The Complex Subtype-Dependent Role of Connexin 43 (GJA1) in Breast Cancer
Abstract
:1. Introduction
2. Results
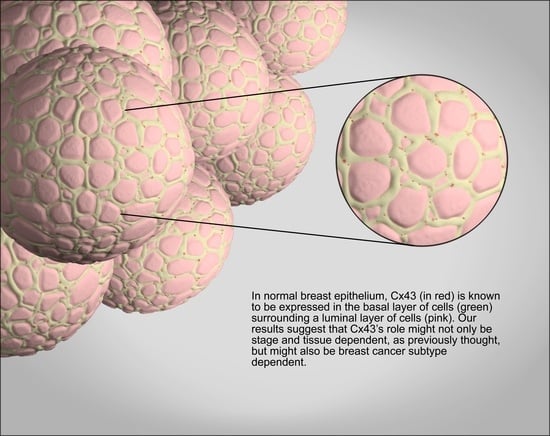
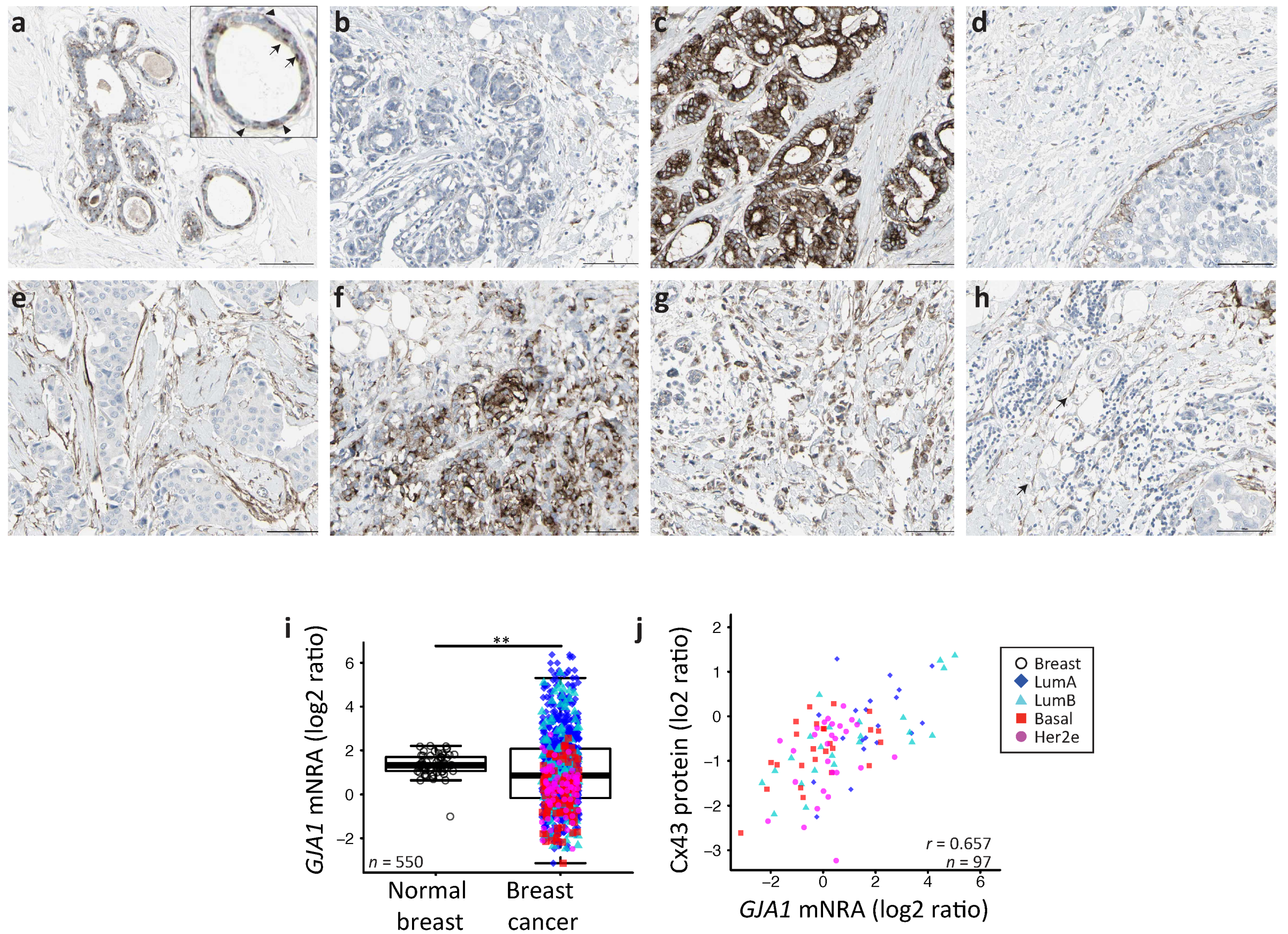
2.1. GJA1 Expression and Localization in the Breast
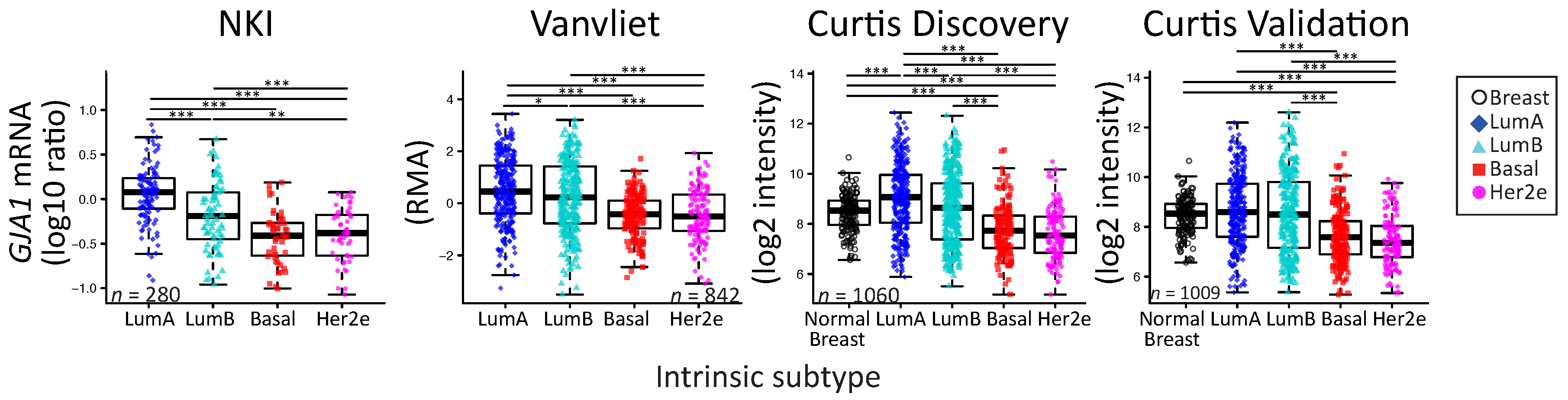
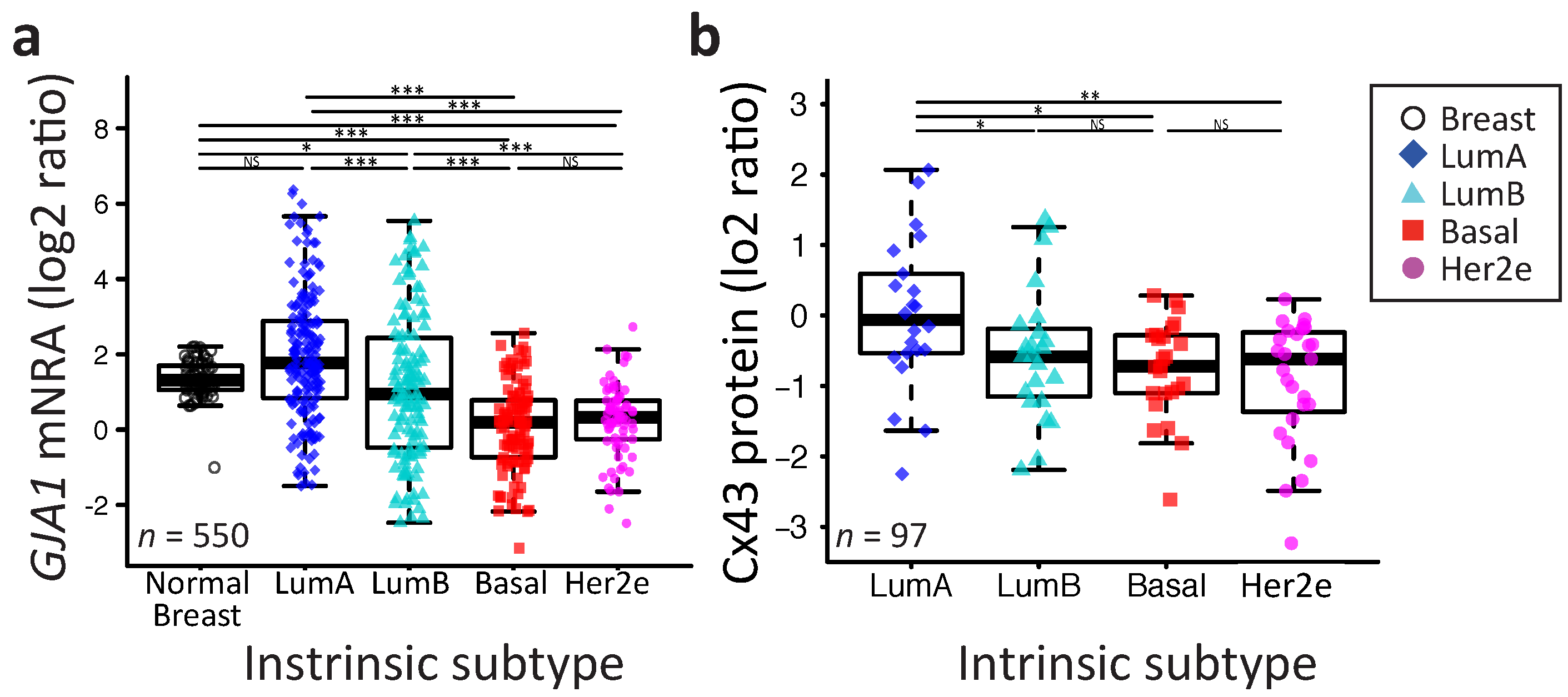
2.2. GJA1 Expression Varies with Breast Cancer Subtype
2.3. Somatic DNA-Level Events of GJA1 Do Not Drive Expression Changes of GJA1 in Breast Cancer
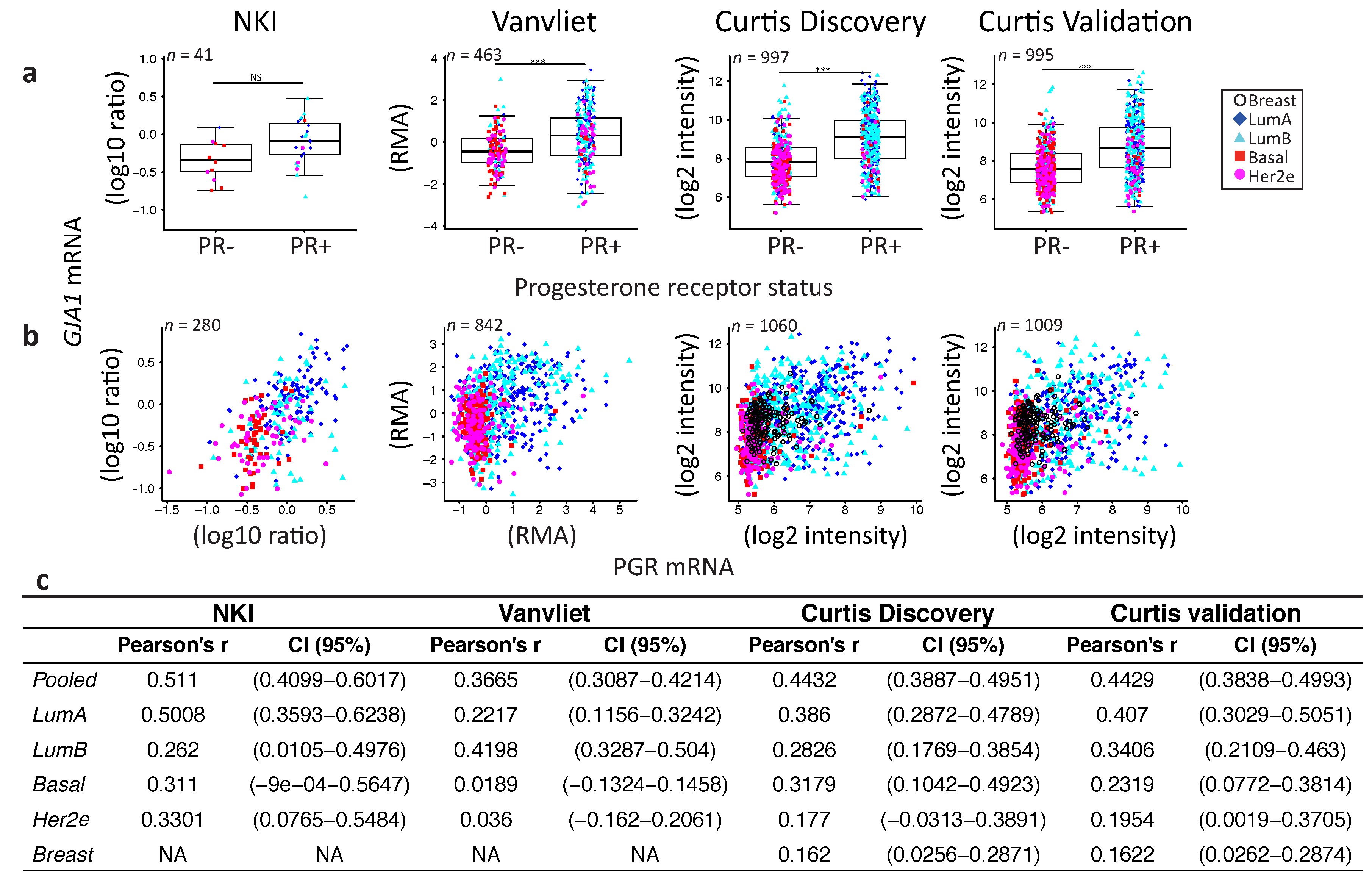
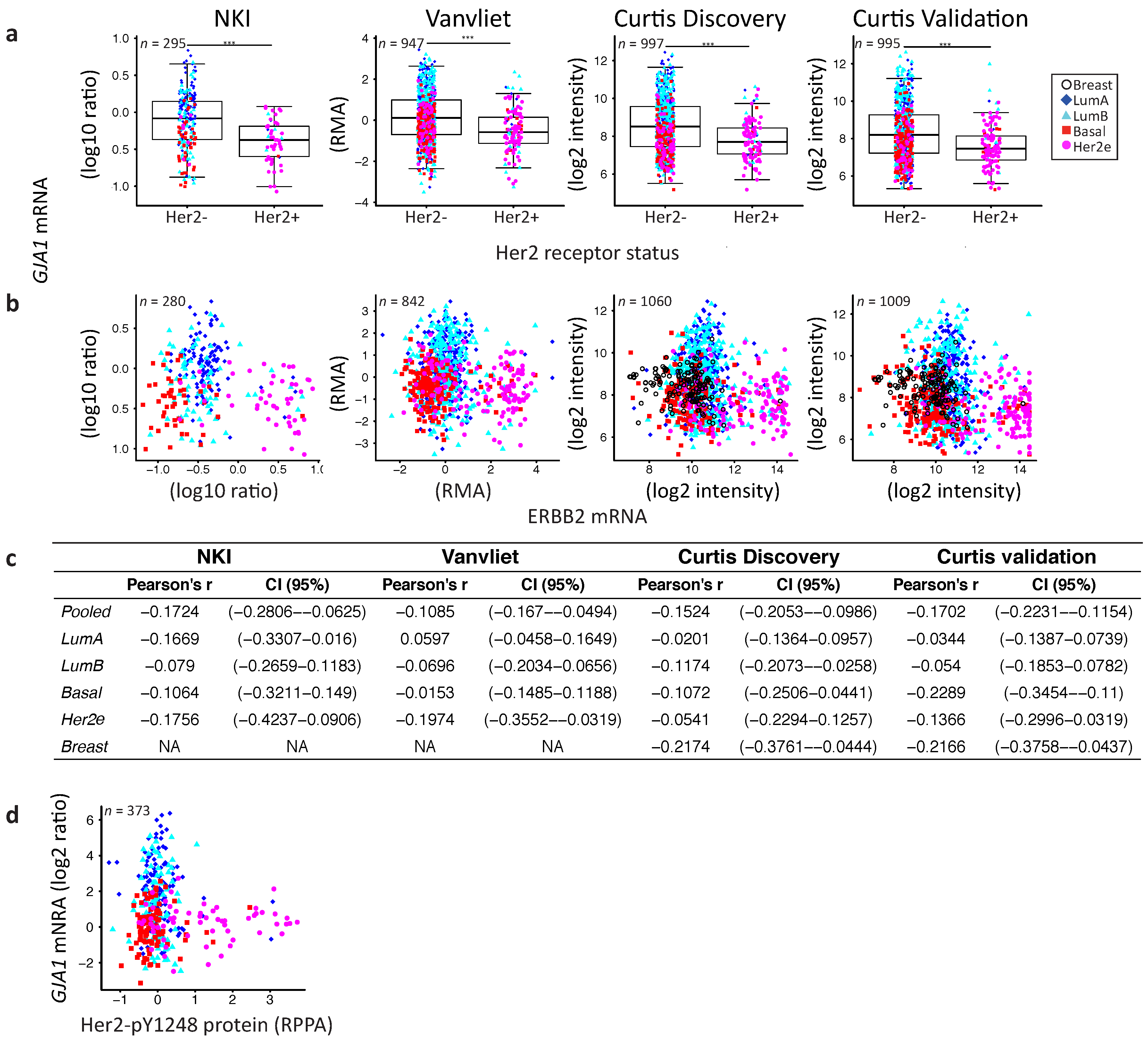
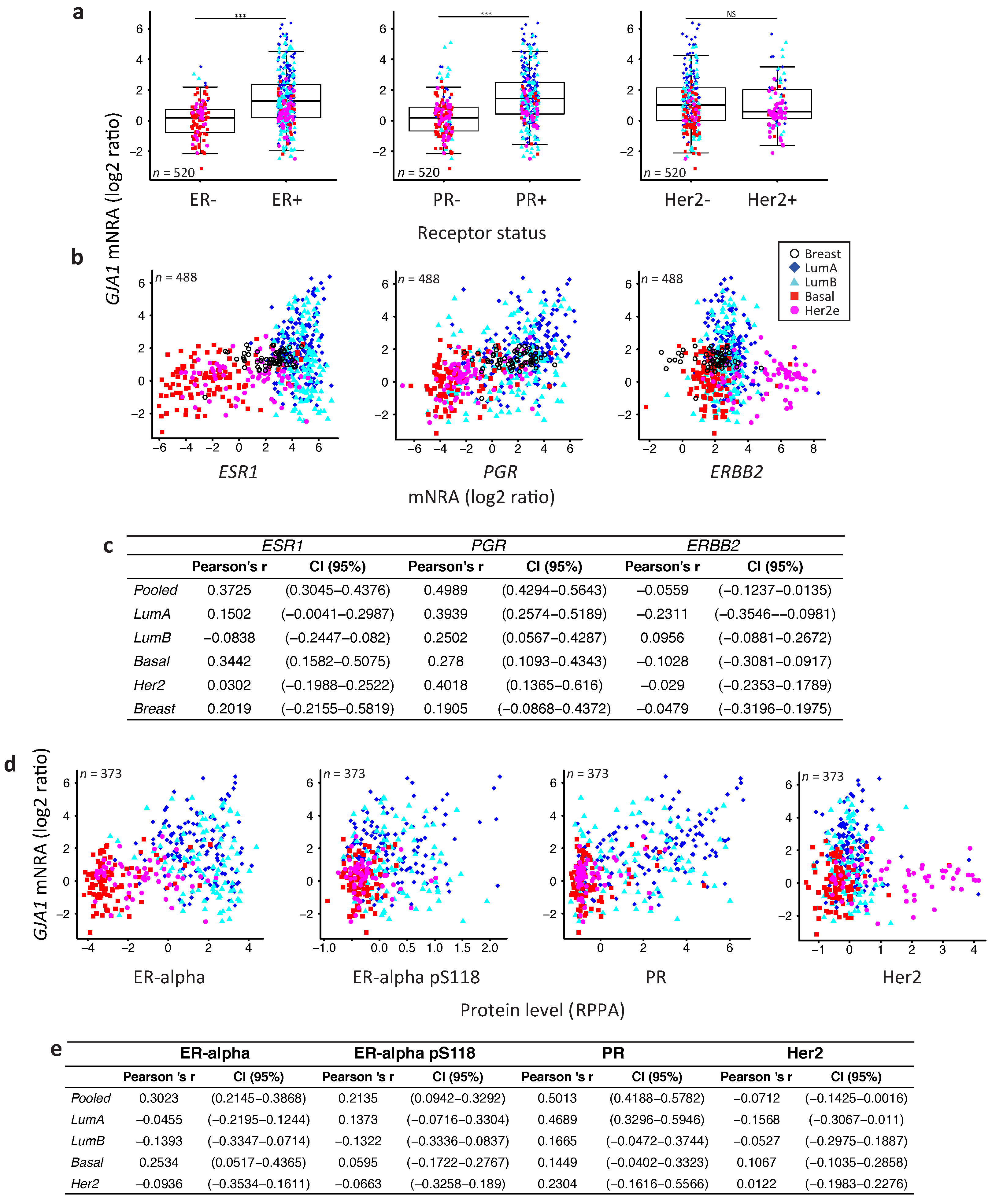
2.4. GJA1 Level Is Dependent on Hormonal Receptor Status
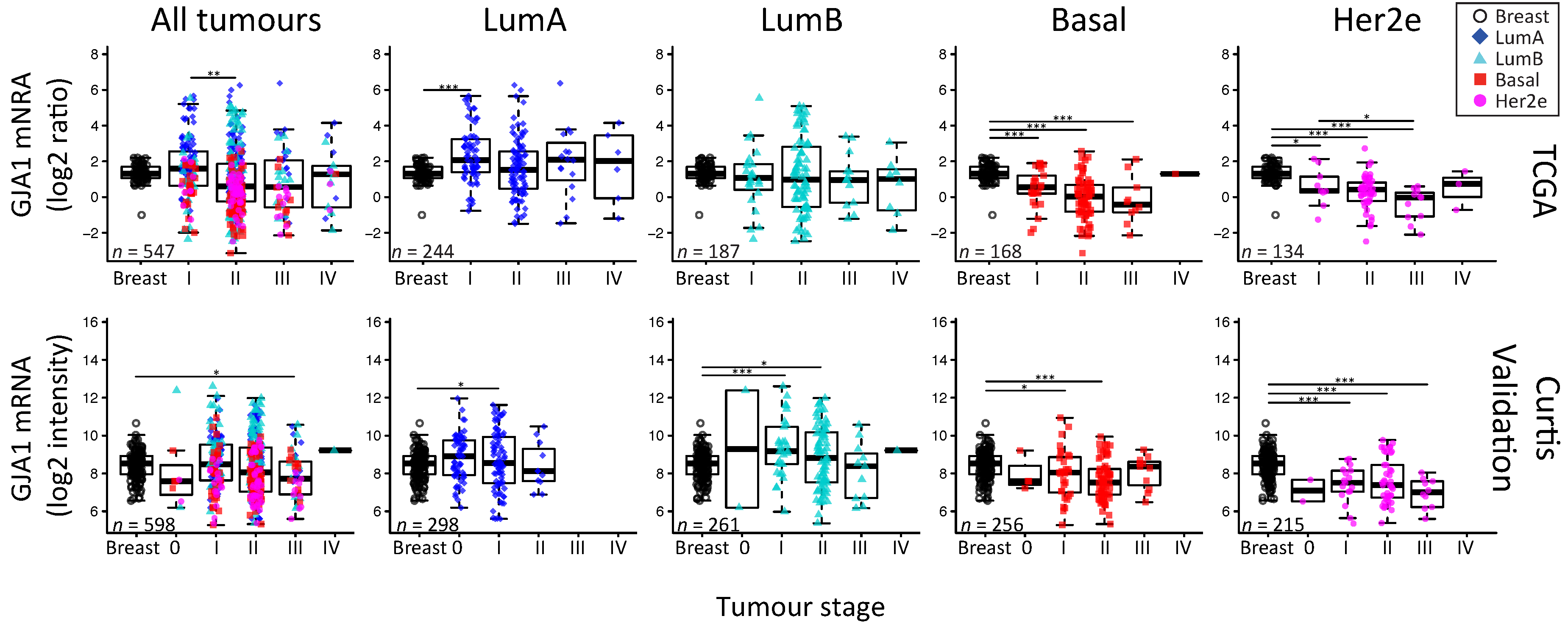
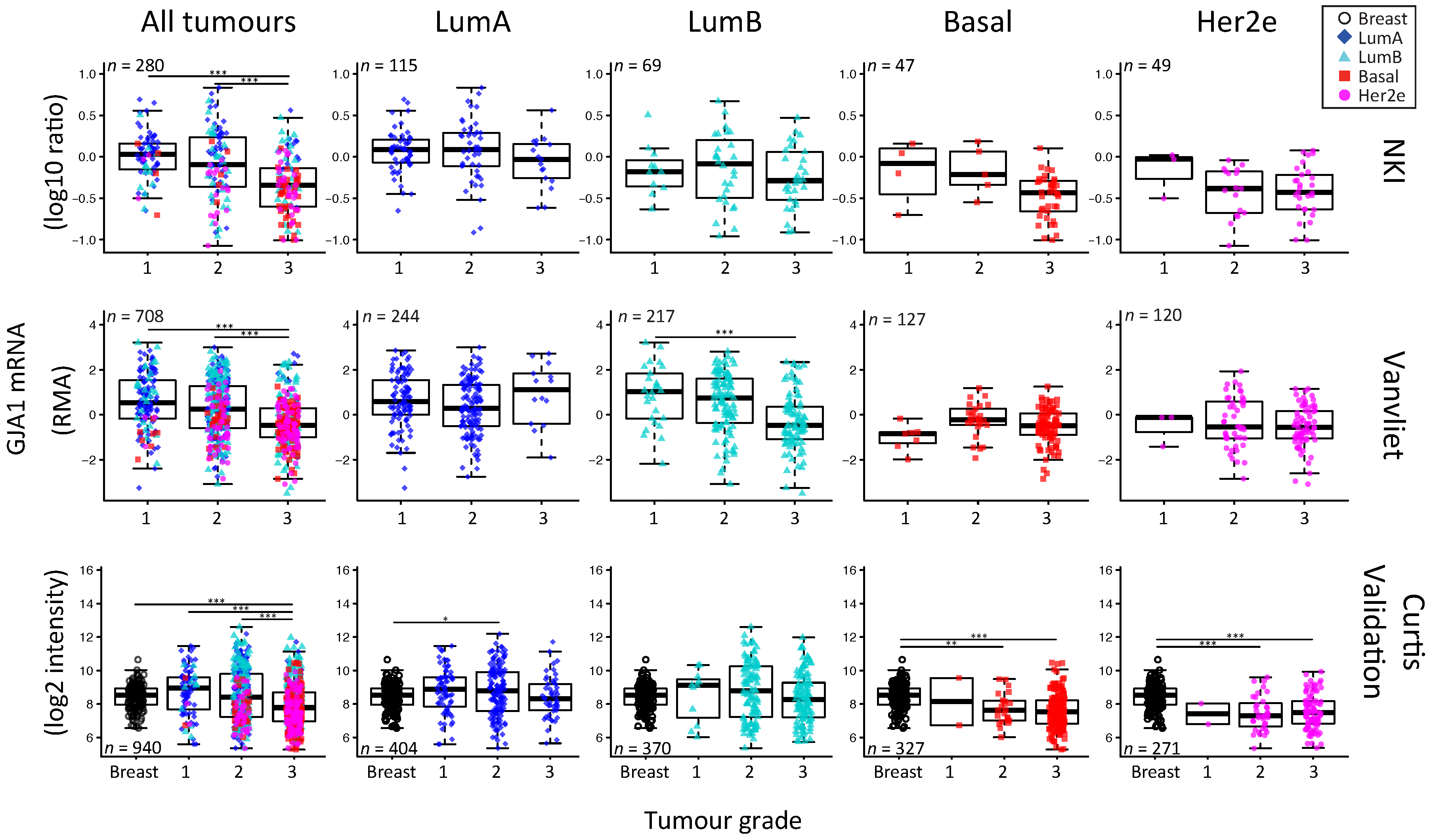
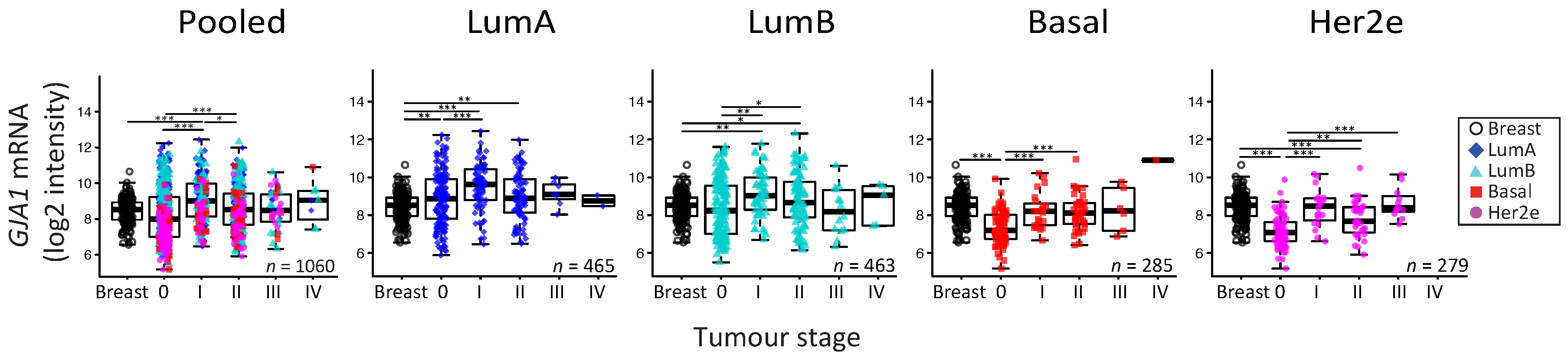
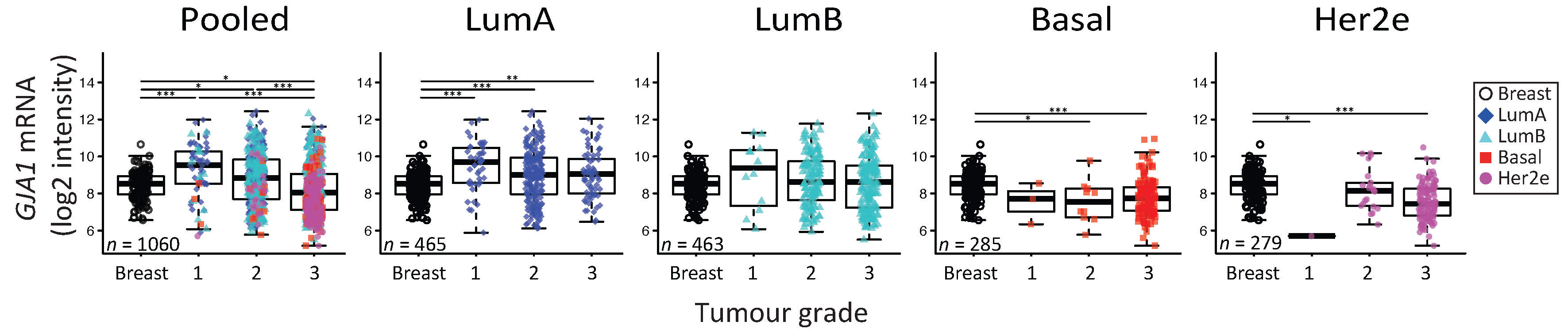
2.5. GJA1 mRNA Is Dysregulated at the Early Stages of Breast Cancer and Is Reduced with Grade When Tumors Are Pooled
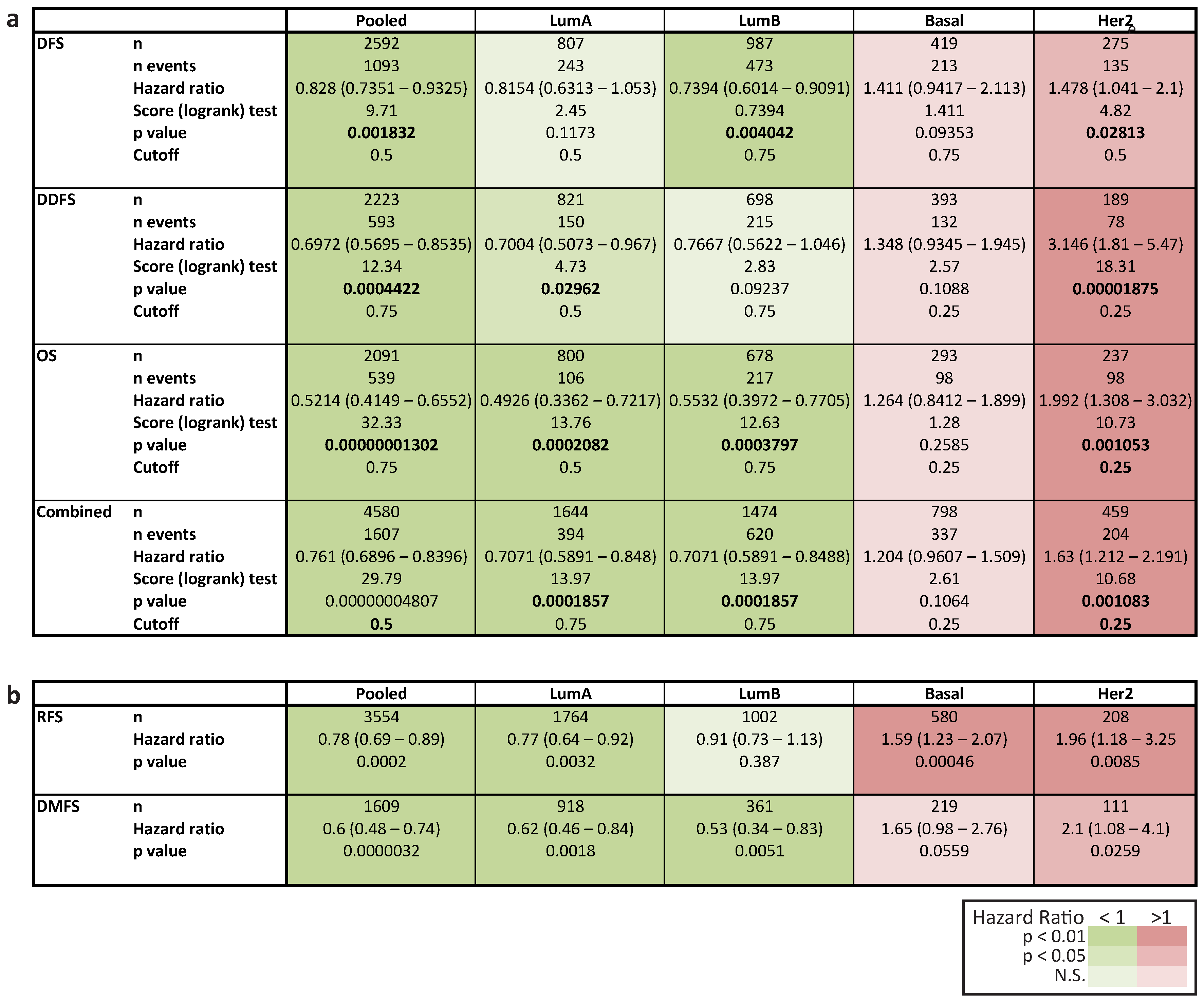
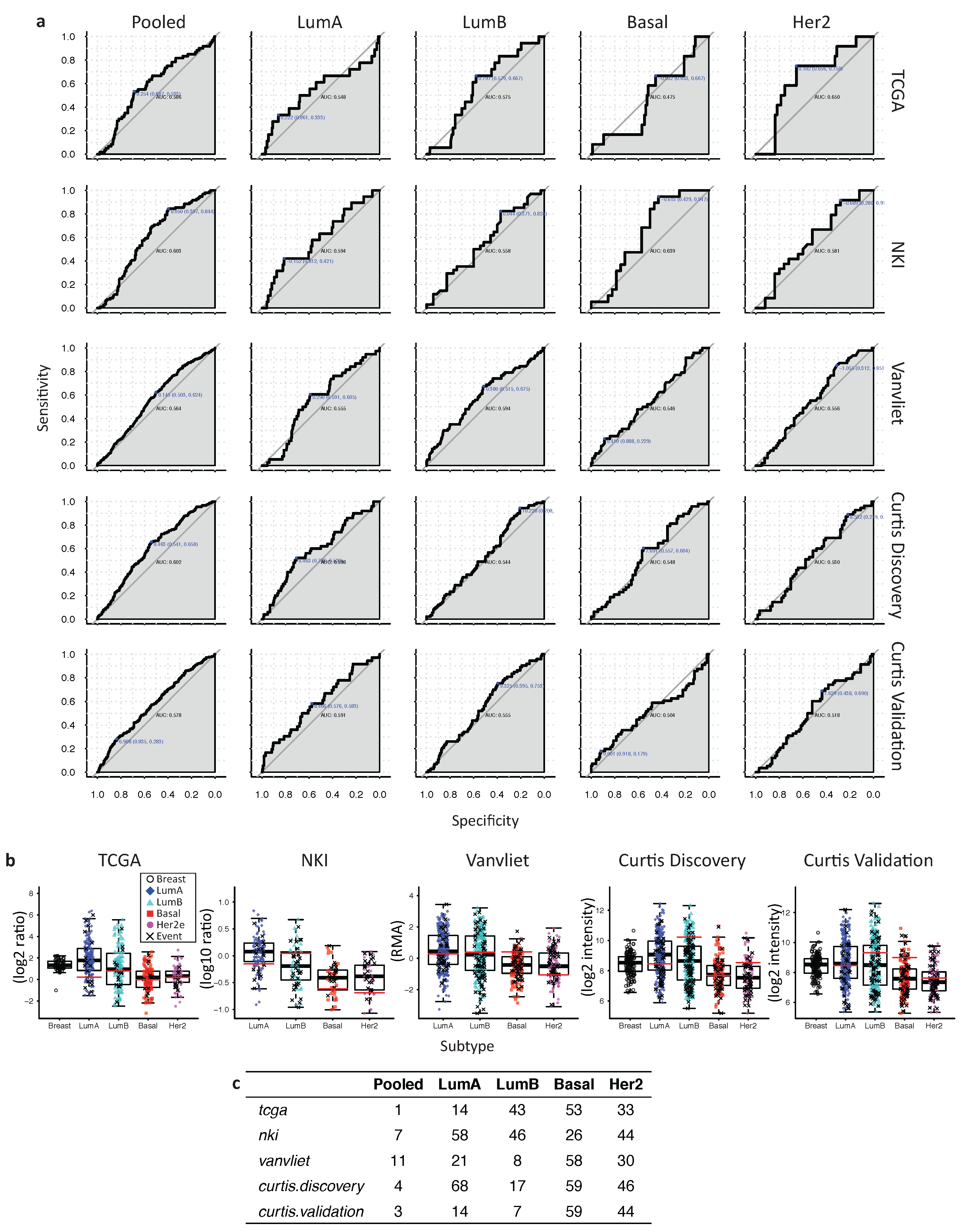
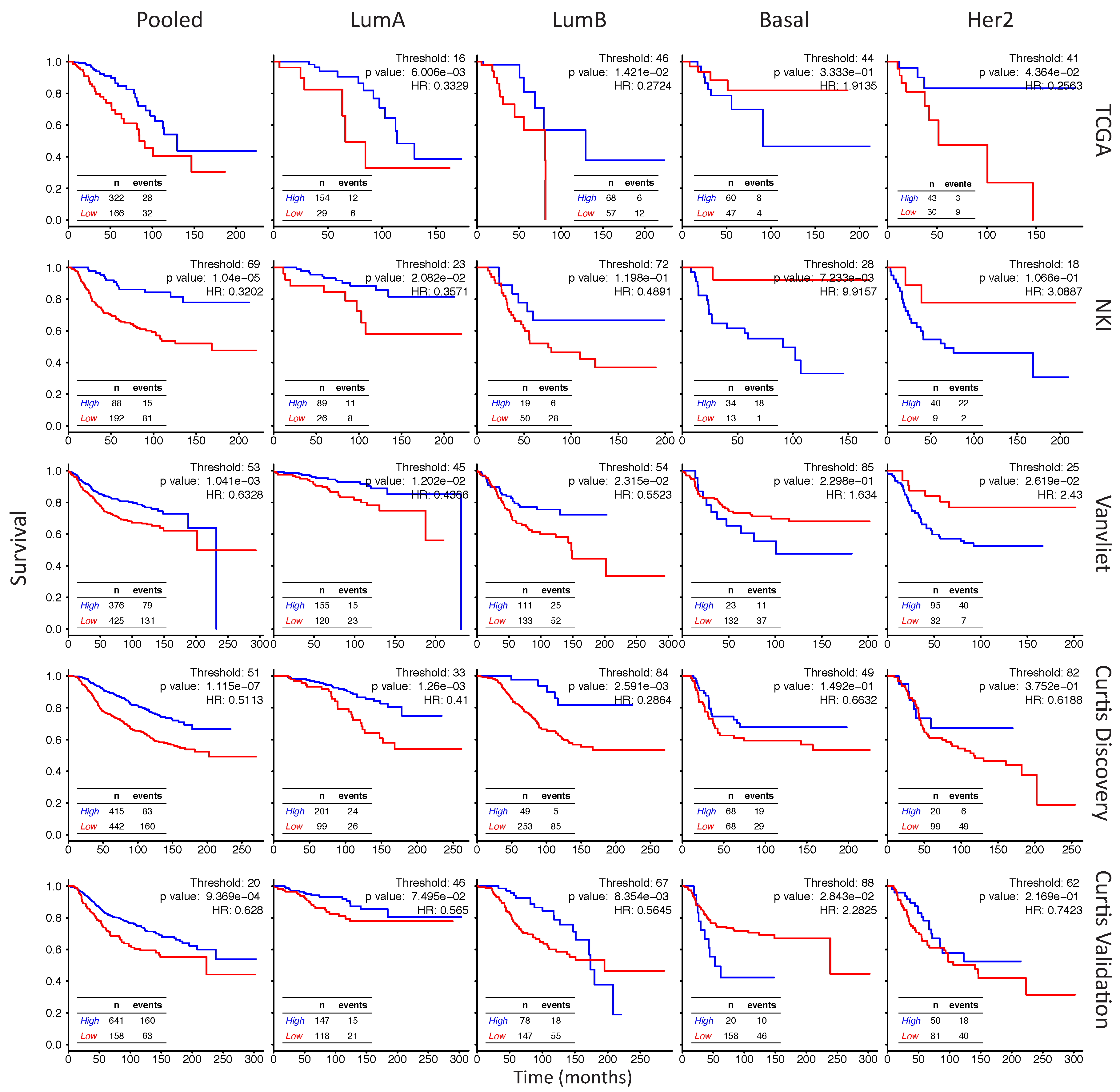
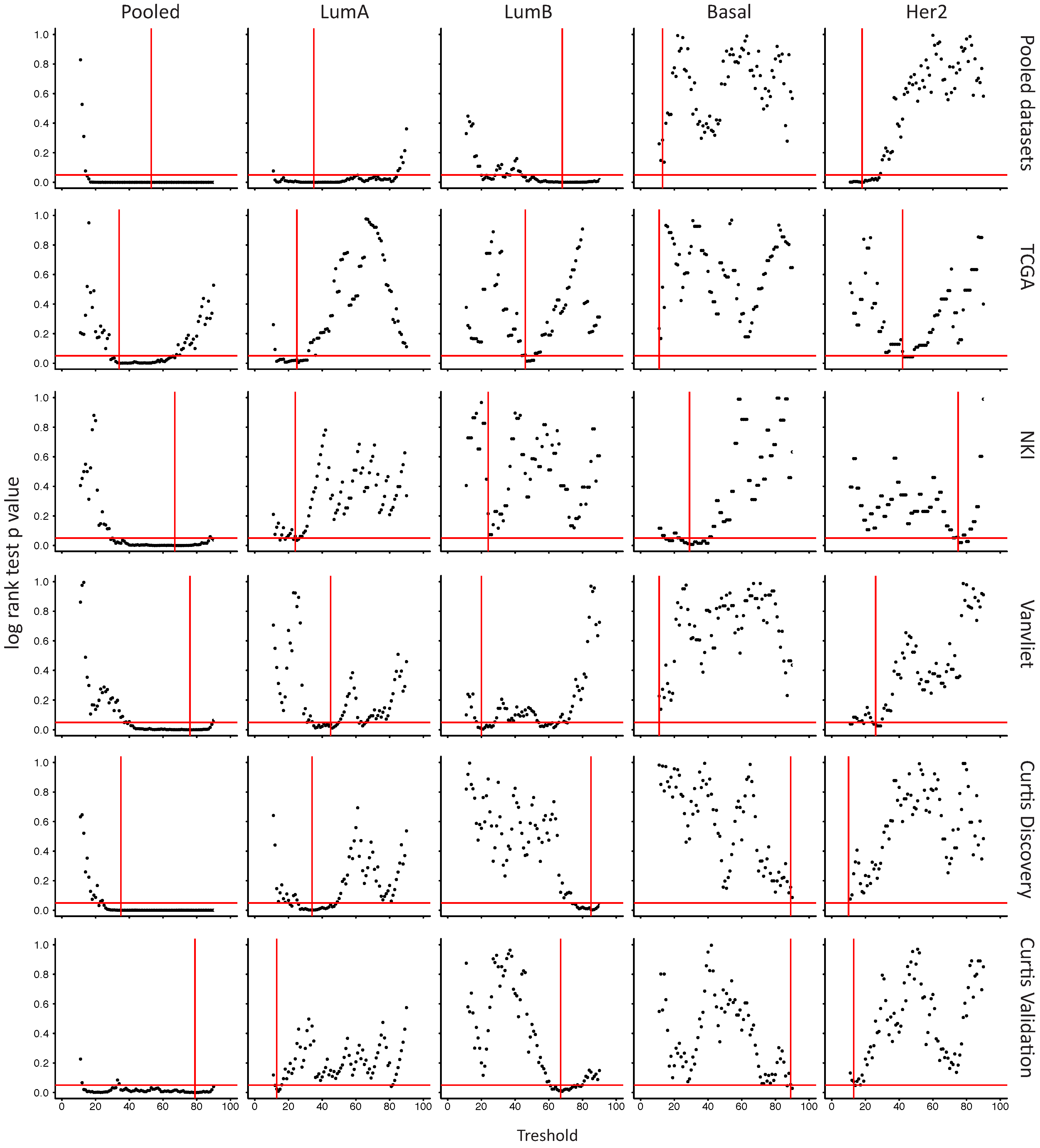
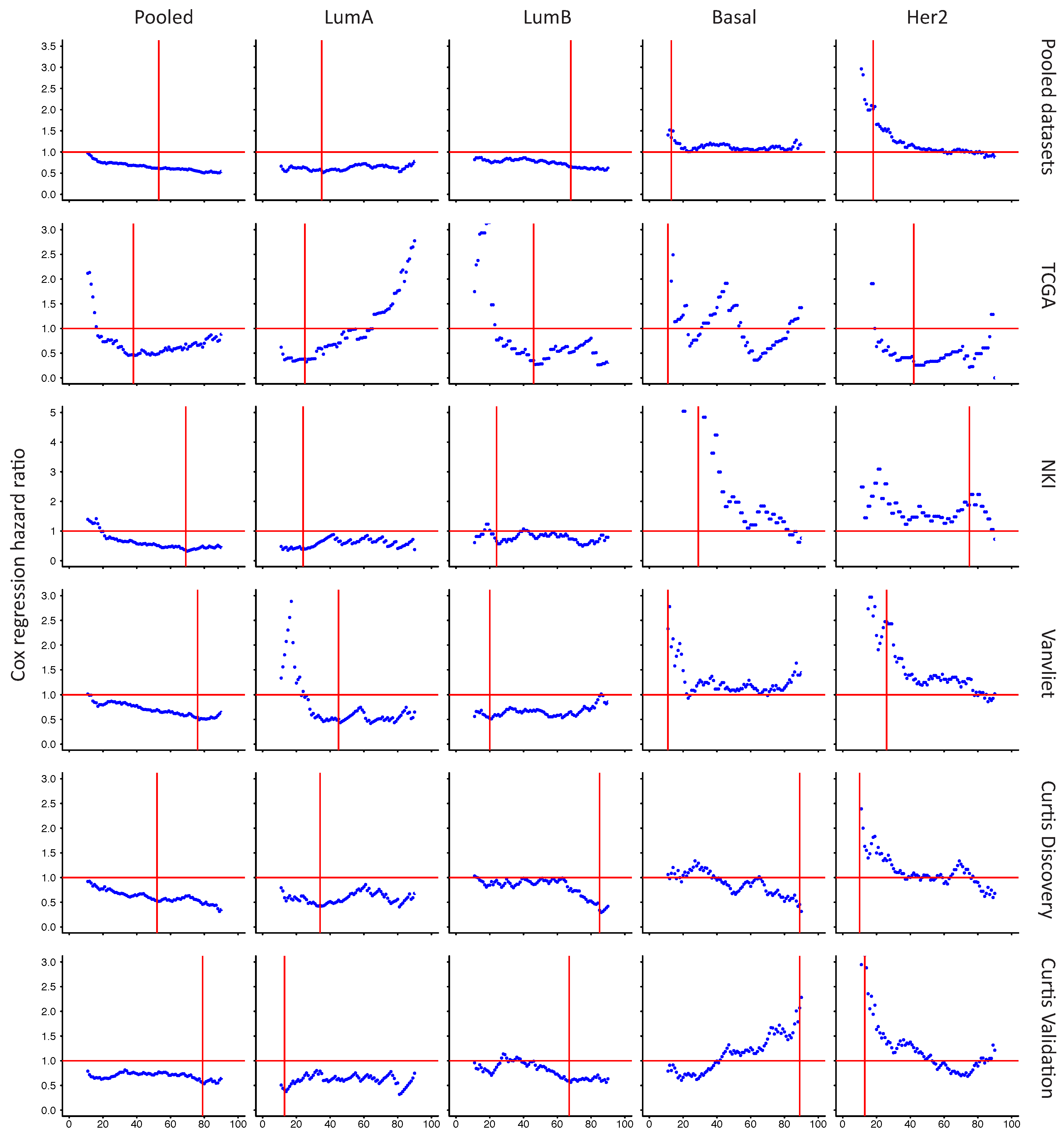
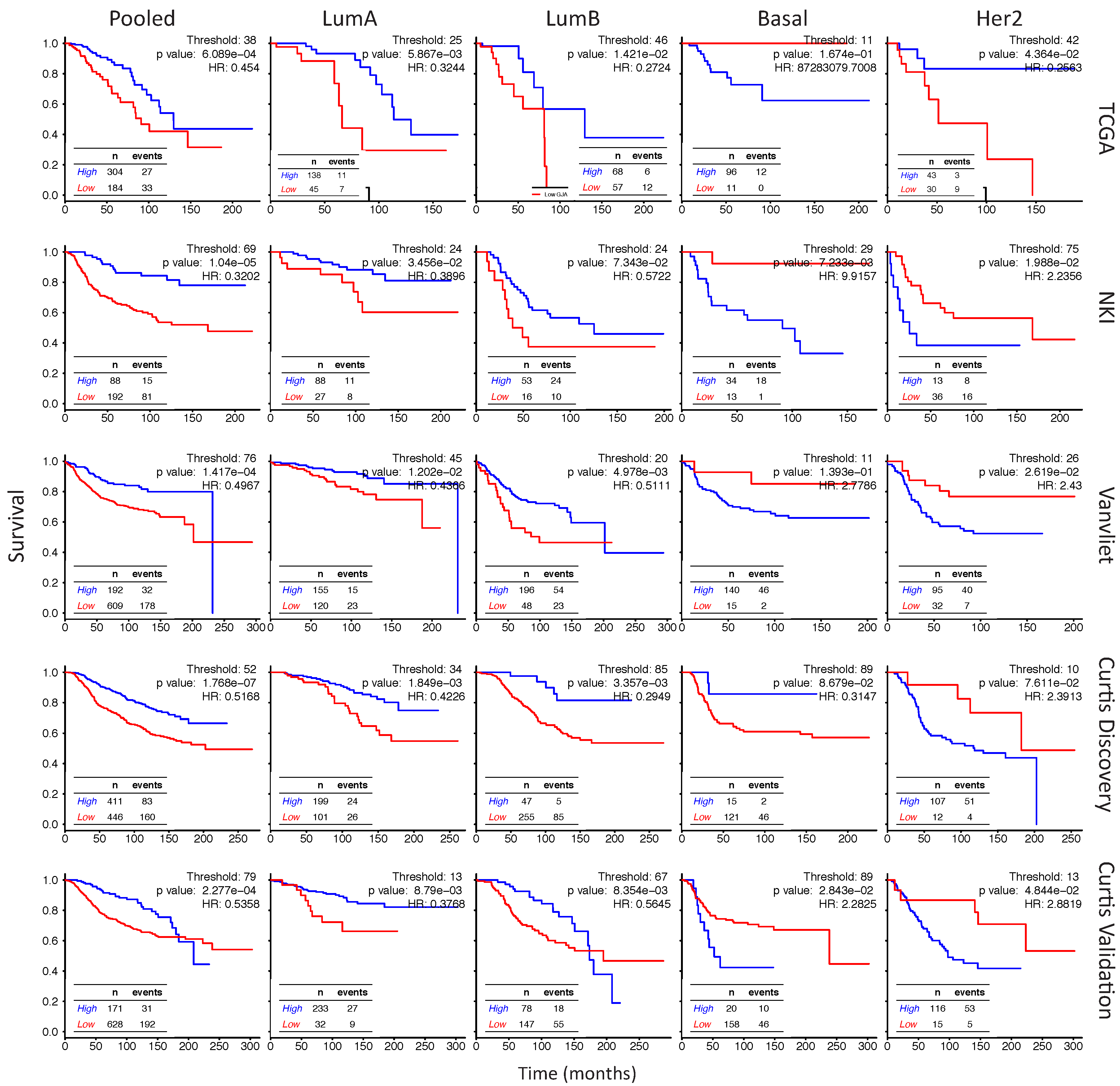
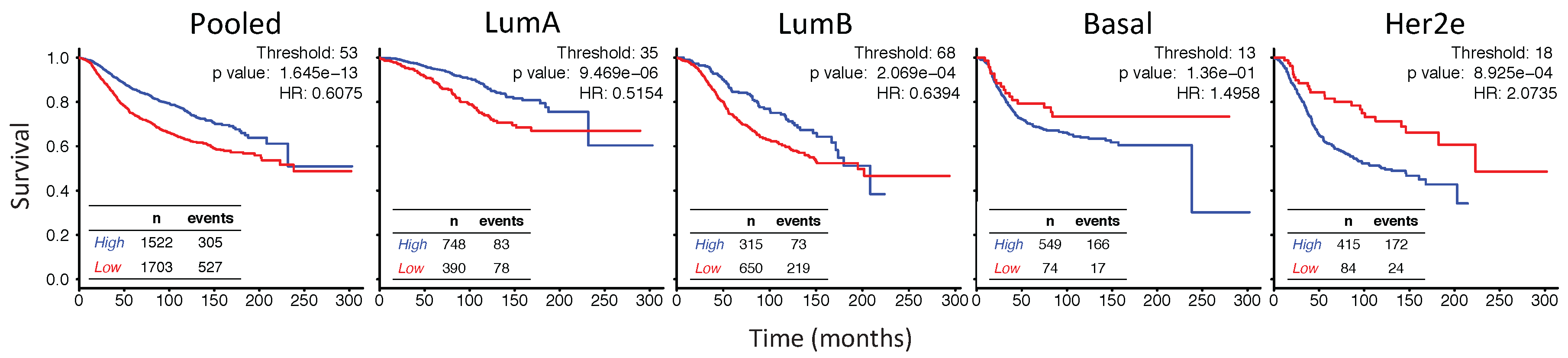
2.6. In Her2e Breast Tumors, a Low Expression of GJA1 Is Associated with a Better Prognosis
3. Discussion
3.1. Cx43 Expression Is Dysregulated in Breast Cancer
3.2. Dysregulation of Cx43 Is Linked to Hormonal Receptor Status and Tumor Subtype
3.3. Upregulation of Cx43 mRNA Is Not Driven by DNA Amplification in Breast Cancer
3.4. Cx43 mRNA Level Is Dysregulated at the Early Stages of Breast Cancer
3.5. The Apparent Grade-Dependent Decrease in Cx43 Is Linked to Its Low Expression in More Aggressive Subtypes
3.6. High Expression of Cx43 Is Associated with a Good Prognostic in Luminal Subtypes, but with a Worse Prognostic in Her2e Tumors
4. Materials and Methods
4.1. Gene Expression
4.2. DNA Alteration
4.3. Survival Analysis
4.4. Statistical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| CNA | Copy number alterations |
| Cx43 | Connexin 43 |
| DCIS | Ductal carcinoma in situ |
| ER | Estrogen receptor alpha |
| ERBB2 | erb-b2 receptor tyrosine kinase 2 |
