Biochemical and MALDI-TOF Mass Spectrometric Characterization of a Novel Native and Recombinant Cystine Knot Miniprotein from Solanum tuberosum subsp. andigenum cv. Churqueña
Abstract
:1. Introduction
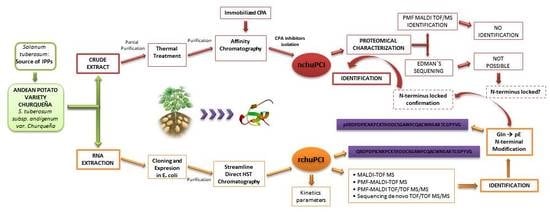
2. Results and Discussion
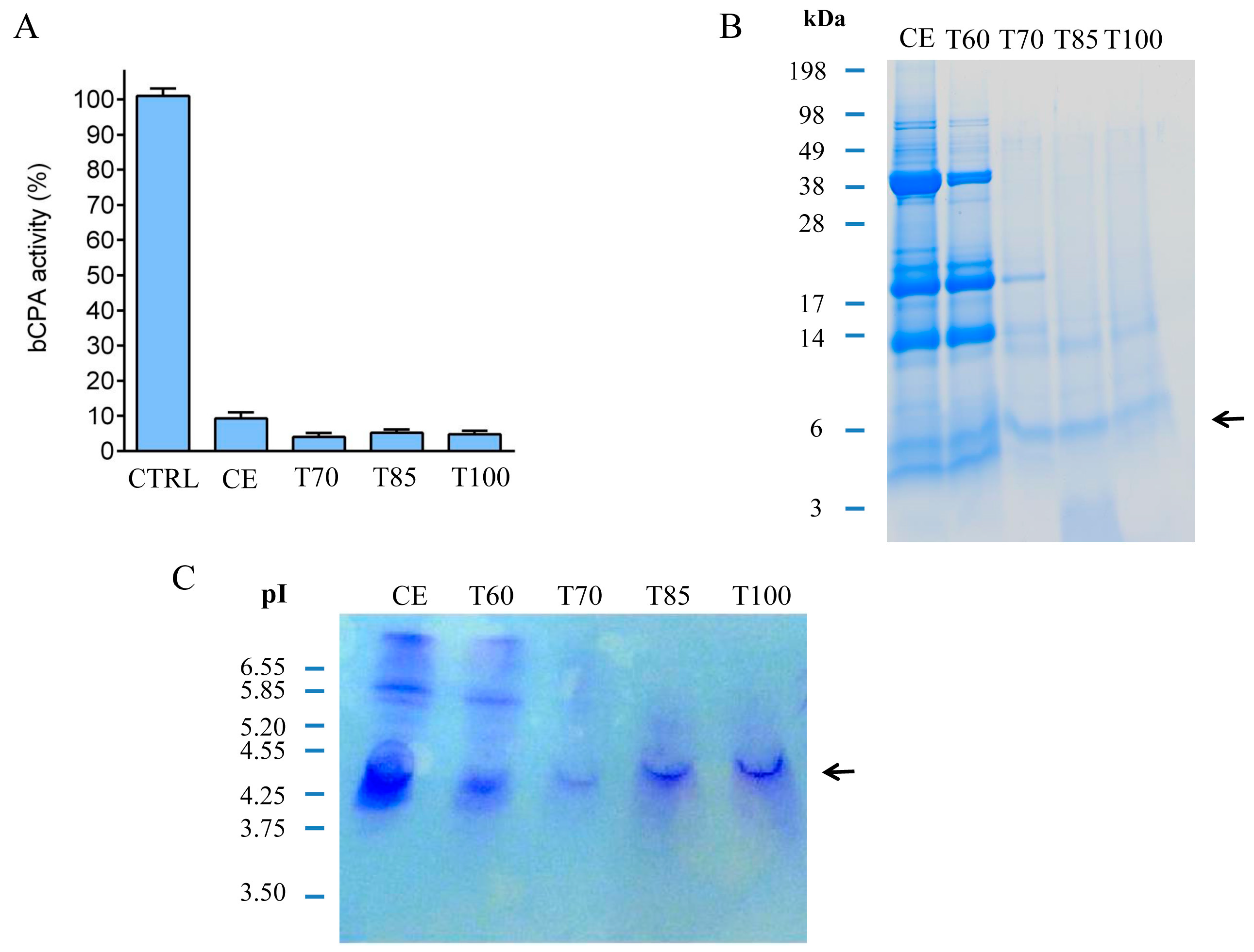
2.1. Identification and Initial Characterization of a Native Metallocarboxypeptidase Inhibitor from S. tuberosum subsp. andigenum cv. Churqueña
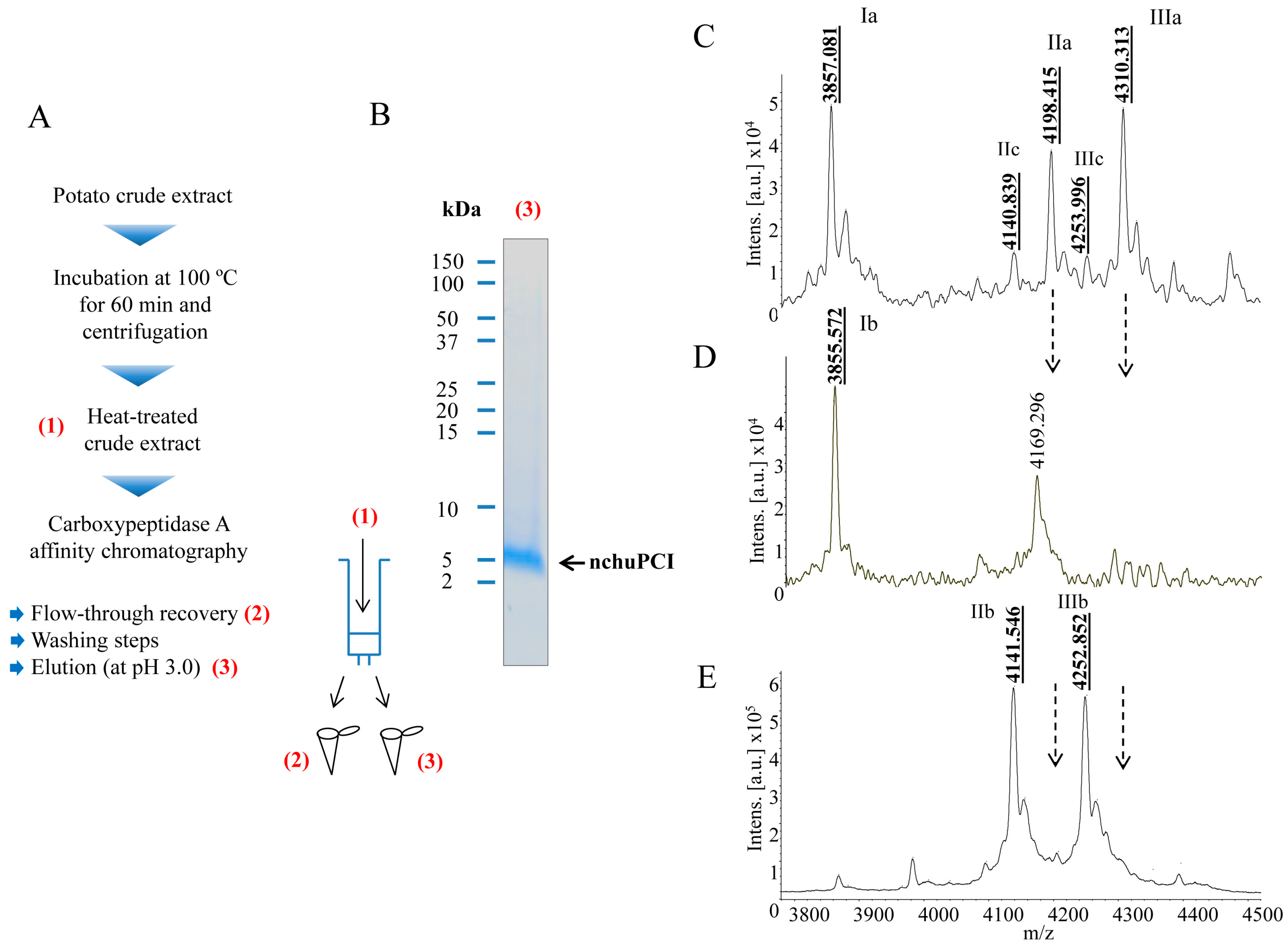
2.2. Purification and MALDI-TOF Mass Spectrometric Analysis of the Native Carboxypeptidase Inhibitor
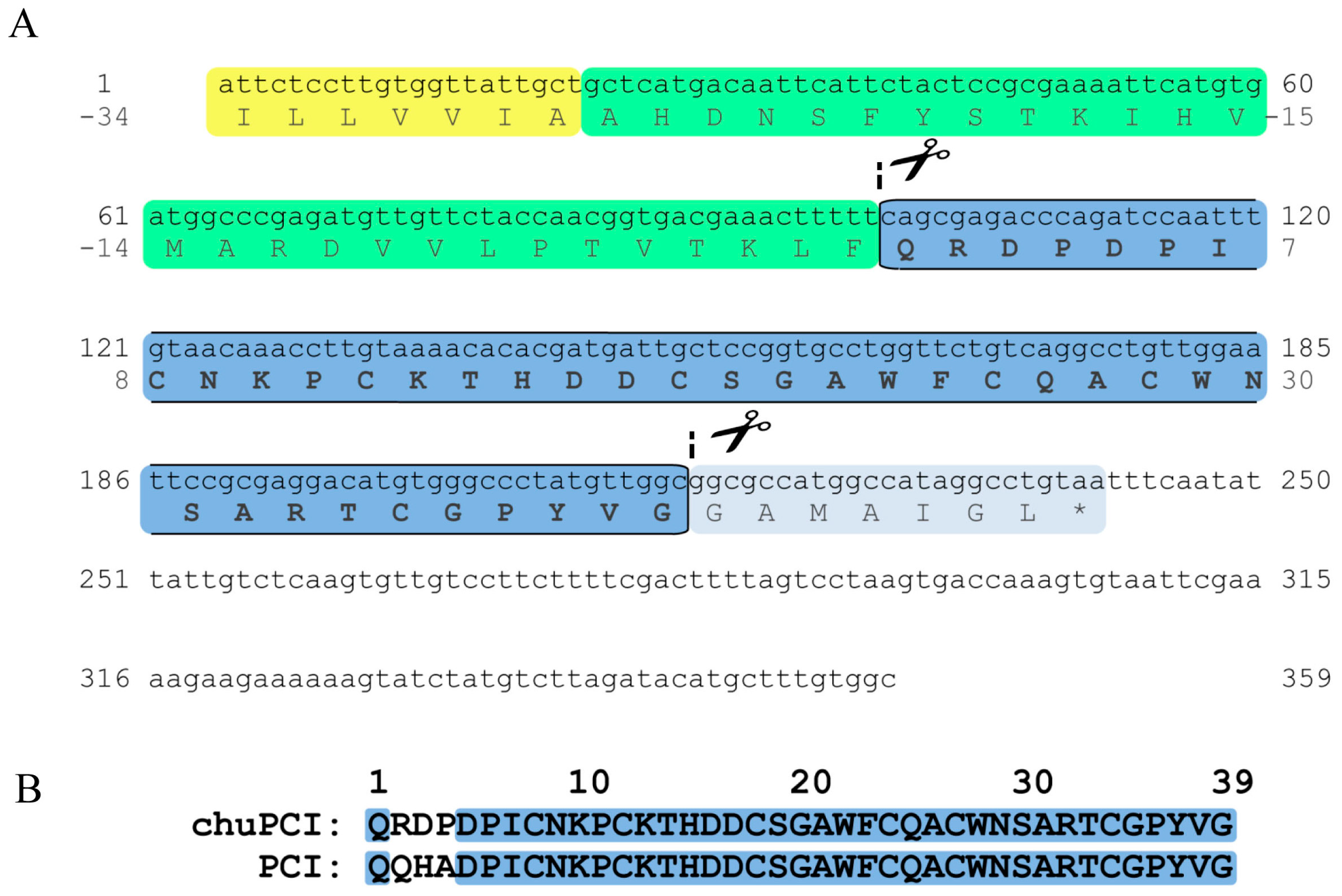
2.3. Cloning and Sequence Analysis of the Novel Cystine-Knot Metallocarboxypeptidase Inhibitor (chuPCI)
2.4. Expression, Purification, and MALDI-TOF Mass Spectrometry Analysis of rchuPCI
2.5. Determination of the Inhibition Kinetic Constants (Ki) against A/B-Type MCPs
2.6. Identification and De Novo Sequencing of the Native chuPCI
3. Materials and Methods
3.1. Materials and Reagents
3.2. Crude Extract Preparation
3.3. Total Protein Quantitation
3.4. Carboxypeptidase A Inhibition Measurements and IC50 Determination
3.5. Thermal Stability Experiments and nchuPCI Purification by Affinity Chromatography
3.5.1. Thermal Stability Experiments
3.5.2. Affinity Chromatography Purification
3.6. Cloning and Expression of rchuPCI
3.6.1. Cloning and Sequence Analysis of chuPCI
3.6.2. Expression and Purification of rchuPCI
3.7. General Characterization of Methods
3.7.1. Tris-Tricine-SDS-PAGE
3.7.2. Isoelectrofocusing (IEF)
3.7.3. MALDI-TOF Mass Spectrometric Analysis
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| CKMPs | Cystine-knot miniproteins |
| MCPIs | Metallocarboxypeptidase inhibitors |
| chuPCI | Potato carboxypeptidase inhibitor isolated from churqueña variety |
| rchuPCI | Recombinant chuPCI |
| MALDI-TOF/MS | Matrix-assisted laser desorption/ionization-time of flight mass spectrometry |
| PMF | Peptide mass fingerprint |
| MCPs | Metallocarboxypeptidases |
| PPIs | Protease peptide inhibitors |
| PCI | Potato carboxypeptidase inhibitor |
| ACI | Ascaris suum carboxypeptidase inhibitor |
| LCI | Hirudo medicinalis carboxypeptidase inhibitor |
