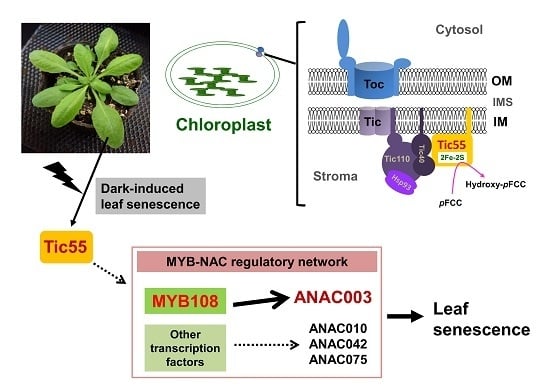
The Direct Involvement of Dark-Induced Tic55 Protein in Chlorophyll Catabolism and Its Indirect Role in the MYB108-NAC Signaling Pathway during Leaf Senescence in Arabidopsis thaliana
Abstract
:1. Introduction
2. Results and Discussion
2.1. Molecular Characterization and Phenotype of T-DNA Insertion Mutant tic55-II
2.2. The Relationship between Tic55 and Other Translocon Proteins
2.3. Possible Novel Biological Function of Tic55 in the Aging of A. thaliana
2.4. Microarray Gene Expression Analysis in IDLs during Leaf Senescence of A. thaliana
2.5. Functional Categorization of log2-Fold Induced and Repressed Genes in the IDLs Based on Microarray Analysis
2.6. Confirmation of Microarray Results by Real-Time qRT-PCR
2.7. Sequence Alignment and Phylogenetic Analysis of ANAC TFs
2.8. MYB-NAC Linked Regulatory Pathway and Leaf Senescence
3. Materials and Methods
3.1. Plant Materials and Growth Conditions
3.2. Preparation of Chloroplasts and Chloroplast Subfractions
3.3. Protein Gel Blot Analysis
3.4. Coimmunoprecipitation (Co-IP) Analysis
3.5. Dark Treatment and Chlorophyll Concentration Analysis
3.6. Hybridization of A. thaliana Microarrays and Data Analysis
3.7. Relative Real-Time qRT-PCR and Microarray Validation
3.8. Transactivation Assays in Yeast Cells
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| Toc and Tic | Translocon at the outer/inner membrane of chloroplasts |
| IDLs | individually darkened leaves |
| ASP3 | aspartate aminotransferase 3 |
| APG7 | autophagy-related 7, ubiquitin-like modifier-activating enzyme ATG7 |
| DIN2 | dark inducible 2, beta-glucosidase 30 |
| DIN11 | dark inducible 11,2-oxoacid-dependent dioxygenase-like protein |
| SAG12 | senescence-associated gene 12, cysteine protease |
| SAG13 | senescence-associated gene 13, senescence-associated protein |
| YLS9 | yellow-leaf-specific gene 9, protein NDR1/HIN1-like 10 |
| ABRC | Arabidopsis Biological Resource Center |
| NBT | nitro blue tetrazolium |
| BCIP | bromochloroindoyl phosphate |
References
- Chou, M.L.; Fitzpatric, L.M.; Tu, S.L.; Budziszewski, G.; Potter-Lewis, S.; Akita, M.; Levin, J.Z.; Keegstra, K.; Li, H.-M. Tic40, a membrane-anchored co-chaperone homolog in the chloroplast protein translocon. EMBO J. 2003, 22, 2970–2980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inaba, T.; Li, M.; Alvarez-Huerta, M.; Kessler, F.; Schnell, D.J. atTic110 functions as a scaffold for coordinating the stromal events of protein import into chloroplasts. J. Biol. Chem. 2003, 278, 38617–38627. [Google Scholar] [CrossRef] [PubMed]
- Chou, M.L.; Chu, C.C.; Chen, L.J.; Akita, M.; Li, H.M. Stimulation of transit-peptide release and ATP hydrolysis by a cochaperone during protein import into chloroplasts. J. Cell Biol. 2006, 175, 893–900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benz, J.P.; Soll, J.; Bölter, B. Protein transport in organelles: The composition, function and regulation of the Tic complex in chloroplast protein import. FEBS J. 2009, 276, 1166–1176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kikuchi, S.; Oishi, M.; Hirabayashi, Y.; Lee, D.W.; Hwang, I.; Nakai, M. A 1-megadalton translocation complex containing Tic20 and Tic21 mediates chloroplast protein import at the inner envelope membrane. Plant Cell 2009, 21, 1781–1797. [Google Scholar] [CrossRef] [PubMed]
- Su, P.H.; Li, H.M. Stromal Hsp70 is important for protein translocation into pea and Arabidopsis chloroplasts. Plant Cell 2010, 22, 1516–1531. [Google Scholar] [CrossRef] [PubMed]
