Expression Profiling of Regulatory and Biosynthetic Genes in Contrastingly Anthocyanin Rich Strawberry (Fragaria × ananassa) Cultivars Reveals Key Genetic Determinants of Fruit Color
Abstract
:1. Introduction
2. Results
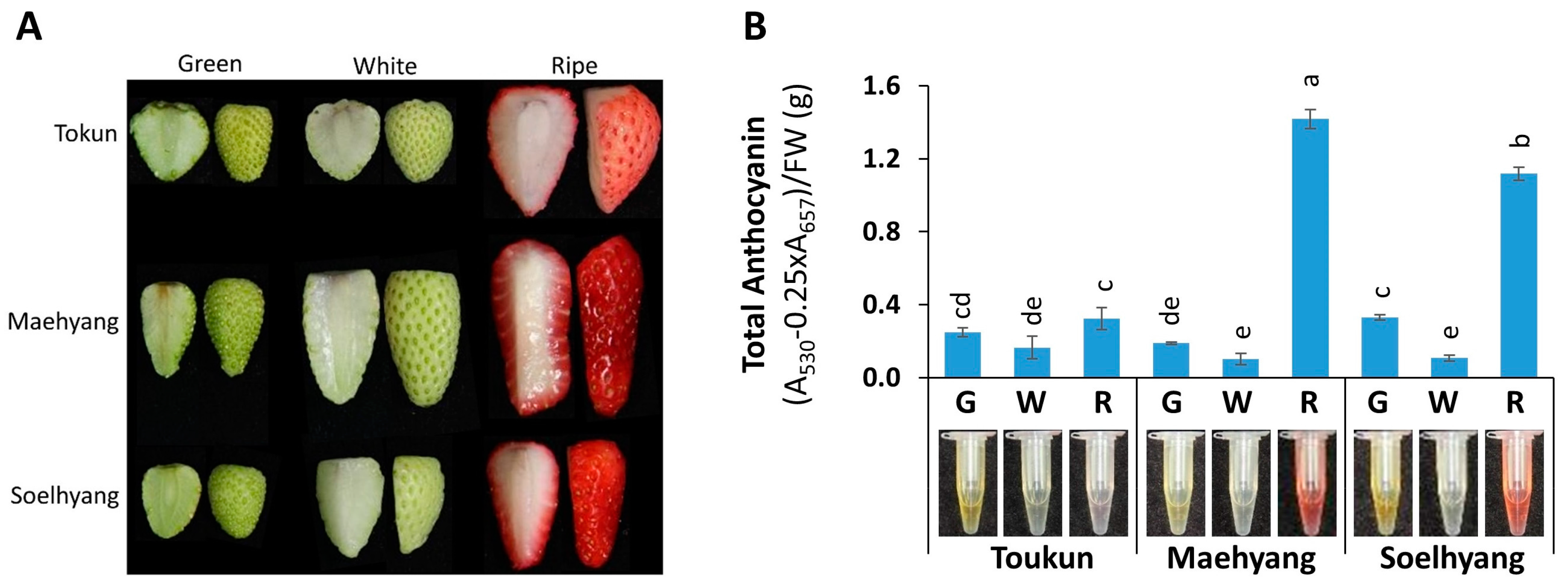
2.1. Total Anthocyanin Varied Across Fruit Developmental Stages and Cultivars of Strawberry
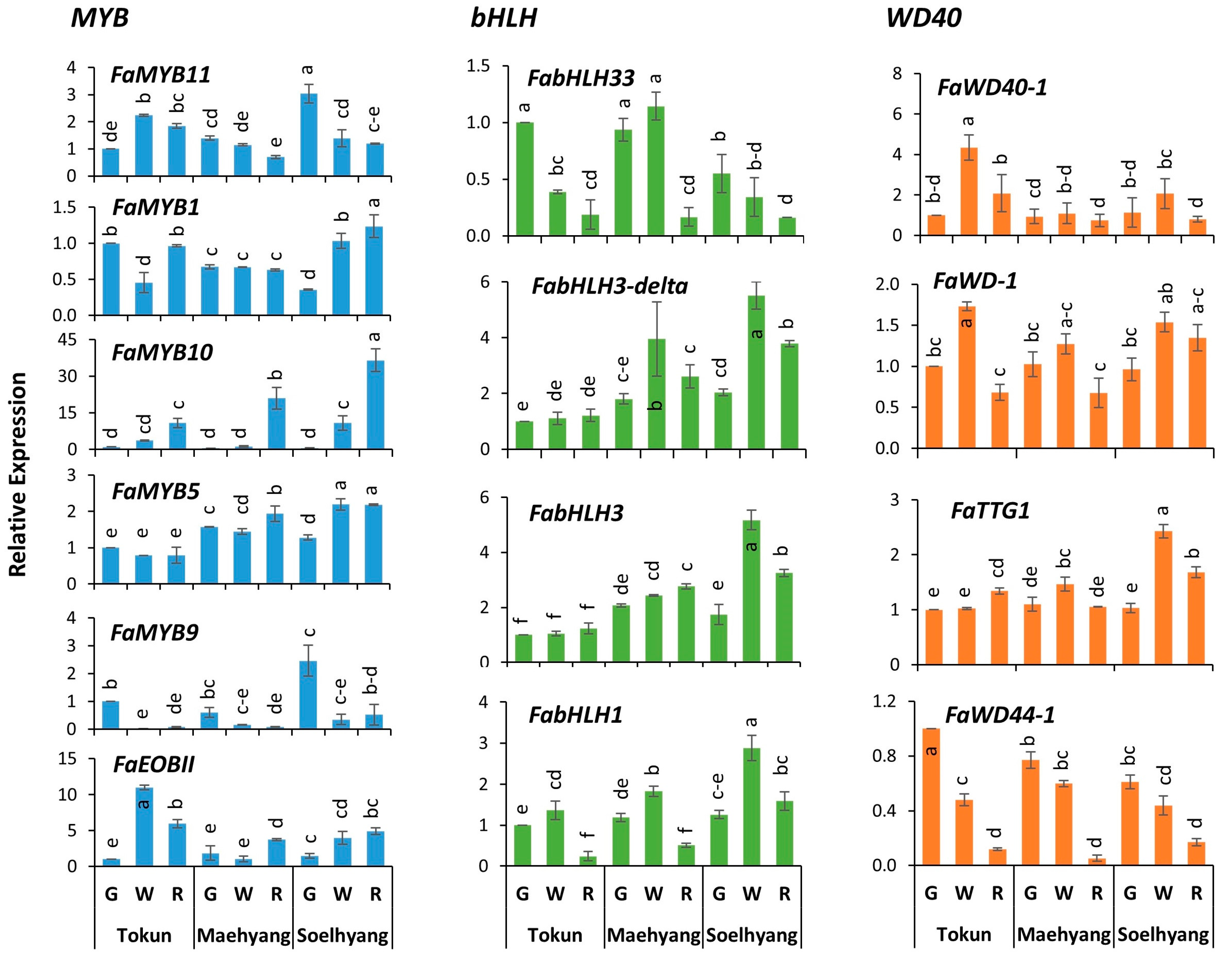
2.2. FaMYB10 Dominated the Anthocyanin Regulatory Complex
2.3. FaMYB11 Is the Potential Negative Regulator of Anthocyanin Biosynthesis
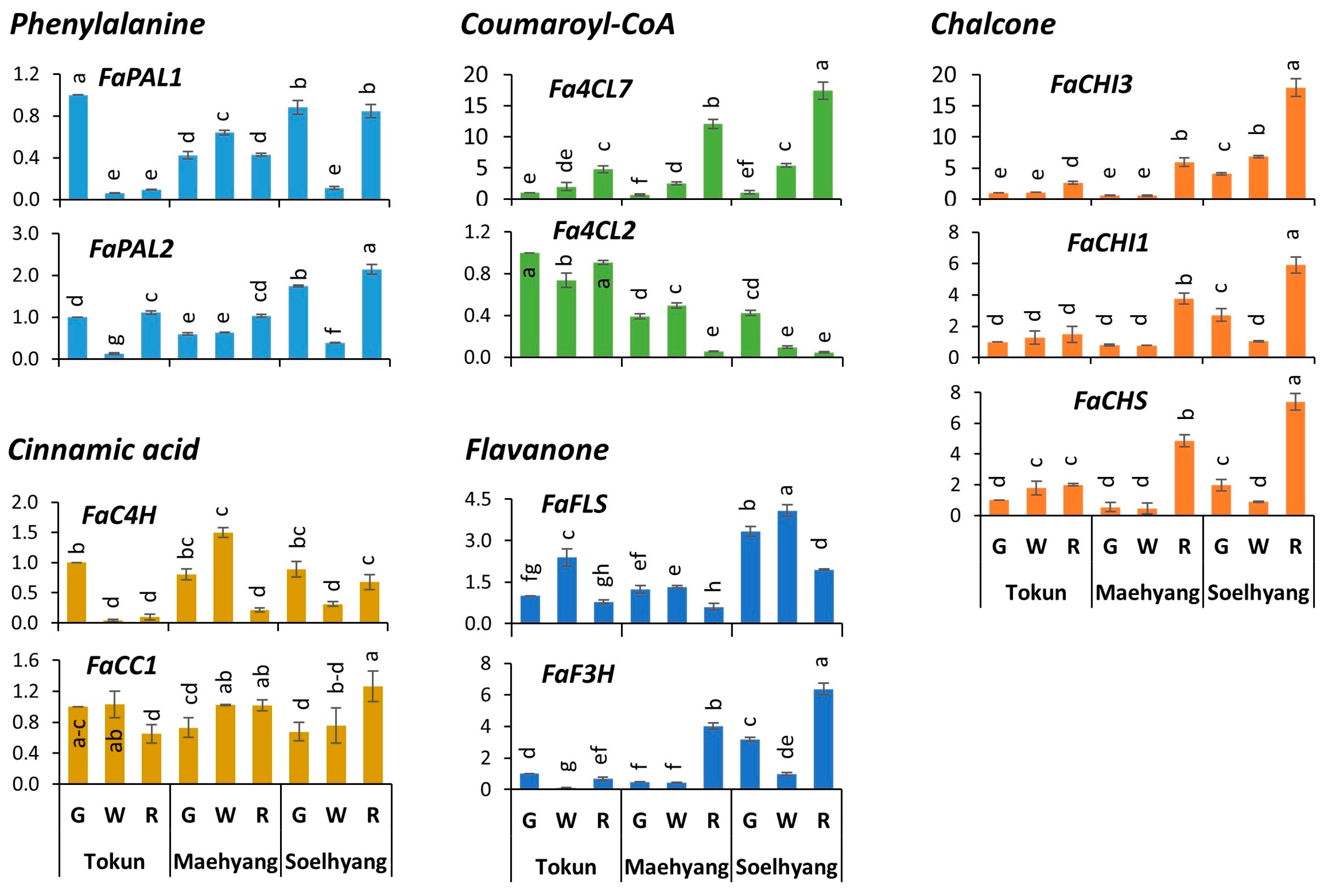
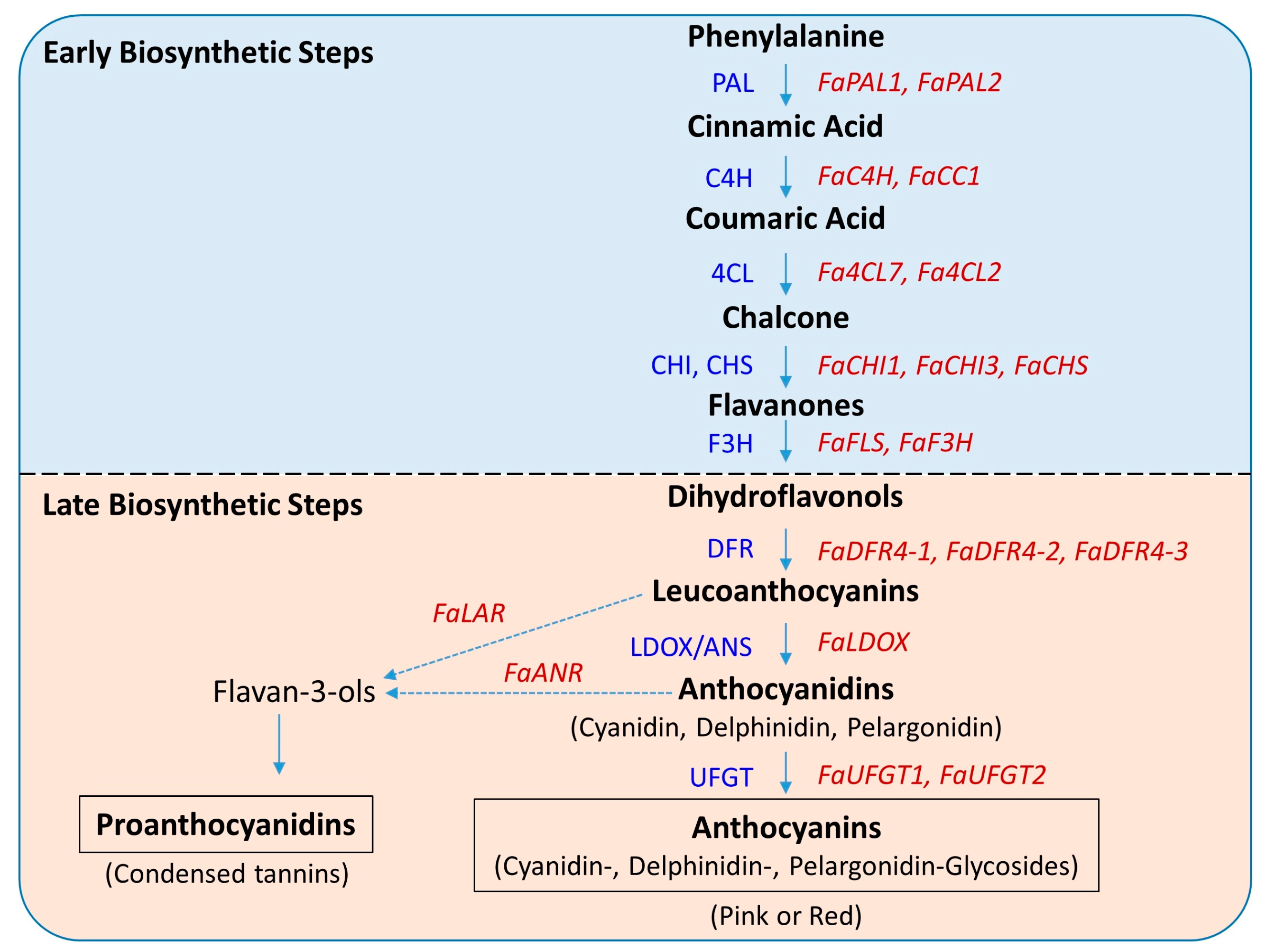
2.4. Expression Profiles of Early Biosynthetic Genes
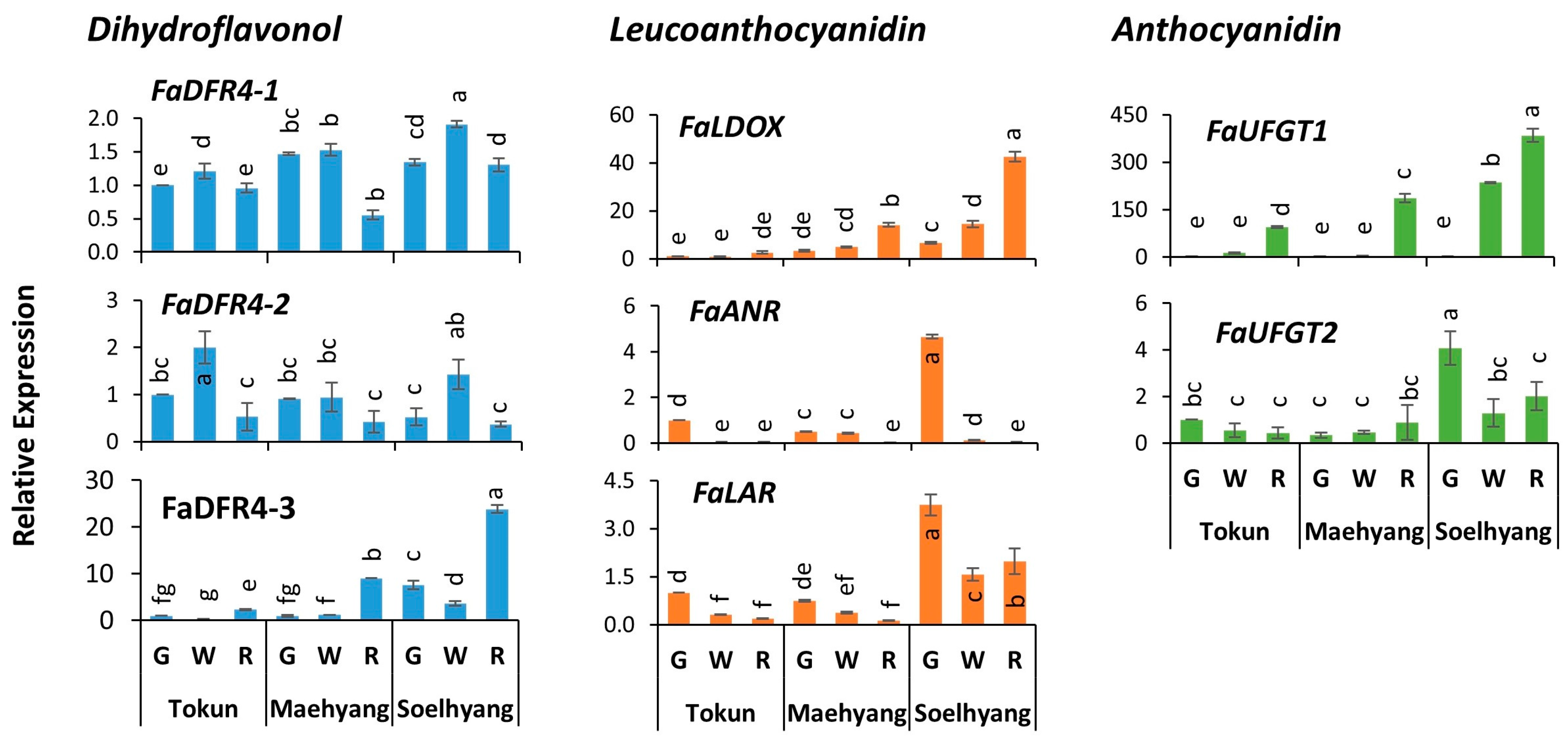
2.5. Key Genes of the Late Biosynthetic Steps
2.6. Late Biosynthetic Genes Show Comparatively Higher Expression
2.7. Association between Contents of Total Anthocyanin and Expressions of Related Regulatory and Biosynthetic Genes
3. Discussion
3.1. Positive Regulators of Anthocyanin Biosynthesis in Korean Strawberry Cultivars
3.2. Potential Repressors of Anthocyanin Biosynthesis
3.3. Anthocyanin and Proanthocyaninid Might Share Few Contrasting Regulatory Genes
3.4. Key Structural Genes and Their Association with Total Anthocyanin and Regulatory Genes
3.5. Progressive Intensification of Pathway Flux May Lead to Higher Anthocyanin Accumulation
4. Methods
4.1. Plant Materials
4.2. Extraction and Photometric Determination of Anthocyanin
4.3. Selection of Anthocyanin Related Genes in Fragaria × ananassa
4.4. RNA Extraction and Expression Analysis via Real-Time qRT-PCR
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgment
Author Contributions
Conflicts of Interest
Abbreviations
| F/P | Phenylapropanoid/flavonoid pathway |
| BGs | Biosynthetic Genes |
| EBGs | Early Biosynthetic Genes |
| LBGs | Late Biosynthetic Genes |
| TF | Transcription Factor |
| MYB | Myeloblastosis family of transcription factors |
| bHLH | Basic helix-loop-helix |
| PAL | Phenylalanine ammonia lyase |
| C4H | Cinnamate-4-hydroxylase |
| 4CL | 4-Coumaroyl-CoA-ligase |
| CHS | Chalcone synthase |
| CHI | Chalcone isomerase |
| F3H | Flavanone 3 hydroxylase |
| FLS | Flavonol synthase |
| DFR | Dihydroflavanol reductase |
| LAR | Leucoanthocyanidin reductase |
| ANR | Anthocyanidin reductase |
References
- Mezzetti, B. Breeding and biotechnology for improving the nutritional quality of strawberry. J. Berry Res. 2013, 3, 127–133. [Google Scholar]
- Schaart, J.G.; Dubos, C.; Romero De La Fuente, I.; van Houwelingen, A.M.M.L.; de Vos, R.C.H.; Jonker, H.H.; Xu, W.; Routaboul, J.M.; Lepiniec, L.; Bovy, A.G. Identification and characterization of MYB-bHLH-WD40 regulatory complexes controlling proanthocyanidin biosynthesis in strawberry (Fragaria × ananassa) fruits. New Phytol. 2013, 197, 454–467. [Google Scholar] [CrossRef] [PubMed]