| TCI | Ticks carboxypeptidase inhibitor |
| H1TCI | Hemaphysalis longicornis carboxypeptidase inhibitor |
| NvCI | Nerita versicolor carboxypeptidase inhibitor |
| ECI | Endogenous carboxypeptidase inhibitor |
| SmCI | Sabellastarte magnifica carboxypeptidase inhibitor |
| CPA | Carboxypeptidase A |
| CPB | Carboxypeptidase B |
| SDS | Sodium dodecyl sulfate |
| βME | β-Mercaptoethanol |
| TEMED | N,N,N′,N′-Tetramethyl ethylene diamine |
| BSA | Bovine serum albumin |
| PCR | Polymerase chain reaction |
References
- Kolmar, H. Biological diversity and therapeutic potential of natural and engineered cystine knot miniproteins. Curr. Opin. Pharmacol. 2009, 9, 608–614. [Google Scholar] [CrossRef] [PubMed]
- Kolmar, H. Alternative binding proteins: Biological activity and therapeutic potential of cystine-knot miniproteins. FEBS J. 2008, 275, 2684–2690. [Google Scholar] [CrossRef] [PubMed]
- Colgrave, M.L.; Craik, D.J. Thermal, chemical, and enzymatic stability of the cyclotide kalata B1: The importance of the cyclic cystine knot. Biochemistry 2004, 43, 5965–5975. [Google Scholar] [CrossRef] [PubMed]
- Mas, J.M.; Aloy, P.; Marti-Renom, M.A.; Oliva, B.; Blanco-Aparicio, C.; Molina, M.A.; de Llorens, R.; Querol, E.; Aviles, F.X. Protein similarities beyond disulphide bridge topology. J. Mol. Biol. 1998, 284, 541–548. [Google Scholar] [CrossRef] [PubMed]
- Cavallini, C.; Trettene, M.; Degan, M.; Delva, P.; Molesini, B.; Minuz, P.; Pandolfini, T. Anti-angiogenic effects of two cystine-knot miniproteins from tomato fruit. Br. J. Pharmacol. 2011, 162, 1261–1273. [Google Scholar] [CrossRef] [PubMed]
- Vitt, U.A.; Hsu, S.Y.; Hsueh, A.J. Evolution and classification of cystine knot-containing hormones and related extracellular signaling molecules. Mol. Endocrinol. 2001, 15, 681–694. [Google Scholar] [CrossRef] [PubMed]
- Norton, R.S.; Pallaghy, P.K. The cystine knot structure of ion channel toxins and related polypeptides. Toxicon 1998, 36, 1573–1583. [Google Scholar] [CrossRef]
- Quilis, J.; Meynard, D.; Vila, L.; Aviles, F.X.; Guiderdoni, E.; San Segundo, B. A potato carboxypeptidase inhibitor gene provides pathogen resistance in transgenic rice. Plant Biotechnol. J. 2007, 5, 537–553. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.A.; Day, M.; Heavner, J.E. Ziconotide: An update and review. Expert Opin. Pharmacother. 2008, 9, 1575–1583. [Google Scholar] [CrossRef] [PubMed]
