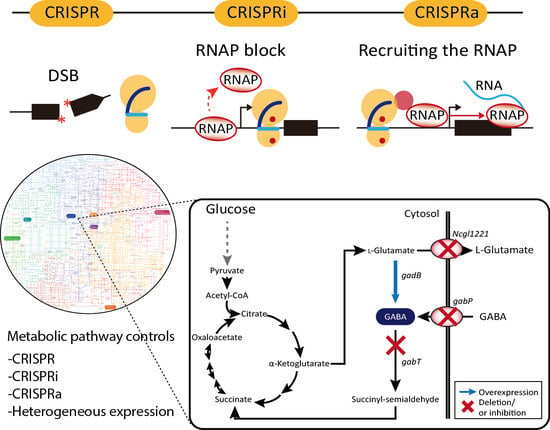
Applications of CRISPR/Cas System to Bacterial Metabolic Engineering
Abstract
:1. Introduction
2. CRISPR/Cas9 System
3. Application of CRISPR/Cas9 Technology to Engineer Bacterial Genomes
3.1. Bacilli
3.2. Clostridia
3.3. Corynebacteria
3.4. Streptomycetes
3.5. Escherichia coli
4. Bioinformatics Tools for Guide RNA Design in CRISPR/Cas9 System
5. Concluding Remarks and Future Perspectives
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ishino, Y.; Shinagawa, H.; Makino, K.; Amemura, M.; Nakata, A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 1987, 169, 5429–5433. [Google Scholar] [CrossRef] [PubMed]
- Grissa, I.; Vergnaud, G.; Pourcel, C. CRISPRfinder: A web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007, 35, W52–W57. [Google Scholar] [CrossRef] [PubMed]
- Carroll, D. Genome engineering with zinc-finger nucleases. Genetics 2011, 188, 773–782. [Google Scholar] [CrossRef] [PubMed]
- Urnov, F.D.; Miller, J.C.; Lee, Y.L.; Beausejour, C.M.; Rock, J.M.; Augustus, S.; Jamieson, A.C.; Porteus, M.H.; Gregory, P.D.; Holmes, M.C. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature 2005, 435, 646–651. [Google Scholar] [CrossRef] [PubMed]
- Boch, J.; Scholze, H.; Schornack, S.; Landgraf, A.; Hahn, S.; Kay, S.; Lahaye, T.; Nickstadt, A.; Bonas, U. Breaking the code of DNA binding specificity of Tal-type III effectors. Science 2009, 326, 1509–1512. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.L.; Mullis, A.S.; Leenay, R.T.; Beisel, C.L. Repurposing endogenous type i CRISPR-Cas systems for programmable gene repression. Nucleic Acids Res. 2015, 43, 674–681. [Google Scholar] [CrossRef] [PubMed]
- Senturk, S.; Shirole, N.H.; Nowak, D.G.; Corbo, V.; Pal, D.; Vaughan, A.; Tuveson, D.A.; Trotman, L.C.; Kinney, J.B.; Sordella, R. Rapid and tunable method to temporally control gene editing based on conditional cas9 stabilization. Nat. Commun. 2017, 8, 14370. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.S.; Larson, M.H.; Gilbert, L.A.; Doudna, J.A.; Weissman, J.S.; Arkin, A.P.; Lim, W.A. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 2013, 152, 1173–1183. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Wolf, Y.I.; Alkhnbashi, O.S.; Costa, F.; Shah, S.A.; Saunders, S.J.; Barrangou, R.; Brouns, S.J.; Charpentier, E.; Haft, D.H.; et al. An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 2015, 13, 722–736. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.Y.; Bikard, D.; Cox, D.; Zhang, F.; Marraffini, L.A. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 2013, 31, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhu, S.; Cai, C.; Yuan, P.; Li, C.; Huang, Y.; Wei, W. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature 2014, 509, 487–491. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Qian, F.; Yang, J.; Liu, Y.; Dong, F.; Xu, C.; Sun, B.; Chen, B.; Xu, X.; Li, Y.; et al. CRISPR-Cpf1 assisted genome editing of Corynebacterium glutamicum. Nat. Commun. 2017, 8, 15179. [Google Scholar] [CrossRef] [PubMed]
- Kleinstiver, B.P.; Tsai, S.Q.; Prew, M.S.; Nguyen, N.T.; Welch, M.M.; Lopez, J.M.; McCaw, Z.R.; Aryee, M.J.; Joung, J.K. Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells. Nat. Biotechnol. 2016, 34, 869–874. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; La Russa, M.; Qi, L.S. CRISPR/Cas9 in genome editing and beyond. Annu. Rev. Biochem. 2016, 85, 227–264. [Google Scholar] [CrossRef] [PubMed]
- Doudna, J.A.; Charpentier, E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.D.; Lander, E.S.; Zhang, F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 2014, 157, 1262–1278. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Tong, Y.; Charusanti, P.; Zhang, L.; Weber, T.; Lee, S.Y. CRISPR-Cas9 based engineering of actinomycetal genomes. ACS Synth. Biol. 2015, 4, 1020–1029. [Google Scholar] [CrossRef] [PubMed]
- Chae, T.U.; Choi, S.Y.; Kim, J.W.; Ko, Y.S.; Lee, S.Y. Recent advances in systems metabolic engineering tools and strategies. Curr. Opin. Biotechnol. 2017, 47, 67–82. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.S.; Wu, H.; Ji, X.; Stelzer, Y.; Wu, X.B.; Czauderna, S.; Shu, J.; Dadon, D.; Young, R.A.; Jaenisch, R. Editing DNA methylation in the mammalian genome. Cell 2016, 167, 233–247. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.H.; Gilbert, L.A.; Cimini, B.A.; Schnitzbauer, J.; Zhang, W.; Li, G.W.; Park, J.; Blackburn, E.H.; Weissman, J.S.; Qi, L.S.; et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 2013, 155, 1479–1491. [Google Scholar] [CrossRef] [PubMed]
- Savic, D.; Partridge, E.C.; Newberry, K.M.; Smith, S.B.; Meadows, S.K.; Roberts, B.S.; Mackiewicz, M.; Mendenhall, E.M.; Myers, R.M. Cetch-seq: CRISPR epitope tagging chip-seq of DNA-binding proteins. Genome Res. 2015, 25, 1581–1589. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, N.; Tamura, T.; Good, L. Paired termini stabilize antisense RNAs and enhance conditional gene silencing in Escherichia coli. Nucleic Acids Res. 2006, 34, e138. [Google Scholar] [CrossRef] [PubMed]