- Jaakola, L. New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 2013, 18, 477–483. [Google Scholar] [CrossRef] [PubMed]
- Kayesh, E.; Shangguan, L.; Korir, N.K.; Sun, X.; Bilkish, N.; Zhang, Y.; Han, J.; Song, C.; Cheng, Z.M.; Fang, J. Fruit skin color and the role of anthocyanin. Acta Physiol. Plant. 2013, 35, 2879–2890. [Google Scholar] [CrossRef]
- Tanaka, Y.; Sasaki, N.; Ohmiya, A. Biosynthesis of plant pigments: Anthocyanins, betalains and carotenoids. Plant J. 2008, 54, 733–749. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Garcia, S.N.; Guevara-Gonzalez, R.G.; Miranda-Lopez, R.; Feregrino-Perez, A.A.; Torres-Pacheco, I.; Vazquez-Cruz, M.A. Functional properties and quality characteristics of bioactive compounds in berries: Biochemistry, biotechnology, and genomics. Food Res. Int. 2013, 54, 1195–1207. [Google Scholar] [CrossRef]
- Lin-Wang, K.; Liu, Y.; Espley, R.V.; Karunairetnam, S.; McGhie, T.K.; Hellens, R.P.; Allan, A.C. Regulation of anthocyanin biosynthesis in strawberry (Fragaria sp.) by over-expression of a key transcription factor. Acta Hortic. 2014, 1048, 137–142. [Google Scholar] [CrossRef]
- Schaefer, H.M.; Schaefer, V.; Levey, D.J. How plant–animal interactions signal new insights in communication. Trends Ecol. Evol. 2004, 19, 577–584. [Google Scholar] [CrossRef]
- Park, K.-I.; Ishikawa, N.; Morita, Y.; Choi, J.-D.; Hoshino, A.; Iida, S. A bHLH regulatory gene in the common morning glory, Ipomoea purpurea, controls anthocyanin biosynthesis in flowers, proanthocyanidin and phytomelanin pigmentation in seeds, and seed trichome formation. Plant J. 2007, 49, 641–654. [Google Scholar] [CrossRef] [PubMed]
- Pillet, J.; Yu, H.; Chambers, A.H.; Whitaker, V.M.; Folta, K.M. Identification of candidate flavonoid pathway genes using transcriptome correlation network analysis in ripe strawberry (Fragaria × ananassa) fruits. J. Exp. Bot. 2015, 66, 4455–4467. [Google Scholar] [CrossRef] [PubMed]
- Koide, T.; Kamei, H.; Hashimoto, Y.; Kojima, T.; Hasegawa, M. Antitumor effect of hydrolyzed anthocyanin from grape rinds and red rice. Cancer Biother. Radiopharm. 1996, 11, 273–277. [Google Scholar] [CrossRef] [PubMed]
- Cristina, L.; Aizza, B.; Dornelas, M.C. Differential Transcription Factor Networks Orchestrate Flavonoid Biosynthesis. In Pigments in Fruits and Vegetables; Chen, C., Ed.; Springer: New York, NY, 2015; pp. 69–91. [Google Scholar]
- Almeida, J.R.M.; D’Amico, E.; Preuss, A.; Carbone, F.; Deiml, B.; Mourgues, F.; Perrotta, G.; Fischer, T.C.; Bovy, A.G.; Martens, S.; Rosati, C. Characterization of major enzymes and genes involved in flavonoid and proanthocyanidin biosynthesis during fruit development in strawberry (Fragaria × ananassa). Arch. Biochem. Biophys. 2007, 465, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Carbone, F.; Preuss, A.; De Vos, R.C.; Amico, E.D.; Perrotta, G.; Bovy, A.G.; Martens, S.; Rosati, C. Developmental, genetic and environmental factors affect the expression of flavonoid genes, enzymes and metabolites in strawberry fruits. Plant Cell Environ. 2009, 32, 1117–1131. [Google Scholar] [CrossRef] [PubMed]