- Pimentel, C.; Choi, S.J.; Chagot, B.; Guette, C.; Camadro, J.M.; Darbon, H. Solution structure of PcFK1, a spider peptide active against plasmodium falciparum. Protein Sci. 2006, 15, 628–634. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.J.; Parent, R.; Guillaume, C.; Deregnaucourt, C.; Delarbre, C.; Ojcius, D.M.; Montagne, J.J.; Celerier, M.L.; Phelipot, A.; Amiche, M.; et al. Isolation and characterization of Psalmopeotoxin I and II: Two novel antimalarial peptides from the venom of the tarantula psalmopoeus cambridgei. FEBS Lett. 2004, 572, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Shao, F.; Hu, Z.; Xiong, Y.M.; Huang, Q.Z.; Wang, C.G.; Zhu, R.H.; Wang, D.C. A new antifungal peptide from the seeds of phytolacca americana: Characterization, amino acid sequence and cDNA cloning. Biochim. Biophys. Acta 1999, 1430, 262–268. [Google Scholar] [CrossRef]
- Tam, J.P.; Lu, Y.A.; Yang, J.L.; Chiu, K.W. An unusual structural motif of antimicrobial peptides containing end-to-end macrocycle and cystine-knot disulfides. Proc. Natl. Acad. Sci. USA 1999, 96, 8913–8918. [Google Scholar] [CrossRef] [PubMed]
- Osaki, T.; Omotezako, M.; Nagayama, R.; Hirata, M.; Iwanaga, S.; Kasahara, J.; Hattori, J.; Ito, I.; Sugiyama, H.; Kawabata, S. Horseshoe crab hemocyte-derived antimicrobial polypeptides, tachystatins, with sequence similarity to spider neurotoxins. J. Biol. Chem. 1999, 274, 26172–26178. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Pardo, J.; Tanco, S.; Diaz, L.; Dasgupta, S.; Fernandez-Recio, J.; Lorenzo, J.; Aviles, F.X.; Fricker, L.D. Substrate specificity of human metallocarboxypeptidase D: Comparison of the two active carboxypeptidase domains. PLoS ONE 2017, 12, e0187778. [Google Scholar] [CrossRef] [PubMed]
- Arolas, J.L.; Vendrell, J.; Aviles, F.X.; Fricker, L.D. Metallocarboxypeptidases: Emerging drug targets in biomedicine. Curr. Pharm. Des. 2007, 13, 349–366. [Google Scholar] [CrossRef] [PubMed]