- Gagarinova, A.; Emili, A. Genome-scale genetic manipulation methods for exploring bacterial molecular biology. Mol. Biosyst. 2012, 8, 1626–1638. [Google Scholar] [CrossRef] [PubMed]
- Hathaway, N.A.; Bell, O.; Hodges, C.; Miller, E.L.; Neel, D.S.; Crabtree, G.R. Dynamics and memory of heterochromatin in living cells. Cell 2012, 149, 1447–1460. [Google Scholar] [CrossRef] [PubMed]
- Margolin, J.F.; Friedman, J.R.; Meyer, W.K.; Vissing, H.; Thiesen, H.J.; Rauscher, F.J. Kruppel-associated boxes are potent transcriptional repression domains. Proc. Natl. Acad. Sci. USA 1994, 91, 4509–4513. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, L.A.; Larson, M.H.; Morsut, L.; Liu, Z.; Brar, G.A.; Torres, S.E.; Stern-Ginossar, N.; Brandman, O.; Whitehead, E.H.; Doudna, J.A.; et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 2013, 154, 442–451. [Google Scholar] [CrossRef] [PubMed]
- Schreiber-Agus, N.; Chin, L.; Chen, K.; Torres, R.; Rao, G.; Guida, P.; Skoultchi, A.I.; DePinho, R.A. An amino-terminal domain of mxi1 mediates anti-myc oncogenic activity and interacts with a homolog of the yeast transcriptional repressor Sin3. Cell 1995, 80, 777–786. [Google Scholar] [CrossRef] [PubMed]
- Fisher, A.L.; Ohsako, S.; Caudy, M. The WRPW motif of the hairy-related basic helix-loop-helix repressor proteins acts as a 4-amino-acid transcription repression and protein-protein interaction domain. Mol. Cell. Biol. 1996, 16, 2670–2677. [Google Scholar] [CrossRef] [PubMed]
- Beerli, R.R.; Segal, D.J.; Dreier, B.; Barbas, C.F. Toward controlling gene expression at will: Specific regulation of the erbB-2/HER-2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks. Proc. Natl. Acad. Sci. USA 1998, 95, 14628–14633. [Google Scholar] [CrossRef] [PubMed]
- Farzadfard, F.; Perli, S.D.; Lu, T.K. Tunable and multifunctional eukaryotic transcription factors based on CRISPR/Cas. ACS Synth. Biol. 2013, 2, 604–613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perez-Pinera, P.; Kocak, D.D.; Vockley, C.M.; Adler, A.F.; Kabadi, A.M.; Polstein, L.R.; Thakore, P.I.; Glass, K.A.; Ousterout, D.G.; Leong, K.W.; et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat. Methods 2013, 10, 973–976. [Google Scholar] [CrossRef] [PubMed]
- Maeder, M.L.; Linder, S.J.; Cascio, V.M.; Fu, Y.; Ho, Q.H.; Joung, J.K. CRISPR RNA-guided activation of endogenous human genes. Nat. Methods 2013, 10, 977–979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, A.W.; Wang, H.; Yang, H.; Shi, L.; Katz, Y.; Theunissen, T.W.; Rangarajan, S.; Shivalila, C.S.; Dadon, D.B.; Jaenisch, R. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 2013, 23, 1163–1171. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, L.A.; Horlbeck, M.A.; Adamson, B.; Villalta, J.E.; Chen, Y.; Whitehead, E.H.; Guimaraes, C.; Panning, B.; Ploegh, H.L.; Bassik, M.C.; et al. Genome-scale CRISPR-mediated control of gene repression and activation. Cell 2014, 159, 647–661. [Google Scholar] [CrossRef] [PubMed]
- Tanenbaum, M.E.; Gilbert, L.A.; Qi, L.S.; Weissman, J.S.; Vale, R.D. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 2014, 159, 635–646. [Google Scholar] [CrossRef] [PubMed]
- Chavez, A.; Scheiman, J.; Vora, S.; Pruitt, B.W.; Tuttle, M.; Eswar, P.R.I.; Lin, S.; Kiani, S.; Guzman, C.D.; Wiegand, D.J.; et al. Highly efficient Cas9-mediated transcriptional programming. Nat. Methods 2015, 12, 326–328. [Google Scholar] [CrossRef] [PubMed]
- Konermann, S.; Brigham, M.D.; Trevino, A.E.; Joung, J.; Abudayyeh, O.O.; Barcena, C.; Hsu, P.D.; Habib, N.; Gootenberg, J.S.; Nishimasu, H.; et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 2015, 517, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Bikard, D.; Jiang, W.; Samai, P.; Hochschild, A.; Zhang, F.; Marraffini, L.A. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 2013, 41, 7429–7437. [Google Scholar] [CrossRef] [PubMed]
- Mougiakos, I.; Bosma, E.F.; Weenink, K.; Vossen, E.; Goijvaerts, K.; van der Oost, J.; van Kranenburg, R. Efficient genome editing of a facultative thermophile using mesophilic spCas9. ACS Synth. Biol. 2017, 6, 849–861. [Google Scholar] [CrossRef] [PubMed]
- Bernheim, A.; Calvo-Villamanan, A.; Basier, C.; Cui, L.; Rocha, E.P.C.; Touchon, M.; Bikard, D. Inhibition of NHEJ repair by type ii-a CRISPR-Cas systems in bacteria. Nat. Commun. 2017, 8, 2094. [Google Scholar] [CrossRef] [PubMed]
- Altenbuchner, J. Editing of the Bacillus subtilis genome by the CRISPR-Cas9 system. Appl. Environ. Microbiol. 2016, 82, 5421–5427. [Google Scholar] [CrossRef] [PubMed]
- Peters, J.M.; Colavin, A.; Shi, H.; Czarny, T.L.; Larson, M.H.; Wong, S.; Hawkins, J.S.; Lu, C.H.S.; Koo, B.M.; Marta, E.; et al. A comprehensive, CRISPR-based functional analysis of essential genes in bacteria. Cell 2016, 165, 1493–1506. [Google Scholar] [CrossRef] [PubMed]
- Mougiakos, I.; Mohanraju, P.; Bosma, E.F.; Vrouwe, V.; Finger Bou, M.; Naduthodi, M.I.S.; Gussak, A.; Brinkman, R.B.L.; van Kranenburg, R.; van der Oost, J. Characterizing a thermostable Cas9 for bacterial genome editing and silencing. Nat. Commun. 2017, 8, 1647. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Cai, D.; Wang, Z.; He, Z.; Chen, S. Development of an efficient genome editing tool in Bacillus licheniformis using CRISPR-Cas9 nickase. Appl. Environ. Microbiol. 2018. [Google Scholar] [CrossRef] [PubMed]