- Inoue, H.; Li, M.; Schnell, D.J. An essential role for chloroplast heat shock protein 90 (Hsp90C) in protein import into chloroplasts. Proc. Natl. Acad. Sci. USA 2013, 110, 3173–3178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bölter, B.; Soll, J.; Schwenkert, S. Redox meets protein trafficking. Biochim. Biophys. Acta 2015, 1847, 949–956. [Google Scholar] [CrossRef] [PubMed]
- Nakai, M. The Tic complex uncovered: The alternative view on the molecular mechanism of protein translocation across the inner envelope membrane of chloroplasts. Biochim. Biophys. Acta 2015, 1847, 957–967. [Google Scholar] [CrossRef] [PubMed]
- Li, H.M.; Chiu, C.C. Protein transport into chloroplasts. Annu. Rev. Plant Biol. 2010, 61, 157–180. [Google Scholar] [CrossRef] [PubMed]
- Kovács-Bogdán, E.; Benz, J.P.; Soll, J.; Bölter, B. Tic20 forms a channel independent of Tic110 in chloroplasts. BMC Plant Biol. 2011, 11, 133–148. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, S.; Bedard, J.; Hirano, M.; Hirabayashi, Y.; Oishi, M.; Imai, M.; Takase, M.; Ide, T.; Nakai, M. Uncovering the protein translocon at the chloroplast inner envelope membrane. Science 2013, 339, 571–574. [Google Scholar] [CrossRef] [PubMed]
- Balsera, M.; Goetze, T.A.; Kovacs-Bogdan, E.; Schurmann, P.; Wagner, R.; Buchanan, B.B.; Soll, J.; Bölter, B. Characterization of Tic110, a channel-forming protein at the inner envelope membrane of chloroplasts, unveils a response to Ca2+ and a stromal regulatory disulfide bridge. J. Biol. Chem. 2009, 284, 2603–2616. [Google Scholar] [CrossRef] [PubMed]
- Hirohashi, T.; Hase, T.; Nakai, M. Maize non-photosynthetic ferredoxin precursor is mis-sorted to the intermembrane space of chloroplasts in the presence of light. Plant Physiol. 2001, 125, 2154–2163. [Google Scholar] [CrossRef] [PubMed]
- Stengel, A.; Benz, J.P.; Soll, J.; Bölter, B. Redox-regulation of protein import into chloroplasts and mitochondria: Similarities and differences. Plant Signal. Behav. 2010, 5, 105–109. [Google Scholar] [CrossRef] [PubMed]
- Stengel, A.; Benz, J.P.; Balsera, M.; Soll, J.; Bölter, B. Tic62 redox-regulated translocon composition and dynamics. J. Biol. Chem. 2008, 283, 6656–6667. [Google Scholar] [CrossRef] [PubMed]
- Chigri, F.; Hörmann, F.; Stamp, A.; Stammers, D.K.; Bölter, B.; Soll, J.; Vothknecht, U.C. Calcium regulation of chloroplast translocation is mediated by calmodulin binding to Tic32. Proc. Natl. Acad. Sci. USA 2006, 103, 16051–16056. [Google Scholar] [CrossRef] [PubMed]
- Hormann, F.; Kuchler, M.; Sveshnikov, D.; Oppermann, U.; Li, Y.; Soll, J. Tic32, an essential component in chloroplast biogenesis. J. Biol. Chem. 2004, 279, 34756–34762. [Google Scholar] [CrossRef] [PubMed]
- Bartsch, S.; Monnet, J.; Selbach, K.; Quigley, F.; Gray, J.; von Wettstein, D.; Reinbothe, S.; Reinbothe, C. Three thioredoxin targets in the inner envelope membrane of chloroplasts function in protein import and chlorophyll metabolism. Proc. Natl. Acad. Sci. USA 2008, 105, 4933–4938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benz, J.P.; Stengel, A.; Lintala, M.; Lee, Y.H.; Weber, A.; Philippar, K.; Gügel, I.L.; Kaieda, S.; Ikegami, T.; Mulo, P.; et al. Arabidopsis Tic62 and ferredoxin-NADP(H) oxidoreductase form light-regulated complexes that are integrated into the chloroplast redox poise. Plant Cell 2009, 21, 3965–3983. [Google Scholar] [CrossRef] [PubMed]
- Boij, P.; Patel, R.; Garcia, C.; Jarvis, P.; Aronsson, H. In vivo studies on the roles of Tic55-related proteins in chloroplast protein import in Arabidopsis thaliana. Mol. Plant 2009, 2, 1397–1409. [Google Scholar] [CrossRef] [PubMed]