- Rees, D.C.; Lipscomb, W.N. Refined crystal structure of the potato inhibitor complex of carboxypeptidase A at 2.5 a resolution. J. Mol. Biol. 1982, 160, 475–498. [Google Scholar] [CrossRef]
- Hass, G.M.; Hermodson, M.A. Amino acid sequence of a carboxypeptidase inhibitor from tomato fruit. Biochemistry 1981, 20, 2256–2260. [Google Scholar] [CrossRef] [PubMed]
- Hass, G.M.; Nau, H.; Biemann, K.; Grahn, D.T.; Ericsson, L.H.; Neurath, H. The amino acid sequence of a carboxypeptidase inhibitor from potatoes. Biochemistry 1975, 14, 1334–1342. [Google Scholar] [CrossRef] [PubMed]
- Lufrano, D.; Cotabarren, J.; Garcia-Pardo, J.; Fernandez-Alvarez, R.; Tort, O.; Tanco, S.; Aviles, F.X.; Lorenzo, J.; Obregon, W.D. Biochemical characterization of a novel carboxypeptidase inhibitor from a variety of andean potatoes. Phytochemistry 2015, 120, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Homandberg, G.A.; Litwiller, R.D.; Peanasky, R.J. Carboxypeptidase inhibitors from ascaris suum: The primary structure. Arch. Biochem. Biophys. 1989, 270, 153–161. [Google Scholar] [CrossRef]
- Homandberg, G.A.; Peanasky, R.J. Characterization of proteins from ascaris lumbricoides which bind specifically to carboxypeptidase. J. Biol. Chem. 1976, 251, 2226–2233. [Google Scholar] [PubMed]
- Reverter, D.; Vendrell, J.; Canals, F.; Horstmann, J.; Aviles, F.X.; Fritz, H.; Sommerhoff, C.P. A carboxypeptidase inhibitor from the medical leech hirudo medicinalis. Isolation, sequence analysis, cdna cloning, recombinant expression, and characterization. J. Biol. Chem. 1998, 273, 32927–32933. [Google Scholar] [CrossRef] [PubMed]
- Arolas, J.L.; Lorenzo, J.; Rovira, A.; Castella, J.; Aviles, F.X.; Sommerhoff, C.P. A carboxypeptidase inhibitor from the tick rhipicephalus bursa: Isolation, cdna cloning, recombinant expression, and characterization. J. Biol. Chem. 2005, 280, 3441–3448. [Google Scholar] [CrossRef] [PubMed]
- Gong, H.; Zhou, J.; Liao, M.; Hatta, T.; Harnnoi, T.; Umemiya, R.; Inoue, N.; Xuan, X.; Fujisaki, K. Characterization of a carboxypeptidase inhibitor from the tick haemaphysalis longicornis. J. Insect Physiol. 2007, 53, 1079–1087. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Yu, L.; Gao, J.; Fu, Q.; Zhang, J.; Zhang, P.; Chen, J.; Zhao, S. Cloning, tissue expression pattern and genomic organization of latexin, a human homologue of rat carboxypeptidase A inhibitor. Mol. Biol. Rep. 2000, 27, 241–246. [Google Scholar] [CrossRef] [PubMed]