- McAllister, K.N.; Bouillaut, L.; Kahn, J.N.; Self, W.T.; Sorg, J.A. Using CRISPR-Cas9-mediated genome editing to generate C. difficile mutants defective in selenoproteins synthesis. Sci. Rep. 2017, 7, 14672. [Google Scholar] [CrossRef] [PubMed]
- Negahdaripour, M.; Nezafat, N.; Hajighahramani, N.; Rahmatabadi, S.S.; Ghasemi, Y. Investigating CRISPR-Cas systems in Clostridium botulinum via bioinformatics tools. Infect. Genet. Evol. 2017, 54, 355–373. [Google Scholar] [CrossRef] [PubMed]
- Wasels, F.; Jean-Marie, J.; Collas, F.; Lopez-Contreras, A.M.; Ferreira, N.L. A two-plasmid inducible CRISPR/Cas9 genome editing tool for Clostridium acetobutylicum. J. Microbiol. Methods 2017, 140, 5–11. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.H.; Dong, S.; Wang, P.X.; Tao, Y.; Wang, Y. Genome editing in Clostridium saccharoperbutylacetonicum n1-4 with the CRISPR-Cas9 system. Appl. Environ. Microbiol. 2017, 83, e00233-17. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, Z.T.; Seo, S.O.; Lynn, P.; Lu, T.; Jin, Y.S.; Blaschek, H.P. Gene transcription repression in Clostridium beijerinckii using CRISPR-dCas9. Biotechnol. Bioeng. 2016, 113, 2739–2743. [Google Scholar] [CrossRef] [PubMed]
- Nagaraju, S.; Davies, N.K.; Walker, D.J.F.; Kopke, M.; Simpson, S.D. Genome editing of Clostridium autoethanogenum using CRISPR/Cas9. Biotechnol. Biofuels 2016, 9, 219. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Chen, J.; Minton, N.P.; Zhang, Y.; Wen, Z.Q.; Liu, J.L.; Yang, H.F.; Zeng, Z.; Ren, X.D.; Yang, J.J.; et al. CRISPR-based genome editing and expression control systems in Clostridium acetobutylicum and Clostridium beijerinckii. Biotechnol. J. 2016, 11, 961–972. [Google Scholar] [CrossRef] [PubMed]
- Pyne, M.E.; Bruder, M.R.; Moo-Young, M.; Chung, D.A.; Chou, C.P. Harnessing heterologous and endogenous CRISPR-Cas machineries for efficient markerless genome editing in Clostridium. Sci. Rep. 2016, 6, 25666. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, Z.T.; Seo, S.O.; Lynn, P.; Lu, T.; Jin, Y.S.; Blaschek, H.P. Bacterial genome editing with CRISPR-Cas9: Deletion, integration, single nucleotide modification, and desirable “clean” mutant selection in Clostridium beijerinckii as an example. ACS Synth. Biol. 2016, 5, 721–732. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Shin, H.; Lee, S.M.; Um, Y.; Woo, H.M. RNA-guided single/double gene repressions in Corynebacterium glutamicum using an efficient CRISPR interference and its application to industrial strain. Microb. Cell Fact. 2018, 17, 4. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, Y.; Lu, Y.; Zheng, P.; Sun, J.; Ma, Y. Development of a CRISPR/Cas9 genome editing toolbox for Corynebacterium glutamicum. Microb. Cell Fact. 2017, 16, 205. [Google Scholar] [CrossRef] [PubMed]
- Peng, F.; Wang, X.Y.; Sun, Y.; Dong, G.B.; Yang, Y.K.; Liu, X.X.; Bai, Z.G. Efficient gene editing in Corynebacterium glutamicum using the CRISPR/Cas9 system. Microb. Cell Fact. 2017, 16, 201. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.S.; Choi, K.R.; Prabowo, C.P.S.; Shin, J.H.; Yang, D.; Jang, J.; Lee, S.Y. CRISPR/Cas9-coupled recombineering for metabolic engineering of Corynebacterium glutamicum. Metab. Eng. 2017, 42, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Cleto, S.; Jensen, J.V.; Wendisch, V.F.; Lu, T.K. Corynebacterium glutamicum metabolic engineering with CRISPR interference (CRISPRi). ACS Synth. Biol. 2016, 5, 375–385. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Huang, H.; Xiong, Z.; Ai, L.; Yang, S. CRISPR-Cas9(D10A) nickase-assisted genome editing in Lactobacillus casei. Appl. Environ. Microbiol. 2017, 83. [Google Scholar] [CrossRef] [PubMed]
- Sanozky-Dawes, R.; Selle, K.; O’Flaherty, S.; Klaenhammer, T.; Barrangou, R. Occurrence and activity of a type II CRISPR-Cas system in Lactobacillus gasseri. Microbiology 2015, 161, 1752–1761. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.H.; van Pijkeren, J.P. CRISPR-Cas9-assisted recombineering in Lactobacillus reuteri. Nucleic Acids Res. 2014, 42, e131. [Google Scholar] [CrossRef] [PubMed]
- Rock, J.M.; Hopkins, F.F.; Chavez, A.; Diallo, M.; Chase, M.R.; Gerrick, E.R.; Pritchard, J.R.; Church, G.M.; Rubin, E.J.; Sassetti, C.M.; et al. Programmable transcriptional repression in mycobacteria using an orthogonal CRISPR interference platform. Nat. Microbiol. 2017, 2, 16274. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Carette, X.; Potluri, L.P.; Sharp, J.D.; Xu, R.F.; Prisic, S.; Husson, R.N. Investigating essential gene function in Mycobacterium tuberculosis using an efficient CRISPR interference system. Nucleic Acids Res. 2016, 44. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.Z.; Reisch, C.R.; Prather, K.L.J. A robust CRISPRi gene repression system in Pseudomonas. J. Bacteriol. 2018, 200. [Google Scholar] [CrossRef] [PubMed]
