Identification and Characterization of microRNAs during Retinoic Acid-Induced Regeneration of a Molluscan Central Nervous System
Abstract
1. Introduction
2. Results
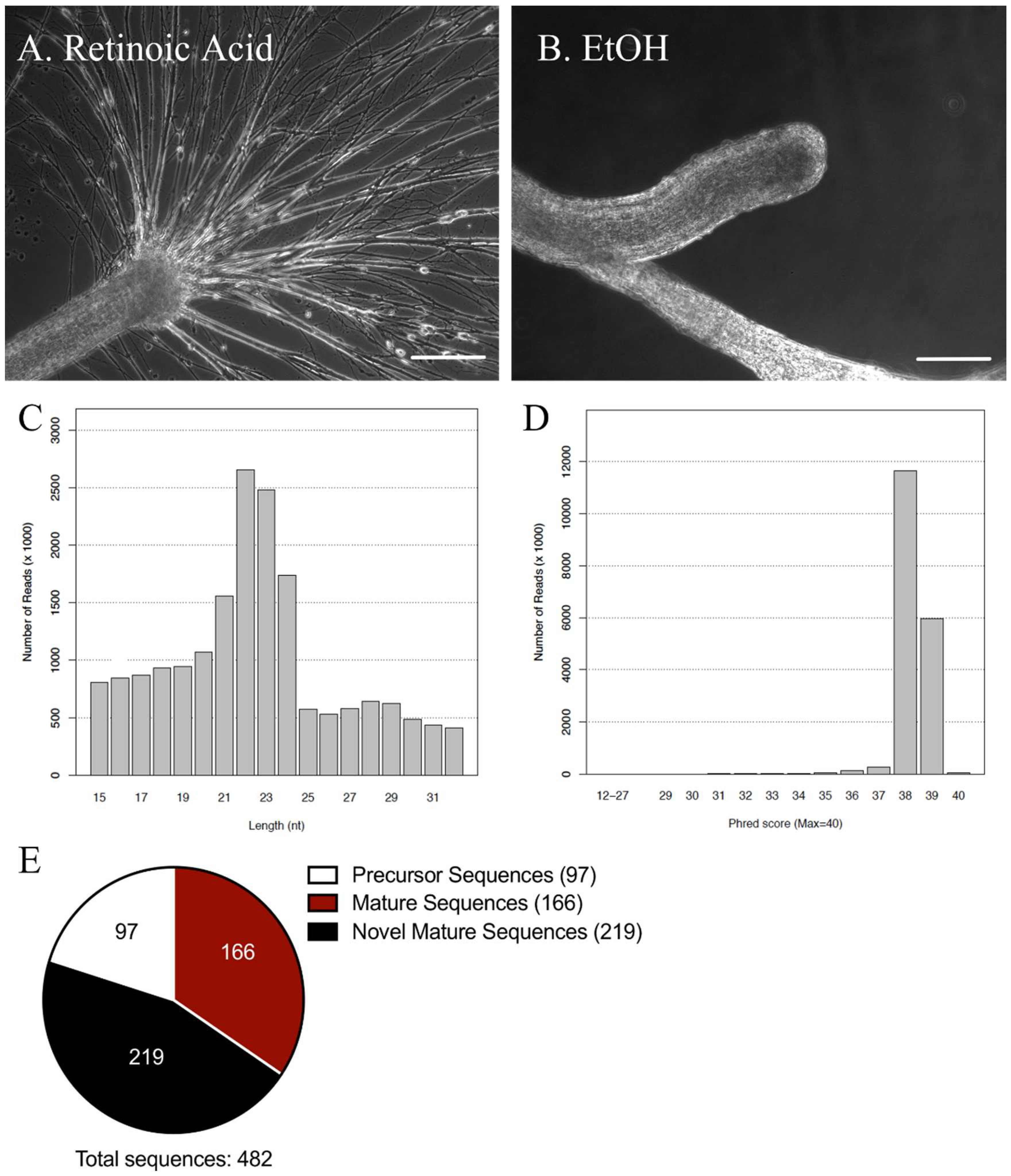
2.1. miRNA Sequencing Analysis of miRNAs in the Lymnaea Central Nervous System (CNS)
2.2. Identification of miRNAs That Were Differentially Regulated during Retinoic Acid (RA)-Induced Regeneration
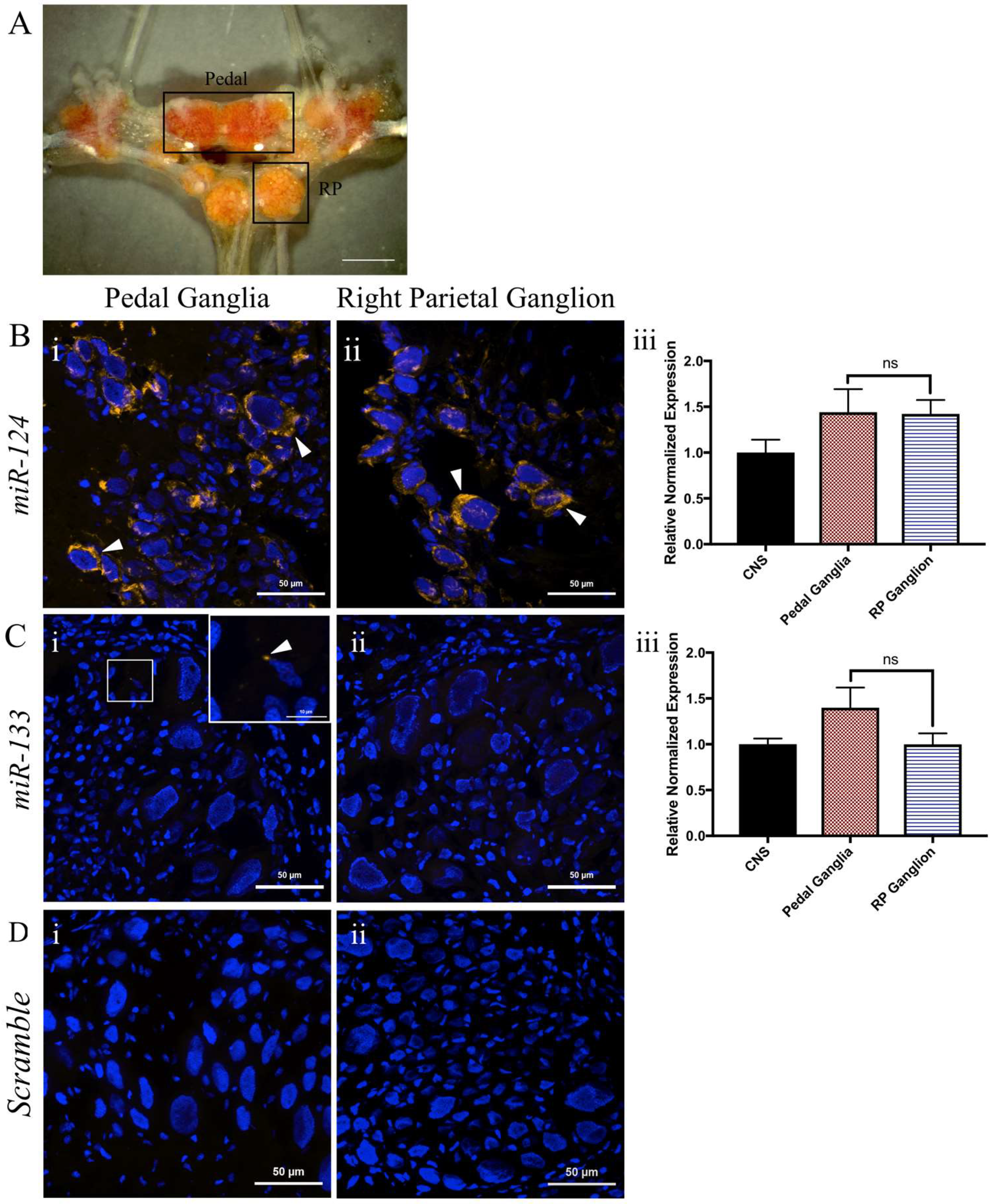
2.3. miR-124 Is Highly Enriched in the Adult Lymnaea CNS
2.4. miR-124 Is Expressed in Both the Pedal and Right Parietal Ganglia
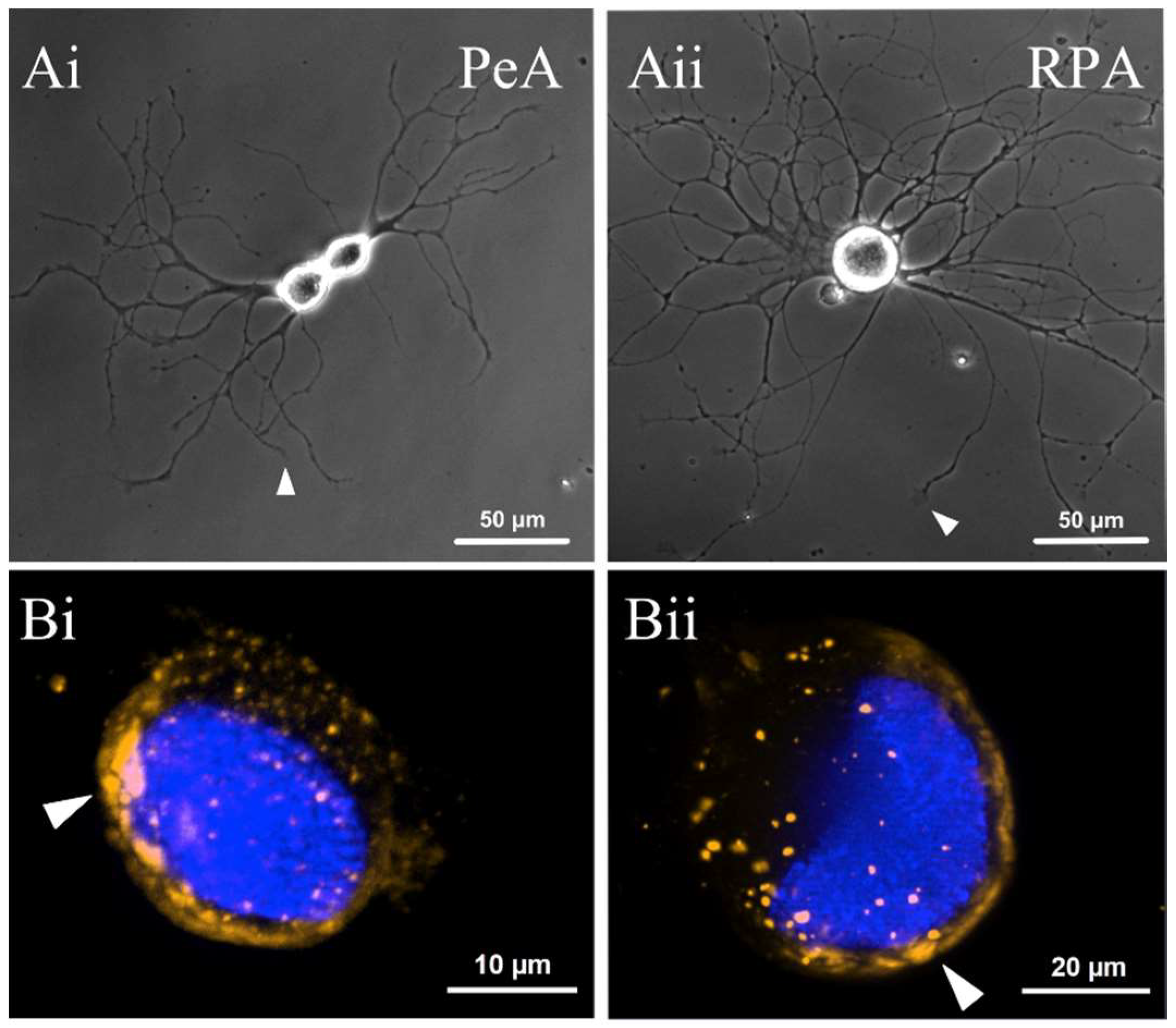
2.5. miR-124 Is Expressed within the Cell Bodies and Neurites of Two Populations of Regenerating Motorneurons
2.6. miR-124 Is Differentially Expressed in the Growth Cones of Different Classes of Motorneurons
3. Discussion
3.1. Lymnaea Stagnalis miRNA Transcriptome
3.2. miR-124 Expression Patterns in Lymnaea CNS
3.3. Expression of miR-124 in Motorneurons