- Alonso-del-Rivero, M.; Trejo, S.A.; Reytor, M.L.; Rodriguez-de-la-Vega, M.; Delfin, J.; Diaz, J.; González-González, Y.; Canals, F.; Chavez, M.A.; Aviles, F.X. Tri-domain bifunctional inhibitor of metallocarboxypeptidases A and serine proteases isolated from marine annelid sabellastarte magnifica. J. Biol. Chem. 2012, 287, 15427–15438. [Google Scholar] [CrossRef] [PubMed]
- Covaleda, G.; del Rivero, M.A.; Chavez, M.A.; Aviles, F.X.; Reverter, D. Crystal structure of novel metallocarboxypeptidase inhibitor from marine mollusk nerita versicolor in complex with human carboxypeptidase A4. J. Biol. Chem. 2012, 287, 9250–9258. [Google Scholar] [CrossRef] [PubMed]
- Cotabarren, J.; Tellechea, M.E.; Aviles, F.X.; Lorenzo Rivera, J.; Obregon, W.D. Biochemical characterization of the ybpci miniprotein, the first carboxypeptidase inhibitor isolated from yellow bell pepper (Capsicum annuum L). A novel contribution to the knowledge of miniproteins stability. Protein Expr. Purif. 2017, 144, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Sanglas, L.; Aviles, F.X.; Huber, R.; Gomis-Ruth, F.X.; Arolas, J.L. Mammalian metallopeptidase inhibition at the defense barrier of ascaris parasite. Proc. Natl. Acad. Sci. USA 2009, 106, 1743–1747. [Google Scholar] [CrossRef] [PubMed]
- Alonso-del-Rivero, M.; Trejo, S.A.; Rodriguez de la Vega, M.; Gonzalez, Y.; Bronsoms, S.; Canals, F.; Delfin, J.; Diaz, J.; Aviles, F.X.; Chavez, M.A. A novel metallocarboxypeptidase-like enzyme from the marine annelid sabellastarte magnifica—A step into the invertebrate world of proteases. FEBS J. 2009, 276, 4875–4890. [Google Scholar] [CrossRef] [PubMed]
- Atanasov, A.G.; Waltenberger, B.; Pferschy-Wenzig, E.M.; Linder, T.; Wawrosch, C.; Uhrin, P.; Temml, V.; Wang, L.; Schwaiger, S.; Heiss, E.H.; et al. Discovery and resupply of pharmacologically active plant-derived natural products: A review. Biotechnol. Adv. 2015, 33, 1582–1614. [Google Scholar] [CrossRef] [PubMed]