- Guan, J.; Wang, W.Y.; Sun, B.L. Chromosomal targeting by the type III-A CRISPR-Cas system can reshape genomes in Staphylococcus aureus. Msphere 2017, 2, e00403–e00417. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.H.; Zhang, Y.F.; Yeo, W.S.; Bae, T.; Ji, Q.J. Rapid and efficient genome editing in Staphylococcus aureus by using an engineered CRISPR/Cas9 system. J. Am. Chem. Soc. 2017, 139, 3790–3795. [Google Scholar] [CrossRef] [PubMed]
- Cobb, R.E.; Wang, Y.; Zhao, H. High-efficiency multiplex genome editing of Streptomyces species using an engineered CRISPR/Cas system. ACS Synth. Biol. 2015, 4, 723–728. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Wang, S.W.; Chen, Z.; Guo, Y.J.; Song, Y. An active type i-e CRISPR-Cas system identified in Streptomyces avermitilis. PLoS ONE 2016, 11, e0149533. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.M.; Wong, F.T.; Wang, Y.; Luo, S.; Lim, Y.H.; Heng, E.; Yeo, W.L.; Cobb, R.E.; Enghiad, B.; Ang, E.L.; et al. CRISPR-Cas9 strategy for activation of silent Streptomyces biosynthetic gene clusters. Nat. Chem. Biol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.Y.; Zhang, L.M.; Wang, T.T.; Han, J.; Tang, H.; Zhang, L.P. Development of a CRISPR/Cas9-mediated gene-editing tool in Streptomyces rimosus. Microbiology 2017, 163, 1148–1155. [Google Scholar] [CrossRef] [PubMed]
- Hao, N.; Shearwin, K.E.; Dodd, I.B. Programmable DNA looping using engineered bivalent dCas9 complexes. Nat. Commun. 2017, 8, 1628. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.Y.; Yan, H.Q.; Ren, G.X.; Zhao, J.P.; Guo, X.P.; Sun, Y.C. CRISPR-Cas12a-assisted recombineering in bacteria. Appl. Environ. Microbiol. 2017, 83. [Google Scholar] [CrossRef] [PubMed]
- Umenhoffer, K.; Draskovits, G.; Nyerges, A.; Karcagi, I.; Bogos, B.; Timar, E.; Csorgo, B.; Herczeg, R.; Nagy, I.; Feher, T.; et al. Genome-wide abolishment of mobile genetic elements using genome shuffling and CRISPR/Cas-assisted MAGE allows the efficient stabilization of a bacterial chassis. ACS Synth. Biol. 2017, 6, 1471–1483. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.J.; Du, G.C.; Chen, J.; Zhou, J.W. Enhancing flavonoid production by systematically tuning the central metabolic pathways based on a CRISPR interference system in Escherichia coli. Sci. Rep. 2015, 5, 13477. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.F.; Lin, Z.Q.; Huang, C.; Zhang, Y.; Wang, Z.W.; Tang, Y.J.; Chen, T.; Zhao, X.M. Metabolic engineering of Escherichia coli using CRISPR-Cas9 meditated genome editing. Metab. Eng. 2015, 31, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Li, S.Y.; Jendresen, C.B.; Grunberger, A.; Ronda, C.; Jensen, S.I.; Noack, S.; Nielsen, A.T. Enhanced protein and biochemical production using CRISPRi-based growth switches. Metab. Eng. 2016, 38, 274–284. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.K.; Han, G.H.; Seong, W.; Kim, H.; Kim, S.W.; Lee, D.H.; Lee, S.G. CRISPR interference-guided balancing of a biosynthetic mevalonate pathway increases terpenoid production. Metab. Eng. 2016, 38, 228–240. [Google Scholar] [CrossRef] [PubMed]
- Lv, L.; Ren, Y.L.; Chen, J.C.; Wu, Q.; Chen, G.Q. Application of CRISPRi for prokaryotic metabolic engineering involving multiple genes, a case study: Controllable p(3hb-co-4hb) biosynthesis. Metab. Eng. 2015, 29, 160–168. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.L.; Guo, L.Q.; Lin, J.F.; He, Z.Q.; Cai, F.J.; Chen, J.F. A novel process for obtaining pinosylvin using combinatorial bioengineering in Escherichia coli. World J. Microbiol. Biotechnol. 2016, 32, 102. [Google Scholar] [CrossRef] [PubMed]
- Cress, B.F.; Leitz, Q.D.; Kim, D.C.; Amore, T.D.; Suzuki, J.Y.; Linhardt, R.J.; Koffas, M.A.G. CRISPRi-mediated metabolic engineering of E. coli for o-methylated anthocyanin production. Microb. Cell Fact. 2017, 16, 10. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.Y.; Sung, L.Y.; Li, H.; Huang, C.H.; Hu, Y.C. Combining CRISPR and CRISPRi systems for metabolic engineering of E. coli and 1,4-bdo biosynthesis. ACS Synth. Biol. 2017, 6, 2350–2361. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Wang, S.; Hu, G.; Guo, L.; Chen, X.; Xu, P.; Liu, L. Engineering Escherichia coli for malate production by integrating modular pathway characterization with CRISPRi-guided multiplexed metabolic tuning. Biotechnol. Bioeng. 2018, 115, 661–672. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.K.; Seong, W.; Han, G.H.; Lee, D.H.; Lee, S.G. CRISPR interference-guided multiplex repression of endogenous competing pathway genes for redirecting metabolic flux in Escherichia coli. Microb. Cell Fact. 2017, 16, 188. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.N.; Zhao, D.D.; Qiu, H.N.; Fan, F.Y.; Man, S.L.; Bi, C.H.; Zhang, X.L. The CRISPR/Cas9-facilitated multiplex pathway optimization (CFPO) technique and its application to improve the Escherichia coli xylose utilization pathway. Metab. Eng. 2017, 43, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.J.; Zhang, X.; Zhu, Y.J.; Tan, Q.Y.; He, J.C.; Dong, M.S. Rational modular design of metabolic network for efficient production of plant polyphenol pinosylvin. Sci. Rep. 2017, 7, 1459. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.J.; Zhou, P.; Zhang, X.; Dong, M.S. Efficient de novo synthesis of resveratrol by metabolically engineered Escherichia coli. J. Ind. Microbiol. Biotechnol. 2017, 44, 1083–1095. [Google Scholar] [CrossRef] [PubMed]