| GJIC | Gap Junction Intercellular Communication |
| GJA1 | Gap Junction protein alpha 1, the gene encoding Connexin 43 |
| Her2e | HER2-enriched |
| IDC | Invasive ductal carcinoma |
| LumA | Luminal A |
| LumB | Luminal B |
| PR | Progesterone Receptor |
| RPPA | Reverse phase protein assay |
| TCGA | The Cancer Genome Atlas |
Appendix A

References
- Arendt, L.M.; Kuperwasser, C. Form and function: how estrogen and progesterone regulate the mammary epithelial hierarchy. J. Mammary Gland Biol. Neoplasia 2015, 20, 9–25. [Google Scholar] [CrossRef] [PubMed]
- Su, V.; Lau, A.F. Connexins: Mechanisms regulating protein levels and intercellular communication. FEBS Lett. 2014, 588, 1212–1220. [Google Scholar] [CrossRef] [PubMed]
- Plante, I.; Stewart, M.K.; Barr, K.; Allan, A.L.; Laird, D.W. Cx43 suppresses mammary tumor metastasis to the lung in a Cx43 mutant mouse model of human disease. Oncogene 2011, 30, 1681–1692. [Google Scholar] [CrossRef] [PubMed]
- Dianati, E.; Poiraud, J.; Weber-Ouellette, A.; Plante, I. Connexins, E-cadherin, Claudin-7 and beta-catenin transiently form junctional nexuses during the post-natal mammary gland development. Dev. Biol. 2016, 416, 52–68. [Google Scholar] [CrossRef] [PubMed]
- Monaghan, P.; Clarke, C.; Perusinghe, N.P.; Moss, D.W.; Chen, X.Y.; Evans, W.H. Gap junction distribution and connexin expression in human breast. Exp. Cell Res. 1996, 223, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Talhouk, R.S.; Elble, R.C.; Bassam, R.; Daher, M.; Sfeir, A.; Mosleh, L.A.; El-Khoury, H.; Hamoui, S.; Pauli, B.U.; El-Sabban, M.E. Developmental expression patterns and regulation of connexins in the mouse mammary gland: Expression of connexin30 in lactogenesis. Cell Tissue Res. 2005, 319, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Pitelka, D.R.; Hamamoto, S.T.; Duafala, J.G.; Nemanic, M.K. Cell contacts in the mouse mammary gland. I. Normal gland in postnatal development and the secretory cycle. J. Cell Biol. 1973, 56, 797–818. [Google Scholar] [CrossRef] [PubMed]
