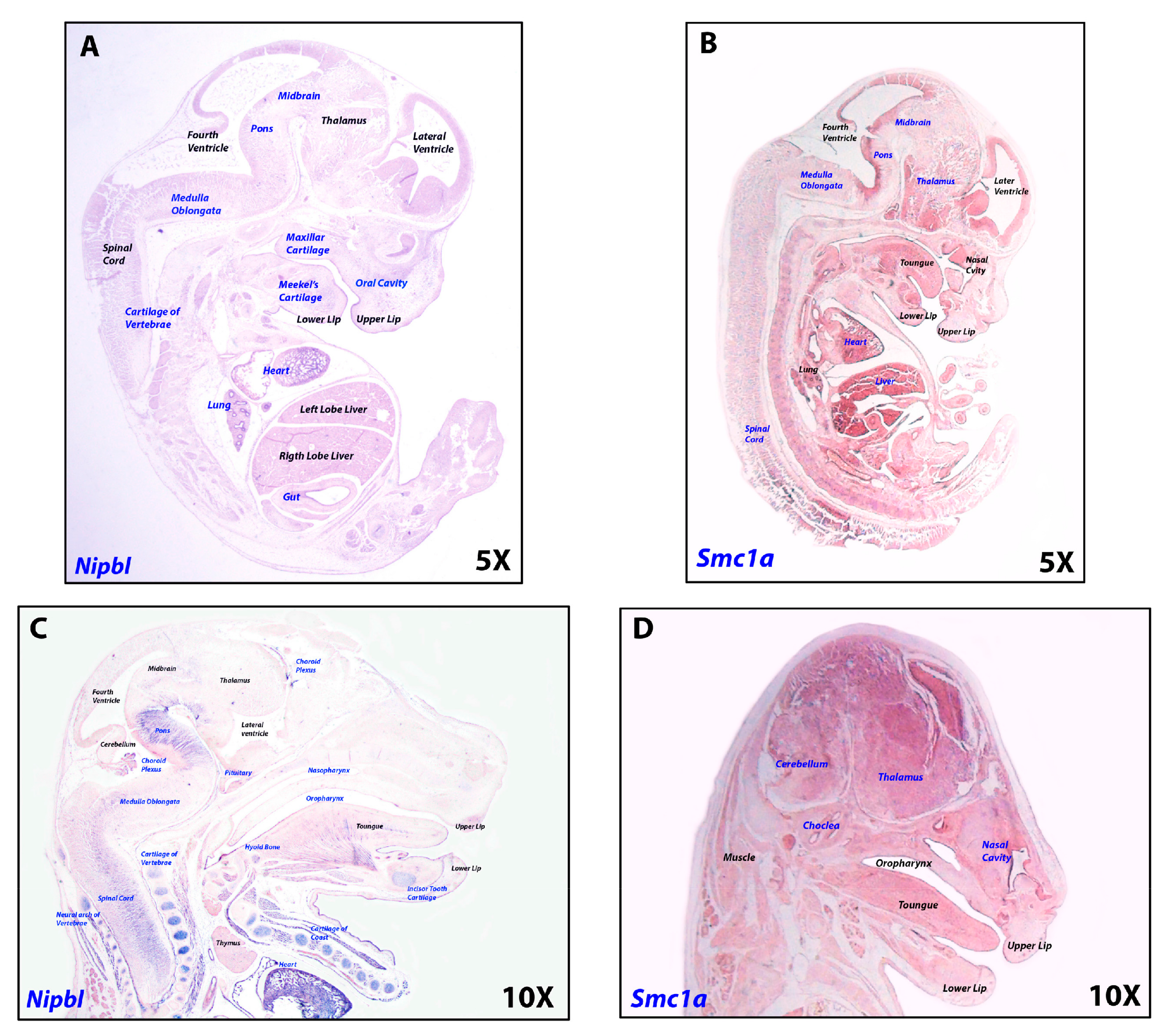
2.4. Cohesins Are Differentially Expressed in Central Nervous System Tissues
3. Discussion
4. Materials and Methods
4.1. Human Tissues
4.2. Antibodies and Flow Cytometry
4.3. Animals
4.4. In Situ Hybridization
4.5. Reverse-Transcription PCR and Real-Time Quantitative-PCR Assays
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Strunnikov, A.V.; Jessberger, R. Structural maintenance of chromosomes (SMC) proteins: Conserved molecular properties for multiple biological functions. Eur. J. Biochem. 1999, 263, 6–13. [Google Scholar] [CrossRef] [PubMed]
- Seitan, V.C.; Banks, P.; Laval, S.; Majid, N.A.; Dorsett, D.; Rana, A.; Smith, J.; Bateman, A.; Krpic, S.; Hostert, A.; et al. Metazoan Scc4 homologs link sister chromatid cohesion to cell and axon migration guidance. PLoS Biol. 2006, 4, 1411–1425. [Google Scholar] [CrossRef] [PubMed]
- White, G.E.; Erickson, H.P. Sequence divergence of coiled coils—Structural rods, myosin filament packing, and the extraordinary conservation of cohesins. J. Struct. Biol. 2006, 154, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Guacci, V.; Koshland, D.; Strunnikov, A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell 1997, 91, 47–57. [Google Scholar] [CrossRef]
- Michaelis, C.; Ciosk, R.; Nasmyth, K. Cohesins: Chromosomal proteins that prevent premature separation of sister chromatids. Cell 1997, 91, 35–45. [Google Scholar] [CrossRef]
- Tóth, A.; Ciosk, R.; Uhlmann, F.; Galova, M.; Schleiffer, A.; Nasmyth, K. Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 1999, 13, 320–333. [Google Scholar] [CrossRef] [PubMed]
- Gruber, S.; Haering, C.H.; Nasmyth, K. Chromosomal cohesin forms a ring. Cell 2003, 112, 765–777. [Google Scholar] [CrossRef]
- Brooker, A.S.; Berkowitz, K.M. The roles of cohesins in mitosis, meiosis, and human health and disease. Methods Mol. Biol. 2014, 1170, 229–266. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, A.R. The role of SMC proteins in the responses to DNA damage. DNA Repair 2005, 4, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Watrin, E.; Peters, J.M. The cohesin complex is required for the DNA damage-induced G2/M checkpoint in mammalian cells. EMBO J. 2009, 28, 2625–2635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dorsett, D. Cohesin, gene expression and development: Lessons from Drosophila. Chromosom. Res. 2009, 17, 185–200. [Google Scholar] [CrossRef] [PubMed]
- Dorsett, D.; Ström, L. The ancient and evolving roles of cohesin in gene expression and DNA repair. Curr. Biol. 2012, 22, R240–R250. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Krantz, I.D. Cornelia de Lange syndrome, cohesin, and beyond. Clin. Genet. 2009, 76, 303–314. [Google Scholar] [CrossRef] [PubMed]