- Afrin, S.; Nuruzzaman, M.; Zhu, J.; Luo, Z. Combinatorial interactions of MYB and bHLH in flavonoid biosynthesis and their function in plants. J. Plant Biol. Res. 2014, 3, 65–77. [Google Scholar]
- Petroni, K.; Tonelli, C. Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci. 2011, 181, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Hichri, I.; Barrieu, F.; Bogs, J.; Kappel, C.; Delrot, S.; Lauvergeat, V. Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J. Exp. Bot. 2011, 62, 2465–2483. [Google Scholar] [CrossRef] [PubMed]
- Allan, A.C.; Hellens, R.P.; Laing, W.A. MYB transcription factors that colour our fruit. Trends Plant Sci. 2008, 13, 99–102. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, A.; Zhao, M.; Leavitt, J.M.; Lloyd, A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/MYB transcriptional complex in Arabidopsis seedlings. Plant J. 2008, 53, 814–827. [Google Scholar] [CrossRef] [PubMed]
- Cutanda-Perez, M.-C.; Ageorges, A.; Gomez, C.; Vialet, S.; Terrier, N.; Romieu, C.; Torregrosa, L. Ectopic expression of VlMYBA1 in grapevine activates a narrow set of genes involved in anthocyanin synthesis and transport. Plant Mol. Biol. 2009, 69, 633–648. [Google Scholar] [CrossRef] [PubMed]
- Borovsky, Y.; Oren-Shamir, M.; Ovadia, R.; De Jong, W.; Paran, I. The A locus that controls anthocyanin accumulation in pepper encodes a MYB transcription factor homologous to Anthocyanin2 of Petunia. Theor. Appl. Genet. 2004, 109, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Grotewold, E.; Sainz, M.B.; Tagliani, L.; Hernandez, J.M.; Bowen, B.; Chandler, V.L. Identification of the residues in the MYB domain of maize C1 that specify the interaction with the bHLH cofactor R. Proc. Natl. Acad. Sci. USA 2000, 97, 13579–13584. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Lin-Wang, K.; Espley, R.V.; Wang, L.; Yang, H.; Yu, B.; Dare, A.; Varkonyi-Gasic, E.; Wang, J.; Zhang, J.; et al. Functional diversification of the potato R2R3 MYB anthocyanin activators AN1, MYBA1, and MYB113 and their interaction with basic helix-loop-helix cofactors. J. Exp. Bot. 2016, 67, 2159–2176. [Google Scholar] [CrossRef] [PubMed]
- Ban, Y.; Honda, C.; Hatsuyama, Y.; Igarashi, M.; Bessho, H.; MoriguCHI, T. Isolation and Functional Analysis of a MYB Transcription Factor Gene that is a Key Regulator for the Development of Red Coloration in Apple Skin. Plant Cell Physiol. 2007, 48, 958–970. [Google Scholar] [CrossRef] [PubMed]