- Pollmann, M.A.; Shao, Q.; Laird, D.W.; Sandig, M. Connexin 43 mediated gap junctional communication enhances breast tumor cell diapedesis in culture. Breast Cancer Res. 2005, 7, R522–R534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomasetto, C.; Neveu, M.J.; Daley, J.; Horan, P.K.; Sager, R. Specificity of gap junction communication among human mammary cells and connexin transfectants in culture. J. Cell Biol. 1993, 122, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Woodward, T.L.; Sia, M.A.; Blaschuk, O.W.; Turner, J.D.; Laird, D.W. Deficient epithelial-fibroblast heterocellular gap junction communication can be overcome by co-culture with an intermediate cell type but not by E-cadherin transgene expression. J. Cell Sci. 1998, 111, 3529–3539. [Google Scholar] [PubMed]
- Naus, C.C.; Laird, D.W. Implications and challenges of connexin connections to cancer. Nat. Rev. Cancer 2010, 10, 435–441. [Google Scholar] [CrossRef] [PubMed]
- Laird, D.W.; Fistouris, P.; Batist, G.; Alpert, L.; Huynh, H.T.; Carystinos, G.D.; Alaoui-Jamali, M.A. Deficiency of connexin43 gap junctions is an independent marker for breast tumors. Cancer Res. 1999, 59, 4104–4110. [Google Scholar] [PubMed]
- Lee, S.W.; Tomasetto, C.; Paul, D.; Keyomarsi, K.; Sager, R. Transcriptional downregulation of gap-junction proteins blocks junctional communication in human mammary tumor cell lines. J. Cell Biol. 1992, 118, 1213–1221. [Google Scholar] [CrossRef] [PubMed]
- Gould, V.E.; Mosquera, J.M.; Leykauf, K.; Gattuso, P.; Durst, M.; Alonso, A. The phosphorylated form of connexin43 is up-regulated in breast hyperplasias and carcinomas and in their neoformed capillaries. Hum. Pathol. 2005, 36, 536–545. [Google Scholar] [CrossRef] [PubMed]
- Jamieson, S.; Going, J.J.; D’Arcy, R.; George, W.D. Expression of gap junction proteins connexin 26 and connexin 43 in normal human breast and in breast tumours. J. Pathol. 1998, 184, 37–43. [Google Scholar] [CrossRef]
- Kanczuga-Koda, L.; Sulkowska, M.; Koda, M.; Reszec, J.; Famulski, W.; Baltaziak, M.; Sulkowski, S. Expression of connexin 43 in breast cancer in comparison with mammary dysplasia and the normal mammary gland. Folia Morphol. (Warsz) 2003, 62, 439–442. [Google Scholar] [PubMed]
- El-Saghir, J.A.; El-Habre, E.T.; El-Sabban, M.E.; Talhouk, R.S. Connexins: A junctional crossroad to breast cancer. Int. J. Dev. Biol. 2011, 55, 773–780. [Google Scholar] [CrossRef] [PubMed]