- Heo, M.J.; Jung, H.M.; Um, J.; Lee, S.W.; Oh, M.K. Controlling citrate synthase expression by CRISPR/Cas9 genome editing for n-butanol production in Escherichia coli. ACS Synth. Biol. 2017, 6, 182–189. [Google Scholar] [CrossRef] [PubMed]
- Della, M.; Palmbos, P.L.; Tseng, H.M.; Tonkin, L.M.; Daley, J.M.; Topper, L.M.; Pitcher, R.S.; Tomkinson, A.E.; Wilson, T.E.; Doherty, A.J. Mycobacterial Ku and ligase proteins constitute a two-component NHEJ repair machine. Science 2004, 306, 683–685. [Google Scholar] [CrossRef] [PubMed]
- Shuman, S.; Glickman, M.S. Bacterial DNA repair by non-homologous end joining. Nat. Rev. Microbiol. 2007, 5, 852–861. [Google Scholar] [CrossRef] [PubMed]
- Gong, C.; Bongiorno, P.; Martins, A.; Stephanou, N.C.; Zhu, H.; Shuman, S.; Glickman, M.S. Mechanism of nonhomologous end-joining in Mycobacteria: A low-fidelity repair system driven by Ku, ligase D and ligase C. Nat. Struct. Mol. Biol. 2005, 12, 304–312. [Google Scholar] [CrossRef] [PubMed]
- Arslan, Z.; Wurm, R.; Brener, O.; Ellinger, P.; Nagel-Steger, L.; Oesterhelt, F.; Schmitt, L.; Willbold, D.; Wagner, R.; Gohlke, H.; et al. Double-strand DNA end-binding and sliding of the toroidal CRISPR-associated protein Csn2. Nucleic Acids Res. 2013, 41, 6347–6359. [Google Scholar] [CrossRef] [PubMed]
- Di Bella, S.; Ascenzi, P.; Siarakas, S.; Petrosillo, N.; di Masi, A. Clostridium difficile toxins A and B: Insights into pathogenic properties and extraintestinal effects. Toxins 2016, 8, 134. [Google Scholar] [CrossRef] [PubMed]
- Bartlett, J.G.; Gerding, D.N. Clinical recognition and diagnosis of Clostridium difficile infection. Clin. Infect. Dis. 2008, 46, S12–S18. [Google Scholar] [CrossRef] [PubMed]
- Rossi, F.; Rizzotti, L.; Felis, G.E.; Torriani, S. Horizontal gene transfer among microorganisms in food: Current knowledge and future perspectives. Food Microbiol. 2014, 42, 232–243. [Google Scholar] [CrossRef] [PubMed]
- Pirazzini, M.; Rossetto, O.; Eleopra, R.; Montecucco, C. Botulinum neurotoxins: Biology, pharmacology, and toxicology. Pharmacol. Rev. 2017, 69, 200–235. [Google Scholar] [CrossRef] [PubMed]
- Pidot, S.; Ishida, K.; Cyrulies, M.; Hertweck, C. Discovery of clostrubin, an exceptional polyphenolic polyketide antibiotic from a strictly anaerobic bacterium. Angew. Chem. Int. Ed. 2014, 53, 7856–7859. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Blaschek, H.P. Optimization of butanol production from tropical maize stalk juice by fermentation with Clostridium beijerinckii NCIMB 8052. Bioresour. Technol. 2011, 102, 9985–9990. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, Z.T.; Seo, S.O.; Choi, K.J.; Lu, T.; Jin, Y.S.; Blaschek, H.P. Markerless chromosomal gene deletion in Clostridium beijerinckii using CRISPR/Cas9 system. J. Biotechnol. 2015, 200, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, P.; Deb, J.K. Gene expression systems in Corynebacteria. Protein Expr. Purif. 2005, 40, 221–229. [Google Scholar] [CrossRef] [PubMed]
- Becker, J.; Wittmann, C. Advanced biotechnology: Metabolically engineered cells for the bio-based production of chemicals and fuels, materials, and health-care products. Angew. Chem. Int. Ed. Engl. 2015, 54, 3328–3350. [Google Scholar] [CrossRef] [PubMed]
- Lindner, S.N.; Petrov, D.P.; Hagmann, C.T.; Henrich, A.; Kramer, R.; Eikmanns, B.J.; Wendisch, V.F.; Seibold, G.M. Phosphotransferase system-mediated glucose uptake is repressed in phosphoglucoisomerase-deficient Corynebacterium glutamicum strains. Appl. Environ. Microbiol. 2013, 79, 2588–2595. [Google Scholar] [CrossRef] [PubMed]
- Sawada, K.; Zen-in, S.; Wada, M.; Yokota, A. Metabolic changes in a pyruvate kinase gene deletion mutant of Corynebacterium glutamicum ATCC 13032. Metab. Eng. 2010, 12, 401–407. [Google Scholar] [CrossRef] [PubMed]
- Riedel, C.; Rittmann, D.; Dangel, P.; Mockel, B.; Petersen, S.; Sahm, H.; Eikmanns, B.J. Characterization of the phosphoenolpyruvate carboxykinase gene from Corynebacterium glutamicum and significance of the enzyme for growth and amino acid production. J. Mol. Microbiol. Biotechnol. 2001, 3, 573–583. [Google Scholar] [PubMed]
- Berdy, J. Bioactive microbial metabolites. J. Antibiot. 2005, 58, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Bibb, M.J. Regulation of secondary metabolism in Streptomycetes. Curr. Opin. Microbiol. 2005, 8, 208–215. [Google Scholar] [CrossRef] [PubMed]
- Seto, H.; Imai, S.; Tsuruoka, T.; Satoh, A.; Kojima, M.; Inouye, S.; Sasaki, T.; Otake, N. Studies on the biosynthesis of bialaphos (SF-1293). 1. Incorporation of 13C- and 2H-labeled precursors into bialaphos. J. Antibiot. 1982, 35, 1719–1721. [Google Scholar] [CrossRef] [PubMed]
- Otten, S.L.; Stutzman-Engwall, K.J.; Hutchinson, C.R. Cloning and expression of daunorubicin biosynthesis genes from Streptomyces peucetius and S. peucetius subsp. caesius. J. Bacteriol. 1990, 172, 3427–3434. [Google Scholar] [CrossRef] [PubMed]