- Fischer, M.; Kuckenberg, M.; Kastilan, R.; Muth, J.; Gebhardt, C. Novel in vitro inhibitory functions of potato tuber proteinaceous inhibitors. Mol. Genet. Genom. 2015, 290, 387–398. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Smith, P.L.; Hsu, M.Y.; Ogletree, M.L.; Schumacher, W.A. Murine model of ferric chloride-induced vena cava thrombosis: Evidence for effect of potato carboxypeptidase inhibitor. J. Thromb. Haemost. 2006, 4, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Aparicio, C.; Molina, M.A.; Fernandez-Salas, E.; Frazier, M.L.; Mas, J.M.; Querol, E.; Aviles, F.X.; de Llorens, R. Potato carboxypeptidase inhibitor, a T-knot protein, is an epidermal growth factor antagonist that inhibits tumor cell growth. J. Biol. Chem. 1998, 273, 12370–12377. [Google Scholar] [CrossRef] [PubMed]
- Tellechea, M.; Garcia-Pardo, J.; Cotabarren, J.; Lufrano, D.; Bakas, L.; Aviles, F.; Obregon, W.; Lorenzo, J.; Tanco, S. Microplate assay to study carboxypeptidase A inhibition in andean potatoes. Bio-Protocol 2016, 6, 1–12. [Google Scholar] [CrossRef]
- Obregón, W.D.; Ghiano, N.; Tellechea, M.; Cisneros, J.S.; Lazza, C.M.; López, L.M.I.; Avilés, F.X. Detection and characterisation of a new metallocarboxypeptidase inhibitor from solanum tuberosum cv. Desirèe using proteomic techniques. Food Chem. 2012, 133, 1163–1168. [Google Scholar]
- Villanueva, J.; Canals, F.; Prat, S.; Ludevid, D.; Querol, E.; Aviles, F.X. Characterization of the wound-induced metallocarboxypeptidase inhibitor from potato. Cdna sequence, induction of gene expression, subcellular immunolocalization and potential roles of the c-terminal propeptide. FEBS Lett. 1998, 440, 175–182. [Google Scholar] [CrossRef]
- Hass, G.M.; Ryan, C.A. Cleavage of the carboxypeptidase inhibitor from potatoes by carboxypeptidase A. Biochem. Biophys. Res. Commun. 1980, 97, 1481–1486. [Google Scholar] [CrossRef]
- Marino-Buslje, C.; Venhudova, G.; Molina, M.A.; Oliva, B.; Jorba, X.; Canals, F.; Aviles, F.X.; Querol, E. Contribution of C-tail residues of potato carboxypeptidase inhibitor to the binding to carboxypeptidase A a mutagenesis analysis. Eur. J. Biochem. 2000, 267, 1502–1509. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped blast and psi-blast: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Rawlings, N.D.; Barrett, A.J.; Finn, R. Twenty years of the merops database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2016, 44, D343–D350. [Google Scholar] [CrossRef] [PubMed]