- Nicolson, G.L.; Dulski, K.M.; Trosko, J.E. Loss of intercellular junctional communication correlates with metastatic potential in mammary adenocarcinoma cells. Proc. Natl. Acad. Sci. USA 1988, 85, 473–476. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Hamada, J.; Takeichi, N.; Fujikawa, S.; Kobayashi, H. Ultrastructural differences in junctional intercellular communication between highly and weakly metastatic clones derived from rat mammary carcinoma. Cancer Res. 1990, 50, 358–362. [Google Scholar] [PubMed]
- Li, Z.; Zhou, Z.; Welch, D.R.; Donahue, H.J. Expressing connexin 43 in breast cancer cells reduces their metastasis to lungs. Clin. Exp. Metastasis 2008, 25, 893–901. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.; Shao, Q.; Curtis, H.; Galipeau, J.; Belliveau, D.J.; Wang, T.; Alaoui-Jamali, M.A.; Laird, D.W. Retroviral delivery of connexin genes to human breast tumor cells inhibits in vivo tumor growth by a mechanism that is independent of significant gap junctional intercellular communication. J. Biol. Chem. 2002, 277, 29132–29138. [Google Scholar] [CrossRef] [PubMed]
- El Sabban, M.E.; Pauli, B.U. Adhesion-mediated gap junctional communication between lung-metastatatic cancer cells and endothelium. Invasion Metastasis 1994, 14, 164–176. [Google Scholar] [PubMed]
- Elzarrad, M.K.; Haroon, A.; Willecke, K.; Dobrowolski, R.; Gillespie, M.N.; Al-Mehdi, A.B. Connexin-43 upregulation in micrometastases and tumor vasculature and its role in tumor cell attachment to pulmonary endothelium. BMC Med. 2008, 6, 20–36. [Google Scholar] [CrossRef] [PubMed]
- Stoletov, K.; Strnadel, J.; Zardouzian, E.; Momiyama, M.; Park, F.D.; Kelber, J.A.; Pizzo, D.P.; Hoffman, R.; Vandenberg, S.R.; Klemke, R.L. Role of connexins in metastatic breast cancer and melanoma brain colonization. J. Cell Sci. 2013, 126, 904–913. [Google Scholar] [CrossRef] [PubMed]
- Kanczuga-Koda, L.; Sulkowski, S.; Lenczewski, A.; Koda, M.; Wincewicz, A.; Baltaziak, M.; Sulkowska, M. Increased expression of connexins 26 and 43 in lymph node metastases of breast cancer. J. Clin. Pathol. 2006, 59, 429–433. [Google Scholar] [CrossRef] [PubMed]
- Sorlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H.; Hastie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 2001, 98, 10869–10874. [Google Scholar] [CrossRef] [PubMed]
- Tofigh, A.; Suderman, M.; Paquet, E.R.; Livingstone, J.; Bertos, N.; Saleh, S.M.; Zhao, H.; Souleimanova, M.; Cory, S.; Lesurf, R.; et al. The prognostic ease and difficulty of invasive breast carcinoma. Cell Rep. 2014, 9, 129–142. [Google Scholar] [CrossRef] [PubMed]
- Uhlen, M.; Bjorling, E.; Agaton, C.; Szigyarto, C.A.; Amini, B.; Andersen, E.; Andersson, A.C.; Angelidou, P.; Asplund, A.; Asplund, C.; et al. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol. Cell Proteom. 2005, 4, 1920–1932. [Google Scholar] [CrossRef] [PubMed]
- Gendoo, D.M.; Ratanasirigulchai, N.; Schroder, M.S.; Pare, L.; Parker, J.S.; Prat, A.; Haibe-Kains, B. Genefu: An R/Bioconductor package for computation of gene expression-based signatures in breast cancer. Bioinformatics 2016, 32, 1097–1099. [Google Scholar] [CrossRef] [PubMed]
- Mitra, S.; Annamalai, L.; Chakraborty, S.; Johnson, K.; Song, X.H.; Batra, S.K.; Mehta, P.P. Androgen-regulated formation and degradation of gap junctions in androgen-responsive human prostate cancer cells. Mol. Biol. Cell 2006, 17, 5400–5416. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Wang, X.H.; Wang, G.C.; Wu, J.H. 17beta estradiol regulation of connexin 43-based gap junction and mechanosensitivity through classical estrogen receptor pathway in osteocyte-like MLO-Y4 cells. Bone 2013, 53, 587–596. [Google Scholar] [CrossRef] [PubMed]
- Stewart, M.; Simek, J.; Laird, D. Insights into the role of Connexins in Mammary Gland Morphogenesis and Function. Reproduction 2015, 149, R279–R290. [Google Scholar] [CrossRef] [PubMed]
- Teleki, I.; Szasz, A.M.; Maros, M.E.; Gyorffy, B.; Kulka, J.; Meggyeshazi, N.; Kiszner, G.; Balla, P.; Samu, A.; Krenacs, T. Correlations of differentially expressed gap junction connexins cx26, cx30, cx32, cx43 and cx46 with breast cancer progression and prognosis. PLoS ONE 2014, 9, e112541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeh, E.S.; Williams, C.J.; Williams, C.B.; Bonilla, I.V.; Klauber-DeMore, N.; Phillips, S.L. Dysregulated connexin 43 in HER2-positive drug resistant breast cancer cells enhances proliferation and migration. Oncotarget 2017, 8, 109358–109369. [Google Scholar] [CrossRef] [PubMed]