- Barbero, J.L. Genetic basis of cohesinopathies. Appl. Clin. Genet. 2013, 6, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Kline, A.D.; Krantz, I.D.; Sommer, A.; Kliewer, M.; Jackson, L.G.; FitzPatrick, D.R.; Levin, A.V.; Selicorni, A. Cornelia de Lange syndrome: Clinical review, diagnostic and scoring systems, and anticipatory guidance. Am. J. Med. Genet. A 2007, 143A, 1287–1296. [Google Scholar] [CrossRef] [PubMed]
- Deardorff, M.A.; Noon, S.E.; Krantz, I.D. Cornelia de Lange Syndrome; Academic Press: Cambridge, MA, USA, 1993; ISBN 9780080450469. [Google Scholar]
- Dorsett, D. The Drosophila melanogaster model for Cornelia de Lange syndrome: Implications for etiology and therapeutics. Am. J. Med. Genet. C Semin. Med. Genet. 2016, 172, 129–137. [Google Scholar] [CrossRef] [PubMed]
- Kawauchi, S.; Santos, R.; Muto, A.; Lopez-Burks, M.E.; Schilling, T.F.; Lander, A.D.; Calof, A.L. Using mouse and zebrafish models to understand the etiology of developmental defects in Cornelia de Lange Syndrome. Am. J. Med. Genet. C Semin. Med. Genet. 2016, 172, 138–145. [Google Scholar] [CrossRef] [PubMed]
- Remeseiro, S.; Cuadrado, A.; Kawauchi, S.; Calof, A.L.; Lander, A.D.; Losada, A. Reduction of Nipbl impairs cohesin loading locally and affects transcription but not cohesion-dependent functions in a mouse model of Cornelia de Lange Syndrome. Biochim. Biophys. Acta Mol. Basis Dis. 2013, 1832, 2097–2102. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhang, Z.; Bando, M.; Itoh, T.; Deardorff, M.A.; Clark, D.; Kaur, M.; Tandy, S.; Kondoh, T.; Rappaport, E.; et al. Transcriptional dysregulation in NIPBL and cohesin mutant human cells. PLoS Biol. 2009, 7, e1000119. [Google Scholar] [CrossRef] [PubMed]
- Kawauchi, S.; Calof, A.L.; Santos, R.; Lopez-Burks, M.E.; Young, C.M.; Hoang, M.P.; Chua, A.; Lao, T.; Lechner, M.S.; Daniel, J.A.; et al. Multiple organ system defects and transcriptional dysregulation in the Nipbl+/− mouse, a model of Cornelia de Lange Syndrome. PLoS Genet. 2009, 5, e1000650. [Google Scholar] [CrossRef] [PubMed]
- Dorsett, D. Gene regulation: The cohesin ring connects developmental highways. Curr. Biol. 2010, 20, R886–R888. [Google Scholar] [CrossRef] [PubMed]
- Yuan, B.; Pehlivan, D.; Karaca, E.; Patel, N.; Charng, W.L.; Gambin, T.; Gonzaga-Jauregui, C.; Sutton, V.R.; Yesil, G.; Bozdogan, S.T.; et al. Global transcriptional disturbances underlie Cornelia de Lange syndrome and related phenotypes. J. Clin. Investig. 2015, 125, 636–651. [Google Scholar] [CrossRef] [PubMed]
- Fazio, G.; Gaston-Massuet, C.; Bettini, L.R.; Graziola, F.; Scagliotti, V.; Cereda, A.; Ferrari, L.; Mazzola, M.; Cazzaniga, G.; Giordano, A.; et al. CyclinD1 Down-Regulation and Increased Apoptosis Are Common Features of Cohesinopathies. J. Cell. Physiol. 2016, 231, 613–622. [Google Scholar] [CrossRef] [PubMed]