- Gracy, J.; Le-Nguyen, D.; Gelly, J.C.; Kaas, Q.; Heitz, A.; Chiche, L. Knottin: The knottin or inhibitor cystine knot scaffold in 2007. Nucleic Acids Res. 2008, 36, D314–D319. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Kieras, E.; Sousa, E.; D’Antona, A.; Baber, J.C.; He, T.; Desharnais, J.; Wood, L.; Luxenberg, D.; Stahl, M.; et al. Pyroglutamate and O-linked glycan determine functional production of anti-IL17A and anti-IL22 peptide-antibody bispecific genetic fusions. J. Biol. Chem. 2013, 288, 1409–1419. [Google Scholar] [CrossRef] [PubMed]
- Rink, R.; Arkema-Meter, A.; Baudoin, I.; Post, E.; Kuipers, A.; Nelemans, S.A.; Akanbi, M.H.; Moll, G.N. To protect peptide pharmaceuticals against peptidases. J. Pharmacol. Toxicol. Methods 2010, 61, 210–218. [Google Scholar] [CrossRef] [PubMed]
- Obregon, W.D.; Liggieri, C.S.; Morcelle, S.R.; Trejo, S.A.; Aviles, F.X.; Priolo, N.S. Biochemical and PMF MALDI-TOF analyses of two novel papain-like plant proteinases. Protein Pept. Lett. 2009, 16, 1323–1333. [Google Scholar] [CrossRef] [PubMed]
- Obregon, W.D.; Liggieri, C.S.; Trejo, S.A.; Aviles, F.X.; Vairo-Cavalli, S.E.; Priolo, N.S. Characterization of papain-like isoenzymes from latex of asclepias curassavica by molecular biology validated by proteomic approach. Biochimie 2009, 91, 1457–1464. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Mock, W.L.; Liu, Y.; Stanford, D.J. Arazoformyl peptide surrogates as spectrophotometric kinetic assay substrates for Carboxypeptidase A. Anal. Biochem. 1996, 239, 218–222. [Google Scholar] [CrossRef] [PubMed]
- Morrison, J.F. The slow-binding and slow, tight-binding inhibition of enzyme-catalysed reactions. Trends Biochem. Sci. 1982, 7, 102–105. [Google Scholar] [CrossRef]
- Lian, Q.; Szarka, S.J.; Ng, K.K.; Wong, S.L. Engineering of a staphylokinase-based fibrinolytic agent with antithrombotic activity and targeting capability toward thrombin-rich fibrin and plasma clots. J. Biol. Chem. 2003, 278, 26677–26686. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the expasy server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Shägger, H. Tricine-sds-page. Nat. Protoc. 2006, 1, 16–22. [Google Scholar] [CrossRef] [PubMed]