- Espley, R.V.; Hellens, R.P.; Putterill, J.; Stevenson, D.E.; Kutty-Amma, S.; Allan, A.C. Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 2007, 49, 414–427. [Google Scholar] [CrossRef] [PubMed]
- Jin, W.; Wang, H.; Li, M.; Wang, J.; Yang, Y.; Zhang, X.; Yan, G.; Zhang, H.; Liu, J.; Zhang, K. The R2R3 MYB transcription factor PavMYB10.1 involves in anthocyanin biosynthesis and determines fruit skin colour in sweet cherry (Prunus avium L.). Plant Biotechnol. J. 2016, 14, 2120–2133. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Chen, X.; Zong, X.; Shu, H.; Gao, D. Comparative Transcriptome Analysis of Genes Involved in Anthocyanin Biosynthesis in the Red and Yellow Fruits of Sweet Cherry (Prunus avium L.). PLoS ONE 2015, 10, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Carey, C.C.; Strahle, J.T.; Selinger, D.A.; Chandler, V.L. Mutations in the pale aleurone color1 Regulatory Gene of the Zea mays Anthocyanin Pathway Have Distinct Phenotypes Relative to the Functionally SimiLAR TRANSPARENT TESTA GLABRA1 Gene in Arabidopsis thaliana. Plant Cell 2004, 16, 450–464. [Google Scholar] [CrossRef] [PubMed]
- Lin-wang, K.; Bolitho, K.; Grafton, K.; Kortstee, A.; Karunairetnam, S.; Mcghie, T.K.; Espley, R.V.; Hellens, R.P.; Allan, A.C. An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol. 2010, 10, 50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aharoni, A.; De Vos, C.H.R.; Wein, M.; Sun, Z.; Greco, R.; Kroon, A.; Mol, J.N.M.; O’Connell, A.P. The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J. 2001, 28, 319–332. [Google Scholar] [CrossRef] [PubMed]
- Salvatierra, A.; Pimentel, P.; Moya-León, M.A.; Herrera, R. Increased accumulation of anthocyanins in Fragaria chiloensis fruits by transient suppression of FcMYB1 gene. Phytochemistry 2013, 90, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Lin-Wang, K.; Micheletti, D.; Palmer, J.; Volz, R.; Lozano, L.; Espley, R.; Hellens, R.P.; Chagne, D.; Rowan, D.D.; Troggio, M.; et al. High temperature reduces apple fruit colour via modulation of the anthocyanin regulatory complex. Plant. Cell Environ. 2011, 34, 1176–1190. [Google Scholar] [CrossRef] [PubMed]
- Dubos, C.; Le Gourrierec, J.; Baudry, A.; Huep, G.; Lanet, E.; Debeaujon, I.; Routaboul, J.-M.; Alboresi, A.; Weisshaar, B.; Lepiniec, L. MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J. 2008, 55, 940–953. [Google Scholar] [CrossRef] [PubMed]
- Miao, L.; Zhang, Y.; Yang, X.; Xiao, J.; Zhang, H.; Zhang, Z.; Wang, Y.; Jiang, G. Colored light-quality selective plastic films affect anthocyanin content, enzyme activities, and the expression of flavonoid genes in strawberry (Fragaria × ananassa) fruit. Food Chem. 2016, 207, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Rabino, I.; Mancinelli, A.L. Light, temperature, and anthocyanin production. Plant Physiol. 1986, 81, 922–924. [Google Scholar] [CrossRef] [PubMed]