- Hauenstein, M.; Christ, B.; Das, A.; Aubry, S.; Hörtensteiner, S. A Role for Tic55 as a hydroxylase of phyllobilins, the products of chlorophyll breakdown during plant senescence. Plant Cell 2016, 28, 2510–2527. [Google Scholar] [CrossRef] [PubMed]
- Lim, P.O.; Woo, H.R.; Nam, H.G. Molecular genetics of leaf senescence in Arabidopsis. Trends Plant Sci. 2003, 8, 272–278. [Google Scholar] [CrossRef]
- Huang, Y.; Li, T.; Xu, Z.S.; Wang, F.; Xiong, A.S. Six NAC transcription factors involved in response to TYLCV infection in resistant and susceptible tomato cultivars. Plant Physiol. Biochem. 2017, 120, 61–74. [Google Scholar] [CrossRef] [PubMed]
- Lim, P.O.; Kim, H.J.; Nam, H.G. Leaf senescence. Annu. Rev. Plant Biol. 2007, 58, 115–136. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Gan, S. Convergence and divergence in gene expression profiles induced by leaf senescence and 27 senescence-promoting hormonal, pathological and environmental stress treatments. Plant Cell Environ. 2012, 35, 644–655. [Google Scholar] [CrossRef] [PubMed]
- Sakuraba, Y.; Jeong, J.; Kang, M.Y.; Kim, J.; Paek, N.C.; Choi, G. Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat. Commun. 2014, 5, 4636–4648. [Google Scholar] [CrossRef] [PubMed]
- Thomas, H.; Schellenberg, M.; Vicentini, F.; Matile, P. Gregor Mendel’s green and yellow pea seeds Bot. Acta 1996, 109, 3–4. [Google Scholar]
- Buchanan-Wollaston, V.; Earl, S.; Harrison, E.; Mathas, E.; Navabpour, S.; Page, T.; Pink, D. The molecular analysis of leaf senescence—A genomics approach. Plant Biotechnol. J. 2003, 1, 3–22. [Google Scholar] [CrossRef] [PubMed]
- Kilian, J.; Whitehead, D.; Horak, J.; Wanke, D.; Weinl, S.; Batistic, O.; D’Angelo, C.; Bornberg-Bauer, E.; Kudla, J.; Harter, K. The AtGenExpress global stress expression data set: Protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 2007, 50, 347–363. [Google Scholar] [CrossRef] [PubMed]
- Puranik, S.; Sahu, P.P.; Srivastava, P.S.; Prasad, M. NAC proteins: Regulation and role in stress tolerance. Trends Plant. Sci. 2012, 17, 369–381. [Google Scholar] [CrossRef] [PubMed]
- Hickman, R.; Hill, C.; Penfold, C.A.; Breeze, E.; Bowden, L.; Moore, J.D.; Zhang, P.; Jackson, A.; Cooke, E.; Bewicke-Copley, F.; et al. A local regulatory network around three NAC transcription factors in stress responses and senescence in Arabidopsis leaves. Plant J. 2013, 75, 26–39. [Google Scholar] [CrossRef] [PubMed]
- Ooka, H.; Satoh, K.; Doi, K.; Nagata, T.; Otomo, Y.; Murakami, K.; Matsubara, K.; Osato, N.; Kawai, J.; Carninci, P.; et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 2003, 10, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Fujita, M.; Fujita, Y.; Maruyama, K.; Seki, M.; Hiratsu, K.; Ohme-Takagi, M.; Tran, L.-S.P.; Yamaguchi-Shinozaki, K.; Shinozaki, K. A dehydration- induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 2004, 39, 863–876. [Google Scholar] [CrossRef] [PubMed]
- Tran, L.S.; Nakashima, K.; Sakuma, Y.; Simpson, S.D.; Fujita, Y.; Maruyama, K.; Fujita, M.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Isolation and functional analysis of Arabidopsis stress inducible NAC transcription factors that bind to a drought responsive cis-element in the early responsive to dehydration stress promoter. Plant Cell 2004, 16, 2481–2498. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Gan, S. AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J. 2006, 46, 601–612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, H.; Dai, M.; Yao, J.; Xiao, B.; Li, X.; Zhang, Q.; Xiong, L. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc. Natl. Acad. Sci. USA 2006, 103, 12987–12992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balazadeh, S.; Siddiqui, H.; Allu, A.D.; Matallana-Ramirez, L.P.; Caldana, C.; Mehrnia, M.; Zanor, M.I.; Kohler, B.; Mueller-Roeber, B. A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J. 2010, 62, 250–264. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Takasaki, H.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. NAC transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 2012, 1819, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Aida, M.; Ishida, T.; Fukaki, H.; Fujisawa, H.; Tasaka, M. Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell 1997, 9, 841–857. [Google Scholar] [CrossRef] [PubMed]
- Ernst, H.A.; Olsen, A.N.; Larsen, S.; Lo Leggio, L. Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Rep. 2004, 5, 297–303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jensen, M.K.; Kjaersgaard, T.; Nielsen, M.M.; Galberg, P.; Petersen, K.; O’Shea, C.; Skriver, K. The Arabidopsis thaliana NAC transcription factor family: Structure–function relationships and determinants of ANAC019 stress signalling. Biochem. J. 2010, 426, 183–196. [Google Scholar] [CrossRef] [PubMed]
- Tran, L.S.; Nishiyama, R.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Potential utilization of NAC transcription factors to enhance abiotic stress tolerance in plants by biotechnological approach. GM Crops 2010, 1, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Woo, H.R.; Kim, H.J.; Nam, H.G.; Lim, P.O. Plant leaf senescence and death—Regulation by multiple layers of control and implications for aging in general. J. Cell Sci. 2013, 126, 4823–4833. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Nam, H.G.; Lim, P.O. Regulatory network of NAC transcription factors in leaf senescence. Curr. Opin. Plant Biol. 2016, 33, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Breeze, E.; Harrison, E.; McHattie, S.; Hughes, L.; Hickman, R.; Hill, C.; Kiddle, S.; Kim, Y.S.; Penfold, C.A.; Jenkins, D.; et al. High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 2011, 23, 873–894. [Google Scholar] [CrossRef] [PubMed]
- Penfold, C.A.; Buchanan-Wollaston, V. Modelling transcriptional networks in leaf senescence. J. Exp. Bot. 2014, 65, 3859–3873. [Google Scholar] [CrossRef] [PubMed]
- Leibsch, D.; Keech, O. Dark-induced leaf senescence: New insights into a complex light-dependent regulatory pathway. New Phytol. 2016, 212, 563–570. [Google Scholar] [CrossRef] [PubMed]
- Swarbreck, D.; Wilks, C.; Lamesch, P.; Berardini, T.Z.; Garcia-Hernandez, M.; Foerster, H.; Li, D.; Meyer, T.; Muller, R.; Ploetz, L.; et al. The Arabidopsis Information Resource (TAIR): Gene structure and function annotation. Nucleic Acids Res. 2008, 36, D1009–D1014. [Google Scholar] [CrossRef] [PubMed]
- Lamesch, P.; Berardini, T.Z.; Li, D.; Swarbreck, D.; Wilks, C.; Sasidharan, R.; Muller, R.; Dreher, K.; Alexander, D.L.; Garcia-Hernandez, M.; et al. The Arabidopsis Information Resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res. 2012, 40, D1202–D1210. [Google Scholar] [CrossRef] [PubMed]
- Schultz, C.J.; Coruzzi, G.M. The aspartate aminotransferase gene family of Arabidopsis encodes isoenzymes localized to three distinct subcellular compartments. Plant J. 1995, 7, 61–75. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.D.; Arteca, R.N.; Pell, E.J. Senescence-associated gene expression during ozone-induced leaf senescence in Arabidopsis. Plant Physiol. 1999, 120, 1015–1024. [Google Scholar] [CrossRef] [PubMed]