| Enzyme | rPCI | rchuPCI |
|---|---|---|
| Ki (nM) | Ki (nM) | |
| bCPA a | 4.3 ± 0.46 | 10.8 ± 1.55 |
| pCPB b | 10.7 ± 1.17 | 19.8 ± 1.82 |
| Source | Signal/Peak | Sequence | Theor M | Obs M | Error (ppm) |
|---|---|---|---|---|---|
| CE | ND | QRDPDPICNKPCKTHDDCSGAWFCQACWNSARTCGPYVG | 4325.830 | - | - |
| CE | IIIa | pERDPDPICNKPCKTHDDCSGAWFCQACWNSARTCGPYVG | 4308.799 | 4309.313 | 119 |
| CE | IIIc | pERDPDPICNKPCKTHDDCSGAWFCQACWNSARTCGPYV- | 4251.748 | 4252.996 | 293 |
| CE | IIa | -RDPDPICNKPCKTHDDCSGAWFCQACWNSARTCGPYVG | 4197.699 | 4197.415 | −68 |
| CE | IIc | -RDPDPICNKPCKTHDDCSGAWFCQACWNSARTCGPYV- | 4140.647 | 4139.839 | −195 |
| RP | rIIIa | QRDPDPICNKPCKTHDDCSGAWFCQACWNSARTCGPYVG | 4325.830 | 4325.251 | 97 |
| RP | rIV | pERDPDPICNKPCKTHDDCSGAWFCQACWNSARTCGPYVG | 4308.799 | 4307.870 | −216 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cotabarren, J.; Tellechea, M.E.; Tanco, S.M.; Lorenzo, J.; Garcia-Pardo, J.; Avilés, F.X.; Obregón, W.D. Biochemical and MALDI-TOF Mass Spectrometric Characterization of a Novel Native and Recombinant Cystine Knot Miniprotein from Solanum tuberosum subsp. andigenum cv. Churqueña. Int. J. Mol. Sci. 2018, 19, 678. https://doi.org/10.3390/ijms19030678
Cotabarren J, Tellechea ME, Tanco SM, Lorenzo J, Garcia-Pardo J, Avilés FX, Obregón WD. Biochemical and MALDI-TOF Mass Spectrometric Characterization of a Novel Native and Recombinant Cystine Knot Miniprotein from Solanum tuberosum subsp. andigenum cv. Churqueña. International Journal of Molecular Sciences. 2018; 19(3):678. https://doi.org/10.3390/ijms19030678
Chicago/Turabian StyleCotabarren, Juliana, Mariana Edith Tellechea, Sebastián Martín Tanco, Julia Lorenzo, Javier Garcia-Pardo, Francesc Xavier Avilés, and Walter David Obregón. 2018. "Biochemical and MALDI-TOF Mass Spectrometric Characterization of a Novel Native and Recombinant Cystine Knot Miniprotein from Solanum tuberosum subsp. andigenum cv. Churqueña" International Journal of Molecular Sciences 19, no. 3: 678. https://doi.org/10.3390/ijms19030678