- Takos, A.M.; Jaffe, F.W.; Jacob, S.R.; Bogs, J.; Robinson, S.P.; Walker, A.R. Light-Induced Expression of a MYB Gene Regulates Anthocyanin Biosynthesis in Red Apples. Plant Physiol. 2006, 142, 1216–1232. [Google Scholar] [CrossRef] [PubMed]
- Medina-Puche, L.; Cumplido-Laso, G.; Amil-Ruiz, F.; Hoffmann, T.; Ring, L.; Rodríguez-Franco, A.; Caballero, J.L.; Schwab, W.; Muñoz-Blanco, J.; Blanco-Portales, R. MYB10 plays a major role in the regulation of flavonoid/phenylpropanoid metabolism during ripening of Fragaria × ananassa fruits. J. Exp. Bot. 2014, 65, 401–417. [Google Scholar] [CrossRef] [PubMed]
- Lin-Wang, K.; McGhie, T.K.; Wang, M.; Liu, Y.; Warren, B.; Storey, R.; Espley, R.V.; Allan, A.C. Engineering the anthocyanin regulatory complex of strawberry (Fragaria vesca). Front. Plant Sci. 2014, 5, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, W.; Dou, Y.; Zhang, J.; Jiang, G.; Miao, L.; Han, G.; Liu, Y.; Li, H.; Zhang, Z. Transcript Quantification by RNA-Seq Reveals Differentially Expressed Genes in the Red and Yellow Fruits of Fragaria vesca. PLoS ONE 2015, 10, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Fischer, T.C.; Mirbeth, B.; Rentsch, J.; Sutter, C.; Ring, L.; Flachowsky, H.; Habegger, R.; Hoffmann, T.; Hanke, M.-V.; Schwab, W. Premature and ectopic anthocyanin formation by silencing of anthocyanidin reductase in strawberry (Fragaria × ananassa). New Phytol. 2014, 201, 440–451. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; C Allan, A.; Yi, Q.; Chen, L.; Li, K.; Shu, Q.; Su, J. Differential Gene Expression Analysis of Yunnan Red Pear, Pyrus Pyrifolia, During Fruit Skin Coloration. Plant Mol. Biol. Report. 2011, 29, 305–314. [Google Scholar] [CrossRef]
- Lister, C.E.; Lancaster, J.E.; Walker, J.R.L. Developmental Changes in Enzymes of Flavonoid Biosynthesis in the Skins of Red and Green Apple Cultivars. J. Sci. Food Agric. 1996, 71, 313–320. [Google Scholar] [CrossRef]
- Zhang, H.; Yang, B.; Liu, J.; Guo, D.; Hou, J.; Chen, S. Analysis of structural genes and key transcription factors related to anthocyanin biosynthesis in potato tubers. Sci. Hortic. 2017, 225, 310–316. [Google Scholar] [CrossRef]
- Zhang, L.; Xu, B.; Wu, T.; Yang, Y.; Fan, L.; Wen, M.; Sui, J. Transcriptomic profiling of two Pak Choi varieties with contrasting anthocyanin contents provides an insight into structural and regulatory genes in anthocyanin biosynthetic pathway. BMC Genom. 2017, 18, 288. [Google Scholar] [CrossRef] [PubMed]
- Guo, N.; Cheng, F.; Wu, J.; Liu, B.; Zheng, S.; Liang, J.; Wang, X. Anthocyanin biosynthetic genes in Brassica rapa. BMC Genom. 2014, 15, 426. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Amil-ruiz, F.; Garrido-Gala, J.; Blanco-portales, R.; Folta, K.M.; Munoz-Blanco, J.; Caballero, J.L. Identification and Validation of Reference Genes for Transcript Normalization in Strawberry (Fragaria × ananassa) Defense Responses. PLoS ONE 2013, 8, e70603. [Google Scholar] [CrossRef] [PubMed]