- Noh, Y.S.; Amasino, R.M. Identification of a promoter region responsible for the senescence-specific expression of SAG12. Plant Mol. Biol. 1999, 41, 181–194. [Google Scholar] [CrossRef] [PubMed]
- Fujiki, Y.; Yoshikawa, Y.; Sato, T.; Inada, N.; Ito, M.; Nishida, I.; Watanabe, A. Dark-inducible genes from Arabidopsis thaliana are associated with leaf senescence and repressed by sugars. Physiol. Plant 2001, 111, 345–352. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, S.; Ito, M.; Nishida, I.; Watanabe, A. Isolation and RNA gel blot analysis of genes that could serve as potential molecular markers for leaf senescence in Arabidopsis thaliana. Plant Cell Physiol. 2001, 42, 170–178. [Google Scholar] [CrossRef] [PubMed]
- Doelling, J.H.; Walker, J.M.; Friedman, E.M.; Thompson, A.R.; Vierstra, R.D. The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J. Biol. Chem. 2002, 277, 33105–33114. [Google Scholar] [CrossRef] [PubMed]
- Huynh, L.N.; Vantoai, T.; Streeter, J.; Banowetz, G. Regulation of flooding tolerance of SAG12:ipt Arabidopsis plants by cytokinin. J. Exp. Bot. 2005, 56, 1397–1407. [Google Scholar] [CrossRef] [PubMed]
- Otegui, M.S.; Noh, Y.S.; Martínez, D.E.; Vila Petroff, M.G.; Staehelin, L.A.; Amasino, R.M.; Guiamet, J.J. Senescence-associated vacuoles with intense proteolytic activity develop in leaves of Arabidopsis and soybean. Plant J. 2005, 41, 831–844. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Gan, S. Leaf senescence: Signals, execution, and regulation. Curr. Top. Dev. Biol. 2005, 71, 83–112. [Google Scholar] [PubMed]
- Inaba, T.; Alvarez-Huerta, M.; Li, M.; Bauer, J.; Ewers, C.; Kessler, F.; Schnell, D.J. Arabidopsis Tic110 is essential for the assembly and function of the protein import machinery of plastids. Plant Cell 2005, 17, 1482–1496. [Google Scholar] [CrossRef] [PubMed]
- Caliebe, A.; Grimm, R.; Kaiser, G.; Lubeck, J.; Soll, J.; Heins, L. The chloroplastic protein import machinery contains a Rieske-type iron-sulfur cluster and a mononuclear iron-binding protein. EMBO J. 1997, 16, 7342–7350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Küchler, M.; Decker, S.; Hörmann, F.; Soll, J.; Heins, L. Protein import into chloroplasts involves redox-regulated proteins. EMBO J. 2002, 21, 6136–6145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, R.; Hirashima, M.; Satoh, S.; Tanaka, A. The Arabidopsis-accelerated cell death gene ACD1 is involved in oxygenation of pheophorbide a: Inhibition of the pheophorbide a oxygenase activity does not lead to the “stay-green” phenotype in Arabidopsis. Plant Cell Physiol. 2003, 44, 1266–1274. [Google Scholar] [CrossRef] [PubMed]
- Gray, J.; Close, P.S.; Briggs, S.P.; Johal, G.S. A novel suppressor of cell death in plants encoded by the Lls1 gene of maize. Cell 1997, 89, 25–31. [Google Scholar] [CrossRef]
- Gray, J.; Wardzala, E.; Yang, M.; Reinbothe, S.; Haller, S.; Pauli, F. A small family of LLS1-related non-heme oxygenases in plants with an origin amongst oxygenic photosynthesizers. Plant Mol. Biol. 2004, 54, 39–54. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Ito, Y.; Yamaguchishinozaki, K. Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol. 2009, 149, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Pontier, D.; Gan, S.; Amasino, R.M.; Roby, D.; Lam, E. Markers for hypersensitive response and senescence show distinct patterns of expression. Plant Mol. Biol. 1999, 39, 1243–1255. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.Y.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Podzimska-Sroka, D.; O’Shea, C.; Gregersen, P.L.; Skriver, K. NAC transcription factors in senescence: From molecular structure to function in crops. Plants 2015, 4, 412–448. [Google Scholar] [CrossRef] [PubMed]