- Kagey, M.H.; Newman, J.J.; Bilodeau, S.; Zhan, Y.; Orlando, D.A.; van Berkum, N.L.; Ebmeier, C.C.; Goossens, J.; Rahl, P.B.; Levine, S.S.; et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature 2010, 467, 430–435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pistocchi, A.; Fazio, G.; Cereda, A.; Ferrari, L.; Bettini, L.R.; Messina, G.; Cotelli, F.; Biondi, A.; Selicorni, A.; Massa, V. Cornelia de Lange Syndrome: NIPBL haploinsufficiency downregulates canonical Wnt pathway in zebrafish embryos and patients fibroblasts. Cell Death Dis. 2013, 4, e866. [Google Scholar] [CrossRef] [PubMed]
- Schuldiner, O.; Berdnik, D.; Levy, J.M.; Wu, J.S.; Luginbuhl, D.; Gontang, A.C.; Luo, L. piggyBac-Based Mosaic Screen Identifies a Postmitotic Function for Cohesin in Regulating Developmental Axon Pruning. Dev. Cell 2008, 14, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Panigrahi, A.K.; Pati, D. Higher-order orchestration of hematopoiesis: Is cohesin a new player? Exp. Hematol. 2012, 40, 967–973. [Google Scholar] [CrossRef] [PubMed]
- Mullenders, J.; Aranda-Orgilles, B.; Lhoumaud, P.; Keller, M.; Pae, J.; Wang, K.; Kayembe, C.; Rocha, P.P.; Raviram, R.; Gong, Y.; et al. Cohesin loss alters adult hematopoietic stem cell homeostasis, leading to myeloproliferative neoplasms. J. Exp. Med. 2015, 212, 1833–1850. [Google Scholar] [CrossRef] [PubMed]
- Phillips-Cremins, J.E.; Sauria, M.E.G.; Sanyal, A.; Gerasimova, T.I.; Lajoie, B.R.; Bell, J.S.K.; Ong, C.T.; Hookway, T.A.; Guo, C.; Sun, Y.; et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell 2013, 153, 1281–1295. [Google Scholar] [CrossRef] [PubMed]
- Kon, A.; Shih, L.Y.; Minamino, M.; Sanada, M.; Shiraishi, Y.; Nagata, Y.; Yoshida, K.; Okuno, Y.; Bando, M.; Nakato, R.; et al. Recurrent mutations in multiple components of the cohesin complex in myeloid neoplasms. Nat. Genet. 2013, 45, 1232–1237. [Google Scholar] [CrossRef] [PubMed]
- Mazumdar, C.; Majeti, R. The role of mutations in the cohesin complex in acute myeloid leukemia. Int. J. Hematol. 2017, 105, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Horsfield, J.A.; Anagnostou, S.H.; Hu, J.K.; Cho, K.H.; Geisler, R.; Lieschke, G.; Crosier, K.E.; Crosier, P.S. Cohesin-dependent regulation of Runx genes. Development 2007, 134, 2639–2649. [Google Scholar] [CrossRef] [PubMed]
- Marsman, J.; O’Neill, A.C.; Kao, B.R.; Rhodes, J.M.; Meier, M.; Antony, J.; Monnich, M.; Horsfield, J.A. Cohesin and CTCF differentially regulate spatiotemporal runx1 expression during zebrafish development. Biochim. Biophys. Acta 2014, 1839, 50–61. [Google Scholar] [CrossRef] [PubMed]