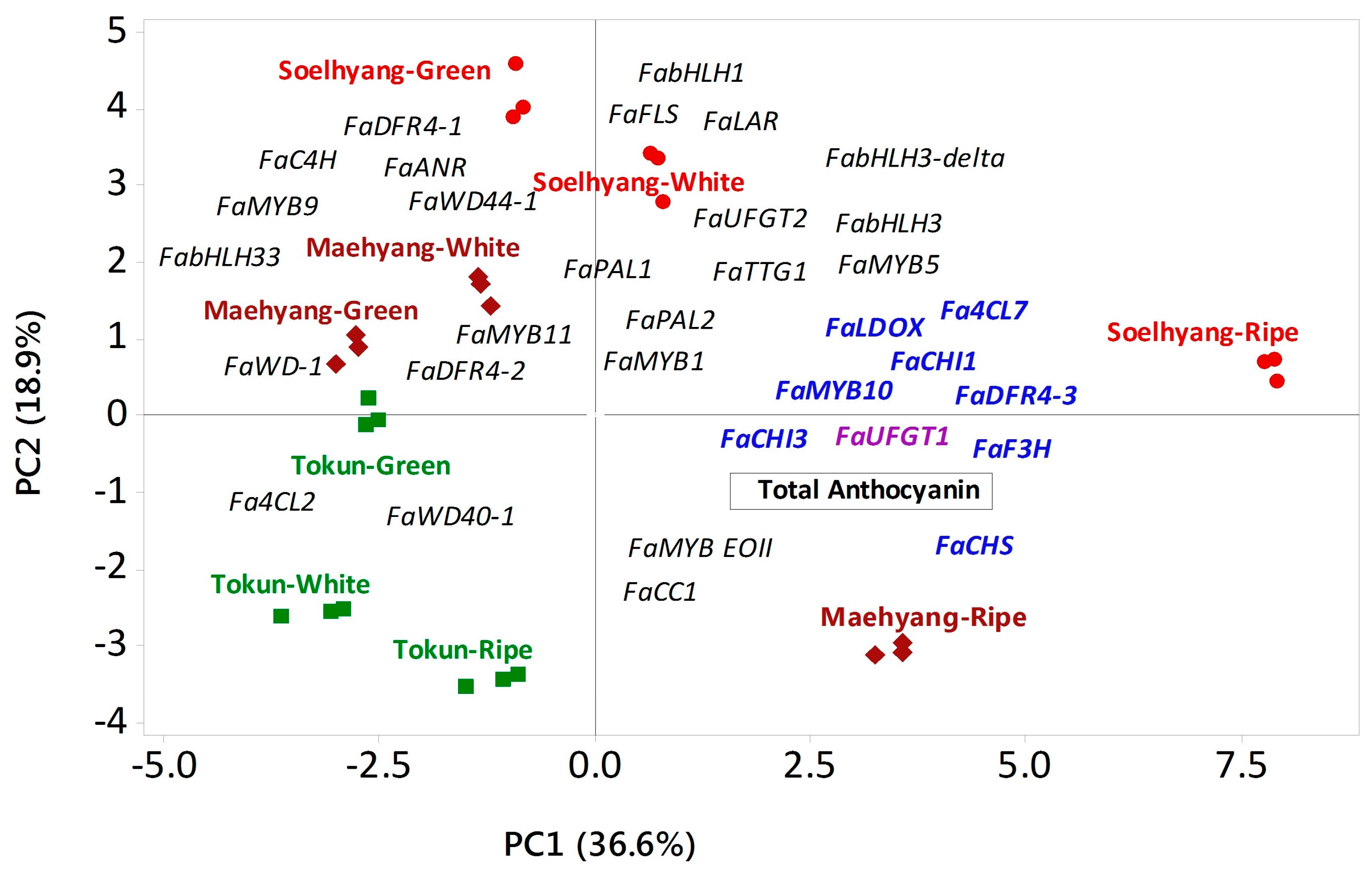
| Variable | PC1 | PC2 | PC3 | |
|---|---|---|---|---|
| Regulatory Genes | ||||
| MYB | FaMYB11 | −0.080 | 0.128 | −0.115 |
| FaMYB1 | 0.132 | −0.020 | 0.139 | |
| FaMYB10 | 0.260 | −0.094 | 0.044 | |
| FaMYB5 | 0.205 | 0.146 | 0.131 | |
| FaMYB9 | −0.012 | 0.253 | −0.271 | |
| FaEOBII | 0.011 | −0.216 | 0.151 | |
| bHLH | FabHLH33 | −0.173 | 0.142 | −0.043 |
| FabHLH3-delta | 0.128 | 0.192 | 0.232 | |
| FabHLH3 | 0.148 | 0.170 | 0.238 | |
| FabHLH1 | 0.018 | 0.271 | 0.267 | |
| WD40 | FaWD40-1 | −0.104 | −0.110 | 0.166 |
| FaWD-1 | −0.033 | 0.068 | 0.239 | |
| FaTTG1 | 0.108 | 0.154 | 0.285 | |
| FaWD44-1 | −0.176 | 0.198 | −0.066 | |
| Early Biosynthetic Genes | ||||
| Phenylalanine | FaPAL1 | 0.064 | 0.171 | −0.264 |
| FaPAL2 | 0.189 | 0.078 | −0.254 | |
| Cinnamic acid | FaC4H | −0.047 | 0.228 | −0.113 |
| FaCC1 | 0.143 | −0.059 | 0.026 | |
| Coumaroyl CoA | Fa4CL7 | 0.268 | −0.094 | 0.036 |
| Fa4CL2 | −0.204 | −0.116 | −0.104 | |
| Chalcone | FaCHI3 | 0.266 | 0.051 | 0.022 |
| FaCHI1 | 0.254 | −0.014 | −0.121 | |
| FaCHS | 0.256 | −0.100 | −0.074 | |
| Flavanone | FaFLS | 0.018 | 0.273 | 0.134 |
| FaF3H | 0.258 | 0.028 | −0.137 | |
| Late Biosynthetic Genes | ||||
| Dihydroflavonol | FaDFR4-1 | −0.035 | 0.294 | 0.222 |
| FaDFR4-2 | −0.136 | 0.014 | 0.216 | |
| FaDFR4-3 | 0.264 | 0.042 | −0.076 | |
| Leucoanthocyanidin | FaLDOX | 0.266 | 0.059 | 0.037 |
| FaANR | −0.049 | 0.233 | −0.265 | |
| FaLAR | 0.074 | 0.300 | −0.157 | |
| Anthocyanidin | FaUFGT1 | 0.260 | −0.015 | 0.126 |
| FaUFGT2 | 0.082 | 0.245 | −0.192 | |
| Total anthocyanin | 0.229 | −0.133 | −0.110 | |
| Eigenvalue | 12.809 | 6.630 | 6.280 | |
| % variation explained | 36.6 | 18.9 | 17.9 | |
| P (Genotype × Fruit developmental stage) | <0.001 | <0.001 | <0.001 | |
| Genotype × Fruit Developmental Stage | Mean PC Scores ± SD | |||
| Tokun | Green | −2.75 ± 0.01 g | −0.06 ± 0.01 d | −2.08±0.01 e |
| White | −3.15 ± 0.37 g | −2.48 ± 0.02 e | 1.87±0.10 b | |
| Ripe | −1.23 ± 0.14 e | −3.51 ± 0.08 f | −0.45±0.12 cd | |
| Maehyang | Green | −2.64 ± 0.17 fg | 0.55 ± 0.26 d | 0.04±0.13 c |
| White | −2.18 ± 0.30 f | 1.54 ± 0.02 c | 1.11±0.16 b | |
| Ripe | 3.57 ± 0.06 b | −3.44 ± 0.04 f | −1.10±0.02 de | |
| Soelhyang | Green | −0.66 ± 0.19 d | 3.94 ± 0.55 a | −4.26±0.41 f |
| White | 0.89 ± 0.13 c | 2.88 ± 0.30 b | 5.01±0.67 a | |
| Ripe | 8.14 ± 0.02 a | 0.588 ± 0.36 d | −0.14±0.64 cd | |
| Gene | Gene ID in Fragaria × ananassa (FANhybrid_r1.2_cds) | CDS (bp) | Annotation | Best Match of F. vesca Gene | e-Value |
|---|---|---|---|---|---|
| Regulatory Genes | |||||
| MYB | |||||