- Gyorffy, B.; Lanczky, A.; Eklund, A.C.; Denkert, C.; Budczies, J.; Li, Q.; Szallasi, Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1809 patients. Breast Cancer Res. Treat. 2010, 123, 725–731. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Lee, H.E.; Li, H.; Shipitsin, M.; Gelman, R.; Polyak, K. Heterogeneity for Stem Cell-Related Markers According to Tumor Subtype and Histologic Stage in Breast Cancer. Clin. Cancer Res. 2010, 16, 876–887. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Shao, Z.M.; He, Q.Z.; Jiang, B.Q.; Wu, Y.; Zhuang, Z.G. Hsa-miR-206 represses the proliferation and invasion of breast cancer cells by targeting Cx43. Eur. Rev. Med. Pharmacol. Sci. 2015, 19, 2091–2104. [Google Scholar] [PubMed]
- Clarke, C.; Madden, S.F.; Doolan, P.; Aherne, S.T.; Joyce, H.; O’Driscoll, L.; Gallagher, W.M.; Hennessy, B.T.; Moriarty, M.; Crown, J.; et al. Correlating transcriptional networks to breast cancer survival: A large-scale coexpression analysis. Carcinogenesis 2013, 34, 2300–2308. [Google Scholar] [CrossRef] [PubMed]
- Yoshihara, K.; Shahmoradgoli, M.; Martinez, E.; Vegesna, R.; Kim, H.; Torres-Garcia, W.; Trevino, V.; Shen, H.; Laird, P.W.; Levine, D.A.; et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013, 4, 2612–2622. [Google Scholar] [CrossRef] [PubMed]
- Grummer, R.; Traub, O.; Winterhager, E. Gap junction connexin genes cx26 and cx43 are differentially regulated by ovarian steroid hormones in rat endometrium. Endocrinology 1999, 140, 2509–2516. [Google Scholar] [CrossRef] [PubMed]
- Gulinello, M.; Etgen, A.M. Sexually dimorphic hormonal regulation of the gap junction protein, CX43, in rats and altered female reproductive function in CX43+/− mice. Brain Res. 2005, 1045, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Nadeem, L.; Shynlova, O.; Matysiak-Zablocki, E.; Mesiano, S.; Dong, X.; Lye, S. Molecular evidence of functional progesterone withdrawal in human myometrium. Nat. Commun. 2016, 7, 11565. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Berga, S.L.; Johnston-MacAnanny, E.B.; Sidell, N.; Bagchi, I.C.; Bagchi, M.K.; Taylor, R.N. Endometrial Stromal Decidualization Responds Reversibly to Hormone Stimulation and Withdrawal. Endocrinology 2016, 157, 2432–2446. [Google Scholar] [CrossRef] [PubMed]