- Yim, H.; Haselbeck, R.; Niu, W.; Pujol-Baxley, C.; Burgard, A.; Boldt, J.; Khandurina, J.; Trawick, J.D.; Osterhout, R.E.; Stephen, R.; et al. Metabolic engineering of Escherichia coli for direct production of 1,4-butanediol. Nat. Chem. Biol. 2011, 7, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.E.; Park, S.J.; Kim, W.J.; Kim, H.J.; Kim, B.J.; Lee, H.; Shin, J.; Lee, S.Y. One-step fermentative production of aromatic polyesters from glucose by metabolically engineered Escherichia coli strains. Nat. Commun. 2018, 9, 79. [Google Scholar] [CrossRef] [PubMed]
- Rude, M.A.; Schirmer, A. New microbial fuels: A biotech perspective. Curr. Opin. Microbiol. 2009, 12, 274–281. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Gu, Q.; Wang, W.; Wong, L.; Bower, A.G.; Collins, C.H.; Koffas, M.A. Modular optimization of multi-gene pathways for fatty acids production in E. coli. Nat. Commun. 2013, 4, 1409. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Li, Y.; Tang, Y. Engineered biosynthesis of bacterial aromatic polyketides in Escherichia coli. Proc. Natl. Acad. Sci. USA 2008, 105, 20683–20688. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Ranganathan, S.; Fowler, Z.L.; Maranas, C.D.; Koffas, M.A.G. Genome-scale metabolic network modeling results in minimal interventions that cooperatively force carbon flux towards malonyl-coA. Metab. Eng. 2011, 13, 578–587. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.K.; Chou, H.; Ham, T.S.; Lee, T.S.; Keasling, J.D. Metabolic engineering of microorganisms for biofuels production: From bugs to synthetic biology to fuels. Curr. Opin. Biotechnol. 2008, 19, 556–563. [Google Scholar] [CrossRef] [PubMed]
- Pleiss, J. The promise of synthetic biology. Appl. Microbiol. Biotechnol. 2006, 73, 735–739. [Google Scholar] [CrossRef] [PubMed]
- Dellomonaco, C.; Fava, F.; Gonzalez, R. The path to next generation biofuels: Successes and challenges in the era of synthetic biology. Microb. Cell Fact. 2010, 9, 3. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Li, L.Y.; Zhang, F.M.; Stephanopoulos, G.; Koffas, M. Improving fatty acids production by engineering dynamic pathway regulation and metabolic control. Proc. Natl. Acad. Sci. USA 2014, 111, 11299–11304. [Google Scholar] [CrossRef] [PubMed]
- Ajikumar, P.K.; Xiao, W.H.; Tyo, K.E.J.; Wang, Y.; Simeon, F.; Leonard, E.; Mucha, O.; Phon, T.H.; Pfeifer, B.; Stephanopoulos, G. Isoprenoid pathway optimization for taxol precursor overproduction in Escherichia coli. Science 2010, 330, 70–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maimone, T.J.; Baran, P.S. Modern synthetic efforts toward biologically active terpenes. Nat. Chem. Biol. 2007, 3, 396–407. [Google Scholar] [CrossRef] [PubMed]
- Pateraki, I.; Heskes, A.M.; Hamberger, B. Cytochromes p450 for terpene functionalisation and metabolic engineering. Adv. Biochem. Eng. Biotechnol. 2015, 148, 107–139. [Google Scholar] [CrossRef] [PubMed]
- Dahl, R.H.; Zhang, F.; Alonso-Gutierrez, J.; Baidoo, E.; Batth, T.S.; Redding-Johanson, A.M.; Petzold, C.J.; Mukhopadhyay, A.; Lee, T.S.; Adams, P.D.; et al. Engineering dynamic pathway regulation using stress-response promoters. Nat. Biotechnol. 2013, 31, 1039–1046. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.Z.; Rodriguez, S.; Keasling, J.D. Metabolic engineering of microbial pathways for advanced biofuels production. Curr. Opin. Biotechnol. 2011, 22, 775–783. [Google Scholar] [CrossRef] [PubMed]
- Castelli, G.; Bruno, F.; Vitale, F.; Roberti, M.; Colomba, C.; Giacomini, E.; Guidotti, L.; Cascio, A.; Tolomeo, M. In vitro antileishmanial activity of trans-stilbene and terphenyl compounds. Exp. Parasitol. 2016, 166, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Riviere, C.; Pawlus, A.D.; Merillon, J.M. Natural stilbenoids: Distribution in the plant kingdom and chemotaxonomic interest in Vitaceae. Nat. Prod. Rep. 2012, 29, 1317–1333. [Google Scholar] [CrossRef] [PubMed]
- Jancinova, V.; Perecko, T.; Nosal, R.; Harmatha, J.; Smidrkal, J.; Drabikova, K. The natural stilbenoid pinosylvin and activated neutrophils: Effects on oxidative burst, protein kinase C, apoptosis and efficiency in adjuvant arthritis. Acta Pharmacol. Sin. 2012, 33, 1285–1292. [Google Scholar] [CrossRef] [PubMed]
- Koskela, A.; Reinisalo, M.; Hyttinen, J.M.T.; Kaarniranta, K.; Karjalainen, R.O. Pinosylvin-mediated protection against oxidative stress in human retinal pigment epithelial cells. Mol. Vis. 2014, 20, 760–769. [Google Scholar] [PubMed]
- Date, A.A.; Destache, C.J. Natural polyphenols: Potential in the prevention of sexually transmitted viral infections. Drug Discov. Today 2016, 21, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Stemmer, M.; Thumberger, T.; Del Sol Keyer, M.; Wittbrodt, J.; Mateo, J.L. Correction: CCTop: An intuitive, flexible and reliable CRISPR/Cas9 target prediction tool. PLoS ONE 2017, 12, e0176619. [Google Scholar] [CrossRef] [PubMed]