| FaMYB11 | FANhyb_icon00023020_a.1.g00001.1/partial | 425 | MYB11.m1; organism = Fragaria × ananassa | gene07416 | 0.00 |
| FaMYB1 | FANhyb_icon00014430_a.1.g00001.1/partial | 272 | MYB1.m1; organism = Fragaria × ananassa | gene09407 | 1 × 10−153 |
| FaMYB10 | FANhyb_icon00002569_a.1.g00001.1/partial | 233 | Transcription factor MYB113 (AtMYB113) (simiLAR to) | gene31413 | 1 × 10−119 |
| FaMYB5 | FANhyb_rscf00000101.1.g00008.1 | 1071 | MYB5.m1; organism = Fragaria × ananassa | gene24821 | 0.00 |
| FaMYB9 | FANhyb_rscf00002302.1.g00001.1/partial | 828 | MYB9.m1; organism = Fragaria × ananassa | gene15392 | 0.00 |
| FaEOBII | FANhyb_rscf00000047.1.g00023.1/TE | 393 | MYB-related protein 305 (putative) | gene28435 | 0.00 |
| bHLH | |||||
| FabHLH33 | FANhyb_rscf00000043.1.g00015.1 | 1962 | BHLH33.m1; organism = Fragaria × ananassa | gene19321 | 0.00 |
| FabHLH3-delta | FANhyb_icon00003421_a.1.g00001.1/partial | 557 | Transcription factor TT8 (bHLH 42) (simiLAR to) | gene27827 | 0.00 |
| FabHLH3 | FANhyb_rscf00003752.1.g00002.1 | 1041 | Transcription factor TT8 (bHLH 42) (simiLAR to) | gene27827 | 0.00 |
| FabHLH1 | FANhyb_icon00000044_a.1.g00001.1 | 1999 | Transcription factor GLABRA 3 (bHLH 1) (putative); FaMYC1 mRNA | gene32494 | 0.00 |
| WD40 | |||||
| FaWD40-1 | FANhyb_rscf00000569.1.g00002.1 | 1539 | WD40 repeat-containing protein SMU1 (putative) | gene27104 | 0.00 |
| FaWD-1 | FANhyb_icon00020056_a.1.g00001.1/partial | 152 | WD repeat-containing protein mio (probable) | gene03735 | 3 × 10−71 |
| FaTTG1 | FANhyb_icon00009619_a.1.g00001.1/partial | 367 | Protein transparent Testa Glabra 1 (simiLAR to); TTG1 | gene12450 | 0.00 |
| FAWD44-1 | FANhyb_rscf00002089.1.g00001.1 | 2067 | Fragaria vesca WD repeat-containing protein 44 | gene17869 | 0.00 |
| Early Biosynthetic Genes | |||||
| Phenylalanine | |||||
| FaPAL1 | FANhyb_rscf00000868.1.g00006.1 | 1674 | Phenylalanine ammonia-lyase 1 | gene23261 | 0.00 |
| FaPAL2 | FANhyb_rscf00000079.1.g00001.1 | 2175 | Phenylalanine ammonia-lyase 2 (putative) | gene09753 | 0.00 |
| Cinnamic Acid | |||||
| FaC4H | FANhyb_rscf00000282.1.g00007.1 | 789 | Trans-cinnamate 4-monooxygenase (CA4H) (putative) | gene28093 | 0.00 |
| FaCC1 | FANhyb_rscf00000685.1.g00004.1 | 6963 | Biotin carboxylase (probable); Acetyl-CoA carboxylase, AtACC1 | gene22077 | 0.00 |
| Coumaroyl-CoA | |||||
| Fa4CL7 | FANhyb_icon00012602_a.1.g00001.1/partial | 352 | 4-coumarate-CoA ligase-like 7 (At4CL6) (putative) | gene09603 | 0.00 |
| Fa4CL2 | FANhyb_rscf00001339.1.g00001.1 | 1644 | 4-coumarate-CoA ligase 2 (4CL 2) (putative) | gene15877 | 0.00 |
| Chalcone | |||||
| FaCHI3 | FANhyb_icon00000880_a.1.g00001.1/partial | 638 | Chalcone-flavonone isomerase (probable) | gene21346 | 0.00 |
| FaCHI1 | FANhyb_icon00004487_a.1.g00001.1/partial | 534 | Chalcone-flavonone isomerase 1; TT5 | gene23367 | 0.00 |