- Conklin, C.; Huntsman, D.; Yorida, E.; Makretsov, N.; Turbin, D.; Bechberger, J.F.; Sin, W.C.; Naus, C.C. Tissue microarray analysis of connexin expression and its prognostic significance in human breast cancer. Cancer Lett. 2007, 255, 284–294. [Google Scholar] [CrossRef] [PubMed]
- Teleki, I.; Krenacs, T.; Szasz, M.A.; Kulka, J.; Wichmann, B.; Leo, C.; Papassotiropoulos, B.; Riemenschnitter, C.; Moch, H.; Varga, Z. The potential prognostic value of connexin 26 and 46 expression in neoadjuvant-treated breast cancer. BMC Cancer 2013, 13, 50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murphy, L.C.; Seekallu, S.V.; Watson, P.H. Clinical significance of estrogen receptor phosphorylation. Endocr. Relat. Cancer 2011, 18, R1–R14. [Google Scholar] [CrossRef] [PubMed]
- Curtis, C.; Shah, S.P.; Chin, S.F.; Turashvili, G.; Rueda, O.M.; Dunning, M.J.; Speed, D.; Lynch, A.G.; Samarajiwa, S.; Yuan, Y.; et al. The genomic and transcriptomic architecture of 2000 breast tumours reveals novel subgroups. Nature 2012, 486, 346–352. [Google Scholar] [PubMed]
- Mesnil, M.; Crespin, S.; Avanzo, J.L.; Zaidan-Dagli, M.L. Defective gap junctional intercellular communication in the carcinogenic process. Biochim. Biophys. Acta 2005, 1719, 125–145. [Google Scholar] [CrossRef] [PubMed]
- Valastyan, S.; Weinberg, R.A. Tumor metastasis: Molecular insights and evolving paradigms. Cell 2011, 147, 275–292. [Google Scholar] [CrossRef] [PubMed]
- Van Vliet, M.H.; Reyal, F.; Horlings, H.M.; van de Vijver, M.J.; Reinders, M.J.; Wessels, L.F. Pooling breast cancer datasets has a synergetic effect on classification performance and improves signature stability. BMC Genom. 2008, 9, 375–397. [Google Scholar] [CrossRef] [PubMed]
- TCGA. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61–70. [Google Scholar]
- Van de Vijver, M.J.; He, Y.D.; van’t Veer, L.J.; Dai, H.; Hart, A.A.; Voskuil, D.W.; Schreiber, G.J.; Peterse, J.L.; Roberts, C.; Marton, M.J.; et al. A gene-expression signature as a predictor of survival in breast cancer. N. Engl. J. Med. 2002, 347, 1999–2009. [Google Scholar] [CrossRef] [PubMed]