- Montague, T.G.; Cruz, J.M.; Gagnon, J.A.; Church, G.M.; Valen, E. CHOPCHOP: A CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 2014, 42, W401–W407. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.D.; Scott, D.A.; Weinstein, J.A.; Ran, F.A.; Konermann, S.; Agarwala, V.; Li, Y.; Fine, E.J.; Wu, X.; Shalem, O.; et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013, 31, 827–832. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Mateos, M.A.; Vejnar, C.E.; Beaudoin, J.D.; Fernandez, J.P.; Mis, E.K.; Khokha, M.K.; Giraldez, A.J. CRISPRscan: Designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat. Methods 2015, 12, 982–988. [Google Scholar] [CrossRef] [PubMed]
- Heigwer, F.; Kerr, G.; Boutros, M. E-CRISP: Fast CRISPR target site identification. Nat. Methods 2014, 11, 122–123. [Google Scholar] [CrossRef] [PubMed]
- Wong, N.; Liu, W.; Wang, X. Wu-CRISPR: Characteristics of functional guide RNAs for the CRISPR/Cas9 system. Genome Biol. 2015, 16, 218. [Google Scholar] [CrossRef] [PubMed]
- Hodgkins, A.; Farne, A.; Perera, S.; Grego, T.; Parry-Smith, D.J.; Skarnes, W.C.; Iyer, V. Wge: A CRISPR database for genome engineering. Bioinformatics 2015, 31, 3078–3080. [Google Scholar] [CrossRef] [PubMed]
- Ronda, C.; Pedersen, L.E.; Hansen, H.G.; Kallehauge, T.B.; Betenbaugh, M.J.; Nielsen, A.T.; Kildegaard, H.F. Accelerating genome editing in CHO cells using CRISPR Cas9 and CRISPy, a web-based target finding tool. Biotechnol. Bioeng. 2014, 111, 1604–1616. [Google Scholar] [CrossRef] [PubMed]
- Perez, A.R.; Pritykin, Y.; Vidigal, J.A.; Chhangawala, S.; Zamparo, L.; Leslie, C.S.; Ventura, A. Guidescan software for improved single and paired CRISPR guide RNA design. Nat. Biotechnol. 2017, 35, 347–349. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.S.; Shen, B.; Zhang, C.B.; Huang, X.X.; Zhang, Y.L. sgRNAcas9: A software package for designing CRISPR sgRNA and evaluating potential off-target cleavage sites. PLoS ONE 2014, 9, e100448. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Pedersen, L.E.; Weber, T.; Lee, S.Y. Crispy-web: An online resource to design sgRNAs for CRISPR applications. Synth. Syst. Biotechnol. 2016, 1, 118–121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Wei, Z.; Dominguez, A.; Li, Y.; Wang, X.; Qi, L.S. CRISPR-era: A comprehensive design tool for CRISPR-mediated gene editing, repression and activation. Bioinformatics 2015, 31, 3676–3678. [Google Scholar] [CrossRef] [PubMed]
- Chari, R.; Yeo, N.C.; Chavez, A.; Church, G.M. SgRNA scorer 2.0: A species-independent model to predict CRISPR/Cas9 activity. ACS Synth. Biol. 2017, 6, 902–904. [Google Scholar] [CrossRef] [PubMed]
- Sidik, S.M.; Huet, D.; Lourido, S. CRISPR-Cas9-based genome-wide screening of Toxoplasma gondii. Nat. Protoc. 2018, 13, 307–323. [Google Scholar] [CrossRef] [PubMed]
- Lonowski, L.A.; Narimatsu, Y.; Riaz, A.; Delay, C.E.; Yang, Z.; Niola, F.; Duda, K.; Ober, E.A.; Clausen, H.; Wandall, H.H.; et al. Genome editing using FACS enrichment of nuclease-expressing cells and indel detection by amplicon analysis. Nat. Protoc. 2017, 12, 581–603. [Google Scholar] [CrossRef] [PubMed]
- Narimatsu, Y.; Joshi, H.J.; Zhang, Y.; Gomes, C.; Chen, Y.H.; Lorenzetti, F.; Furukawa, S.; Schjoldager, K.; Hansen, L.; Clausen, H.; et al. A validated gRNA library for CRISPR/Cas9 targeting of the human glycosyltransferase genome. Glycobiology 2018. [Google Scholar] [CrossRef] [PubMed]
- Pusapati, G.V.; Kong, J.H.; Patel, B.B.; Krishnan, A.; Sagner, A.; Kinnebrew, M.; Briscoe, J.; Aravind, L.; Rohatgi, R. CRISPR screens uncover genes that regulate target cell sensitivity to the morphogen sonic hedgehog. Dev. Cell 2018, 44, 113–129. [Google Scholar] [CrossRef] [PubMed]
- Najm, F.J.; Strand, C.; Donovan, K.F.; Hegde, M.; Sanson, K.R.; Vaimberg, E.W.; Sullender, M.E.; Hartenian, E.; Kalani, Z.; Fusi, N.; et al. Orthologous CRISPR-Cas9 enzymes for combinatorial genetic screens. Nat. Biotechnol. 2018, 36, 179–189. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Choe, D.; Lee, E.; Kim, S.C.; Palsson, B.; Cho, B.K. High-level dcas9 expression induces abnormal cell morphology in Escherichia coli. ACS Synth. Biol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Li, X.T.; Jun, Y.; Erickstad, M.J.; Brown, S.D.; Parks, A.; Court, D.L.; Jun, S. tCRISPRi: Tunable and reversible, one-step control of gene expression. Sci. Rep. 2016, 6, 39076. [Google Scholar] [CrossRef] [PubMed]
- Li, X.T.; Sou, C.; Jun, S. Protocol for construction of a tunable CRISPR interference (tCRISPRi) strain for Escherichia coli. Bio-Protocol 2017, 7. [Google Scholar] [CrossRef] [PubMed]





| Strains | Applications | Year | Ref. |
|---|---|---|---|
| Bacillus | |||
| B. smithii | Genome deletion (90%), knockout (100%), insertion (20%) | 2017 | [41] |
| B. subtilis | DSB, non-homologous end-joining (NHEJ) repair | 2017 | [42] |
| B. subtilis | Deletion, Point mutation | 2016 | [43] |
| B. subtilis | CRISPRi library, Chemical genomics | 2016 | [44] |
| B. smithii | Genome editing and silencing, with ThermoCas9 (active at 55 °C) | 2017 | [45] |
| B. licheniformis | Genome deletion (1 kb) (100%), Two gene deletion (11.6%), Large gene deletion BacABC (42.7 kb) (79.0%), Gene insertion (75.5%) | 2018 | [46] |
| Clostridium | |||
| C. difficile | Genome editing | 2017 | [47] |
| C. botulinum | Genome editing | 2017 | [48] |