- Fujita, M.; Fujita, Y.; Noutoshi, Y.; Takahashi, F.; Narusaka, Y.; Yamaquchi-Shinozaki, K.; Shinozaki, K. Crosstalk between abiotic and biotic stress responses: A current view from the points of convergence in the stress signaling networks. Curr. Opin. Plant Biol. 2006, 9, 436–442. [Google Scholar] [CrossRef] [PubMed]
- Bu, Q.; Jiang, H.; Li, C.B.; Zhai, Q.; Zhang, J.; Wu, X.; Sun, J.; Xie, Q.; Li, C. Role of the Arabidopsis thaliana NAC transcription factors ANAC019 and ANAC055 in regulating jasmonic acid signaled defense responses. Cell Res. 2008, 18, 756–767. [Google Scholar] [CrossRef] [PubMed]
- Solano, R.; Nieto, C.; Avila, J.; Canas, L.; Diaz, I.; Paz-Ares, J. Dual DNA binding specificity of a petal epidermis-specific MYB transcription factor (MYB.Ph3) from Petunia hybrid. EMBO J. 1995, 14, 1773–1784. [Google Scholar] [PubMed]
- Mengiste, T.; Chen, X.; Salmeron, J.; Dietrich, R. The BOTRYTIS SUSCEPTIBLE1 gene encodes an R2R3MYB transcription factor protein that is required for biotic and abiotic stress responses in Arabidopsis. Plant Cell 2003, 15, 2551–2565. [Google Scholar] [CrossRef] [PubMed]
- Mandaokar, A.; Browse, J. MYB108 acts together with MYB24 to regulate jasmonate-mediated stamen maturation in Arabidopsis. Plant Physiol. 2009, 149, 851–862. [Google Scholar] [CrossRef] [PubMed]
- Chow, C.N.; Zheng, H.Q.; Wu, N.Y.; Chien, C.H.; Huang, H.D.; Lee, T.Y.; Chiang-Hsieh, Y.F.; Hou, P.F.; Yang, T.Y.; Chang, W.C. PlantPAN 2.0: An update of plant promoter analysis navigator for reconstructing transcriptional regulatory networks in plants. Nucleic Acids Res. 2016, 44, 1154–1160. [Google Scholar] [CrossRef] [PubMed]
- Budziszewski, G.J.; Lewis, S.P.; Glover, L.W.; Reineke, J.; Jones, G.; Ziemnik, L.S.; Lonowski, J.; Nyfeler, B.; Aux, G.; Zhou, Q.; et al. Arabidopsis genes essential for seedling viability: Isolation of insertional mutants and molecular cloning. Genetics 2001, 159, 1765–1778. [Google Scholar] [PubMed]
- Aronsson, H.; Jarvis, P. A simple method for isolating import-competent Arabidopsis chloroplasts. FEBS Lett. 2002, 529, 215–220. [Google Scholar] [CrossRef]
- Waegemann, K.; Soll, J. Characterization of the protein import apparatus in isolated outer envelopes of chloroplasts. Plant J. 1991, 1, 149–158. [Google Scholar] [CrossRef] [Green Version]
- Fitzpatrick, L.M.; Keegstra, K. A method for isolating a high yield of Arabidopsis chloroplasts capable of efficient import of precursor proteins. Plant J. 2001, 26, 59–66. [Google Scholar] [CrossRef]
- Li, H.M.; Chen, L.J. Protein targeting and integration signal for the chloroplastic outer envelope membrane. Plant Cell 1996, 8, 2117–2126. [Google Scholar] [CrossRef] [PubMed]
- Harlow, E.; Lane, D. Antibodies: A Laboratory Manual; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 1988. [Google Scholar]
- Wada, S.; Ishida, H.; Izumi, M.; Yoshimoto, K.; Ohsumi, Y.; Mae, T.; Makino, A. Autophagy plays a role in chloroplast degradation during senescence in individually darkened leaves. Plant Physiol. 2008, 149, 885–893. [Google Scholar] [CrossRef] [PubMed]
- Porra, R.J. The chequered history of the development and use of simultaneous equations for the accurate determination of chlorophylls a and b. Photosynth. Res. 2002, 73, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]