- Hazan, R.; Margolis, B.; Dombalagian, M.; Ullrich, A.; Zilberstein, A.; Schlessinger, J. Identification of autophosphorylation sites of HER2/neu. Cell Growth Differ. 1990, 1, 3–7. [Google Scholar] [PubMed]
- Mertins, P.; Mani, D.R.; Ruggles, K.V.; Gillette, M.A.; Clauser, K.R.; Wang, P.; Wang, X.; Qiao, J.W.; Cao, S.; Petralia, F.; et al. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature 2016, 534, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Ciriello, G.; Gatza, M.L.; Beck, A.H.; Wilkerson, M.D.; Rhie, S.K.; Pastore, A.; Zhang, H.; McLellan, M.; Yau, C.; Kandoth, C.; et al. Comprehensive Molecular Portraits of Invasive Lobular Breast Cancer. Cell 2015, 163, 506–519. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, 11–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Therneau, T.M.; Grambsch, P.M. Modeling Survival Data: Extending the Cox Model; Springer: New York, NY, USA, 2000. [Google Scholar]
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.C.; Muller, M. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef] [PubMed]
- Madden, S.F.; Clarke, C.; Gaule, P.; Aherne, S.T.; O’Donovan, N.; Clynes, M.; Crown, J.; Gallagher, W.M. BreastMark: An integrated approach to mining publicly available transcriptomic datasets relating to breast cancer outcome. Breast Cancer Res. 2013, 15, R52–R66. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2016. [Google Scholar]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucl. Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Busby, M.; Hallett, M.T.; Plante, I. The Complex Subtype-Dependent Role of Connexin 43 (GJA1) in Breast Cancer. Int. J. Mol. Sci. 2018, 19, 693. https://doi.org/10.3390/ijms19030693
Busby M, Hallett MT, Plante I. The Complex Subtype-Dependent Role of Connexin 43 (GJA1) in Breast Cancer. International Journal of Molecular Sciences. 2018; 19(3):693. https://doi.org/10.3390/ijms19030693
Chicago/Turabian StyleBusby, Mélanie, Michael T. Hallett, and Isabelle Plante. 2018. "The Complex Subtype-Dependent Role of Connexin 43 (GJA1) in Breast Cancer" International Journal of Molecular Sciences 19, no. 3: 693. https://doi.org/10.3390/ijms19030693