- Ichikawa, M.; Yoshimi, A.; Nakagawa, M.; Nishimoto, N.; Watanabe-Okochi, N.; Kurokawa, M. A role for RUNX1 in hematopoiesis and myeloid leukemia. Int. J. Hematol. 2013, 97, 726–734. [Google Scholar] [CrossRef] [PubMed]
- Rocquain, J.; Gelsi-Boyer, V.; Adélaïde, J.; Murati, A.; Carbuccia, N.; Vey, N.; Birnbaum, D.; Mozziconacci, M.J.; Chaffanet, M. Alteration of cohesin genes in myeloid diseases. Am. J. Hematol. 2010, 85, 717–719. [Google Scholar] [CrossRef] [PubMed]
- Avagliano, L.; Grazioli, P.; Mariani, M.; Bulfamante, G.P.; Selicorni, A.; Massa, V. Integrating molecular and structural findings: Wnt as a possible actor in shaping cognitive impairment in Cornelia de Lange syndrome. Orphanet J. Rare Dis. 2017, 12, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Roshan Lal, T.R.; Kliewer, M.A.; Lopes, T.; Rebsamen, S.L.; O’Connor, J.; Grados, M.A.; Kimball, A.; Clemens, J.; Kline, A.D. Cornelia de Lange syndrome: Correlation of brain MRI findings with behavioral assessment. Am. J. Med. Genet. C Semin. Med. Genet. 2016, 172, 190–197. [Google Scholar] [CrossRef] [PubMed]
- Grados, M.A.; Alvi, M.H.; Srivastava, S. Behavioral and psychiatric manifestations in Cornelia de Lange syndrome. Curr. Opin. Psychiatry 2017, 30, 92–96. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆Ct Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Fazio, G.; Bettini, L.R.; Rigamonti, S.; Meta, D.; Biondi, A.; Cazzaniga, G.; Selicorni, A.; Massa, V. Impairment of Retinoic Acid Signaling in Cornelia de Lange Syndrome Fibroblasts. Birth Defects Res. 2017, 1–9. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bettini, L.R.; Graziola, F.; Fazio, G.; Grazioli, P.; Scagliotti, V.; Pasquini, M.; Cazzaniga, G.; Biondi, A.; Larizza, L.; Selicorni, A.; et al. Rings and Bricks: Expression of Cohesin Components is Dynamic during Development and Adult Life. Int. J. Mol. Sci. 2018, 19, 438. https://doi.org/10.3390/ijms19020438
Bettini LR, Graziola F, Fazio G, Grazioli P, Scagliotti V, Pasquini M, Cazzaniga G, Biondi A, Larizza L, Selicorni A, et al. Rings and Bricks: Expression of Cohesin Components is Dynamic during Development and Adult Life. International Journal of Molecular Sciences. 2018; 19(2):438. https://doi.org/10.3390/ijms19020438
Chicago/Turabian StyleBettini, Laura Rachele, Federica Graziola, Grazia Fazio, Paolo Grazioli, Valeria Scagliotti, Mariavittoria Pasquini, Giovanni Cazzaniga, Andrea Biondi, Lidia Larizza, Angelo Selicorni, and et al. 2018. "Rings and Bricks: Expression of Cohesin Components is Dynamic during Development and Adult Life" International Journal of Molecular Sciences 19, no. 2: 438. https://doi.org/10.3390/ijms19020438