| C. acetobutylicum | Gene substitution, Deletion, Insertion up to 3.6 kb | 2017 | [49] |
| C. saccharoper -butylacetonicum | Gene deletion (pta), Butanol production | 2017 | [50] |
| C. beijerinckii | CRISPRi | 2016 | [51] |
| C. autoethanogenum | Genome deletion | 2016 | [52] |
| C. acetobutylicum | CRISPRi, Genome deletion (20 bp) | 2016 | [53] |
| C. beijerinckii | CRISPRi, Genome deletion (20–1149 bp) | 2016 | [53] |
| C. pasteurianum | Genome editing | 2016 | [54] |
| C. beijerinckii | Genome editing | 2016 | [55] |
| Corynebacterium | |||
| C. glutamicum | CRISPRi, pyc, gltA, idsA, glgC, idsA-glgC | 2018 | [56] |
| C. glutamicum | Genome deletion (60.0%), Insertion (62.5%), Modification (80%) | 2017 | [57] |
| C. glutamicum | Deletion (porB, mepA, clpX and NcgI0911), Insertion | 2017 | [58] |
| C. glutamicum | Genome deletion (up to 100%), Gamma-aminobutyric acid (GABA) production | 2017 | [59] |
| C. glutamicum | Genome editing (86–100%) | 2017 | [12] |
| C. glutamicum | CRISPRi (98%) | 2016 | [60] |
| Lactobacillus | |||
| Lactobacillus casei. | Genome editing | 2017 | [61] |
| Lactobacillus gassen | 17 strains, Optimization for CRISPR/Cas activity | 2015 | [62] |
| Lactobacillus reuteri | Site-directed mutagenesis (efficiency 90~100%) | 2014 | [63] |
| Mycobacterium | |||
| M. tuberculosis | CRISPRi | 2017 | [64] |
| M. tuberculosis | CRISPRi | 2016 | [65] |
| Psedomonas | |||
| P. putida | Genome editing, CRISPRi | 2017 | [45] |
| P. aeruginosa | CRISPRi | 2018 | [66] |
| P. putida | CRISPRi | 2018 | [66] |
| P. fluorescens | CRISPRi | 2018 | [66] |
| Staphylococcus | |||
| S. aureus | Type III-A CRISPR/Cas system | 2017 | [67] |
| S. aureus | Genome deletion, Insertion, Single mutation | 2017 | [68] |
| Streptomyces | |||
| S. lividans | Genome deletion (20 bp, 34 bp, 20–34 bp, 31,415 bp) (70 to 100%) | 2015 | [69] |
| S. viridochromogenes | Genome deletion (20 bp) (100%), 23 bp (67%) | 2015 | [69] |
| S. albus | Genome deletion (67 bp (100%) and 13214 bp (67%)) | 2015 | [69] |
| S. coelicolor A3 | Deletion (100%), CRISPRi | 2015 | [19] |
| S. coelicolor A3 | NHEJ with LigD ligase coexpression | 2015 | [19] |
| S. coelicolor A3 | HDR, Gene deletion | 2015 | [19] |
| S. avermitilis | Type I-E system | 2016 | [70] |
| S. albus | Knock in | 2017 | [71] |
| S. lividans | Knock in | 2017 | [71] |
| S. roseosporus | Knock in | 2017 | [71] |
| S. venezuelae | Knock in | 2017 | [71] |
| S. viridochromogenes | Knock in | 2017 | [71] |
| S. rimosus | Antibiotics (Oxytetracycline) production | 2017 | [72] |
| Escherichia | |||
| E. coli | Programmable DNA looping (15%, 4.7 kb loop) | 2017 | [73] |
| E. coli | Point mutations, Deletions, Insertions, Gene replacements | 2017 | [74] |
| E. coli | CRISPR/Cas-assisted MAGE | 2017 | [75] |
| E. coli | CRISPRi, (2S)-naringenin, Carbon flux toward malonyl-CoA, (7.4 fold, 421.6 mg/mL) | 2015 | [76] |
| E. coli | Genome editiong, β-carotene (2.0 g/L) (Central metabolic pathways, MEP pathway) | 2015 | [77] |
| E. coli | CRISPRi, mevalonate production by dnaA (2.1-fold), oriC (1.8 fold), pyrF (3.5 fold), thyA (1.8-fold) deletion | 2016 | [78] |
| E. coli | CRISPRi, lycopen production in MVA pathway (mvaK1, mvaE) (9-fold) | 2016 | [79] |
| E. coli | CRISPRi, Isoprene production in lycopen pathway (ispA) (2.6-fold) | 2016 | [79] |
| E. coli | CRISPRi, 4,4′-dihydroxybiphenyl (4HB) production, P(3HB-co-4HB) biosynthesis pathway (sad1, sucD, sucC, sdhA, sdhB) (13.1-fold) | 2015 | [80] |
| E. coli | CRISPRi, Pinosylvin, malonyl-CoA pathway (fabD) (1.9-fold) | 2016 | [81] |
| E. coli | CRISPRi, O-methylated anthocyanin, Methionine biosynthetic pathway (metJ) (2-fold) | 2017 | [82] |
| E. coli | CRISPRi, 1,4-Butanediol (1,4-BDO) production, 1,4-BDO biosynthesis pathway (gabD, ybgC, tesB) (1.8 g/L) | 2017 | [83] |
| E. coli | CRISPRi, Malate production, Glyoxylate pathway (pyc from A. flavus, gltA, acnB, aceA, aceB from S. coelicolor) (2.3-fold) | 2017 | [84] |
| E. coli | CRISPRi, n-butanol production, pta, frdA, ldhA, and adhE (3.2 fold) | 2017 | [85] |
| E. coli | CRISPR, Xylose production, Xylose pathway (xylA, xylB, tktA, talB) | 2017 | [86] |
| E. coli | CRISPRi, Pinosylvin production, Malonyl-CoA pathway (eno, adhE, fabB, sucC, fumC, fabF) (1.7-fold) | 2017 | [87] |
| E. coli | CRISPRi, Resveratrol production, Malonyl-CoA pathway (fabD, fabH, fabB, fabF, fabI) (6-fold) | 2017 | [88] |
| E. coli | CRISPR, n-butanol production, gltA | 2017 | [89] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cho, S.; Shin, J.; Cho, B.-K. Applications of CRISPR/Cas System to Bacterial Metabolic Engineering. Int. J. Mol. Sci. 2018, 19, 1089. https://doi.org/10.3390/ijms19041089
Cho S, Shin J, Cho B-K. Applications of CRISPR/Cas System to Bacterial Metabolic Engineering. International Journal of Molecular Sciences. 2018; 19(4):1089. https://doi.org/10.3390/ijms19041089
Chicago/Turabian StyleCho, Suhyung, Jongoh Shin, and Byung-Kwan Cho. 2018. "Applications of CRISPR/Cas System to Bacterial Metabolic Engineering" International Journal of Molecular Sciences 19, no. 4: 1089. https://doi.org/10.3390/ijms19041089