| FaCHS | FANhyb_icon00003433_a.1.g00001.1/partial | 992 | Chalcone synthase, TT4; FvCHS | gene26825 | 0.00 |
| Flavanone | |||||
| FaFLS | FANhyb_icon00020196_a.1.g00001.1/partial | 242 | Flavonol synthase/flavanone 3-hydroxylase (FLS) (simiLAR to) | gene11126 | 1 × 10−111 |
| FaF3H | FANhyb_icon00001777_a.1.g00001.1 | 825 | Naringenin,2-oxoglutarate 3-dioxygenase (F3H) (putative) | gene14611 | 0.00 |
| Late Biosynthetic Genes | |||||
| Dihydroflavonol | |||||
| FaDFR4-1 | FANhyb_rscf00006583.1.g00001.1/partial | 578 | Bifunctional dihydroflavonol 4-reductase (DFR) (probable) | gene29482 | 1 × 10−143 |
| FaDFR4-2 | FANhyb_rscf00000482.1.g00004.1 | 2031 | Bifunctional dihydroflavonol 4-reductase (DFR) (putative) | gene15174 | 0.00 |
| FaDFR4-3 | FANhyb_rscf00000482.1.g00003.1 | 1050 | Bifunctional dihydroflavonol 4-reductase (DFR) (simiLAR to) | gene15176 | 0.00 |
| Leucoanthocyanidin | |||||
| FaLDOX | FANhyb_icon00007826_a.1.g00001.1/partial | 794 | Leucoanthocyanidin dioxygenase (LDOX) (putative); FvLDOX, TT18 | gene32347 | 0.00 |
| FaANR | FANhyb_rscf00000390.1.g00012.1/partial | 887 | Leucoanthocyanidin reductase (LAR) (putative) | gene24665 | 0.00 |
| FaLAR | FANhyb_icon00014507_a.1.g00001.1/partial | 497 | Leucoanthocyanidin reductase (simiLAR to) | gene03877 | 0.00 |
| Anthocyanidin | |||||
| FaUFGT1 | FANhyb_rscf00000061.1.g00006.1 | 1347 | Anthocyanidin 3-O-glucosyltransferase 2 (putative) | gene12591 | 0.00 |
| FaUFGT2 | FANhyb_rscf00000877.1.g00006.1 | 1299 | Anthocyanidin 5,3-O-glucosyltransferase (probable) | gene04355 | 0.00 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hossain, M.R.; Kim, H.-T.; Shanmugam, A.; Nath, U.K.; Goswami, G.; Song, J.-Y.; Park, J.-I.; Nou, I.-S. Expression Profiling of Regulatory and Biosynthetic Genes in Contrastingly Anthocyanin Rich Strawberry (Fragaria × ananassa) Cultivars Reveals Key Genetic Determinants of Fruit Color. Int. J. Mol. Sci. 2018, 19, 656. https://doi.org/10.3390/ijms19030656
Hossain MR, Kim H-T, Shanmugam A, Nath UK, Goswami G, Song J-Y, Park J-I, Nou I-S. Expression Profiling of Regulatory and Biosynthetic Genes in Contrastingly Anthocyanin Rich Strawberry (Fragaria × ananassa) Cultivars Reveals Key Genetic Determinants of Fruit Color. International Journal of Molecular Sciences. 2018; 19(3):656. https://doi.org/10.3390/ijms19030656
Chicago/Turabian StyleHossain, Mohammad Rashed, Hoy-Taek Kim, Ashokraj Shanmugam, Ujjal Kumar Nath, Gayatri Goswami, Jae-Young Song, Jong-In Park, and Ill-Sup Nou. 2018. "Expression Profiling of Regulatory and Biosynthetic Genes in Contrastingly Anthocyanin Rich Strawberry (Fragaria × ananassa) Cultivars Reveals Key Genetic Determinants of Fruit Color" International Journal of Molecular Sciences 19, no. 3: 656. https://doi.org/10.3390/ijms19030656