| Locus ID | GO Term | Annotation | Up or Down | Fold Change (tic55-II/WT) |
|---|---|---|---|---|
| NM_180632 | GO:0007568~aging GO:0010149~senescence GO:0010150~leaf senescence | Arabidopsis thaliana epithiospecifier protein mRNA (ESP) | Up | 2.452 |
| NM_124040 | GO:0007568~aging | Arabidopsis thaliana tetraspanin family protein mRNA (TRN2) | Up | 2.017 |
| NM_001036860 | GO:0007568~aging GO:0010149~senescence | Arabidopsis thaliana vegetative storage protein 2 mRNA (VSP2) | Up | 2.009 |
| NM_121190 | GO:0007568~aging GO:0010149~senescence GO:0010150~leaf senescence | Arabidopsis thaliana aspartate 3 aminotransferase mRNA (ASP3) | Down | −2.107 |
| NM_129157 | GO:0007568~aging GO:0010149~senescence GO:0010150~leaf senescence | Arabidopsis thaliana late embryogenesis abundant hydroxyproline-rich glycoprotein mRNA (YLS9) | Down | −2.52 |
| NM_123958 | GO:0007568~aging GO:0010149~senescence GO:0010150~leaf senescence | Arabidopsis thaliana ubiquitin-like modifier-activating enzyme atg7 mRNA (APG7) | Down | −2.533 |
| NM_201829 | GO:0007568~aging | Arabidopsis thaliana senescence-associated protein 13 mRNA (SAG13) | Down | −3.353 |
| NM_115877 | GO:0007568~aging | Arabidopsis thaliana beta-glucosidase 30 mRNA (DIN2) | Down | −3.735 |
| NM_115877 | GO:0007568~aging | Arabidopsis thaliana 2-oxoacid-dependent dioxygenase-like protein DIN11 mRNA (DIN11) | Down | −3.735 |
| NM_123957 | GO:0007568~aging GO:0010149~senescence GO:0010150~leaf senescence | Arabidopsis thaliana cysteine protease mRNA (SAG12) | Down | −7.388 |
| NAC Genes | MYB Binding Sequences (Nucleotides No.) 1 |
|---|---|
| ANAC003 | AGAATATAGT (-222~-231); ATCGTATCTATGT (-329~-341); AGATAACGGA (-356~-365); AAAGATATGTC (-537~-547); TTGATATTT (-614~-622); GGAATATTTT (-658~-667) |
| ANAC010 | TTGGTAGGTC (-33~-42); AAAATATTAT (-191~-200); TATAATCTGTT (-641~-651); TGAGATCTCT (-786~-795); GTGAAGATATGGT (-850~-862) |
| ANAC042 | AGTTATCCTTT (-133~-143); TGTGAATCTTA (-152~-162); GATAATCTGA (-302~-311); TCAGATTCTCT (-643~-653); TAAGATCTTG (-845~-854) |
| ANAC075 | AAGATATTCG (-9~-18); TTCATATCTTCAC (-132~-144); CGGTTAGGT (-218~-0226); TAAGATCTTA (-310~-319); TAAATATTTT (-725~-734); TTCATATCTC (-879~-888); TCTGATATTAT (-903~-913); ATAAAGATACATA (-932~-944) |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chou, M.-L.; Liao, W.-Y.; Wei, W.-C.; Li, A.Y.-S.; Chu, C.-Y.; Wu, C.-L.; Liu, C.-L.; Fu, T.-H.; Lin, L.-F. The Direct Involvement of Dark-Induced Tic55 Protein in Chlorophyll Catabolism and Its Indirect Role in the MYB108-NAC Signaling Pathway during Leaf Senescence in Arabidopsis thaliana. Int. J. Mol. Sci. 2018, 19, 1854. https://doi.org/10.3390/ijms19071854
Chou M-L, Liao W-Y, Wei W-C, Li AY-S, Chu C-Y, Wu C-L, Liu C-L, Fu T-H, Lin L-F. The Direct Involvement of Dark-Induced Tic55 Protein in Chlorophyll Catabolism and Its Indirect Role in the MYB108-NAC Signaling Pathway during Leaf Senescence in Arabidopsis thaliana. International Journal of Molecular Sciences. 2018; 19(7):1854. https://doi.org/10.3390/ijms19071854
Chicago/Turabian StyleChou, Ming-Lun, Wan-Yu Liao, Wan-Chen Wei, Althea Yi-Shan Li, Ching-Ying Chu, Chia-Ling Wu, Chun-Lin Liu, Ting-Han Fu, and Lee-Fong Lin. 2018. "The Direct Involvement of Dark-Induced Tic55 Protein in Chlorophyll Catabolism and Its Indirect Role in the MYB108-NAC Signaling Pathway during Leaf Senescence in Arabidopsis thaliana" International Journal of Molecular Sciences 19, no. 7: 1854. https://doi.org/10.3390/ijms19071854