3.4. Role of miR-124 during RA-Induced CNS Regeneration
4. Methods
4.1. Isolation of CNS
4.2. Regenerating CNS Preparation
4.3. RNA Sequencing of Lymnaea miRNAs
4.4. Isolation of Lymnaea Embryos
4.5. Cell Culture
4.6. RNA Isolation and cDNA Synthesis
4.7. PCR
4.8. RT-qPCR
4.9. LNA-FISH and Tyramide Signal Amplification
4.10. Statistics
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Allison, P.; Benjamin, P.R. Anatomical studies of central regeneration of an identified molluscan interneuron. Proc. R. Soc. Lond. B 1985. [Google Scholar] [CrossRef]
- Cebria, F. Regenerating the central nervous system: How easy for planarians! Dev. Genes Evol. 2007, 217, 733–748. [Google Scholar] [CrossRef] [PubMed]
- Tessier-Lavigne, M.; Goodman, C.S. The molecular biology of axon guidance. Science 1996, 274, 1123–1133. [Google Scholar] [CrossRef] [PubMed]
- Sanchez Alvarado, A.; Tsonis, P.A. Bridging the regeneration gap: Genetic insights from diverse animal models. Nat. Rev. Genet. 2006, 7, 873–884. [Google Scholar] [CrossRef] [PubMed]
- Somorjai, I.M.; Somorjai, R.L.; Garicia-Fernandez, J.; Escriva, H. Vertebrate-like regeneration in the invertebrate chordate amphioxus. Proc. Natl. Acad. Sci. USA 2012, 109, 517–522. [Google Scholar] [CrossRef] [PubMed]
- Maden, M.; Gale, E.; Kostetskii, I.; Zile, M. Vitamin A-deficient quail embryos have half a hindbrain and other neural defects. Curr. Biol. 1996, 6, 417–426. [Google Scholar] [CrossRef]
- Maden, M. Role and distribution of retinoic acid during CNS development. Int. Rev. Cytol. 2001, 209, 1–77. [Google Scholar] [PubMed]
- Maden, M. Retinoic acid in development and regeneration. J. Biosci. 1996, 21, 299–312. [Google Scholar] [CrossRef]
- Dmetrichuk, J.M.; Spencer, G.E.; Carlone, R.L. Retinoic acid-dependent attraction of adult spinal cord axons towards regenerating newt limb blastemas in vitro. Dev. Biol. 2005, 281, 112–120. [Google Scholar] [CrossRef] [PubMed]
- Dmetrichuk, J.M.; Carlone, R.L.; Spencer, G.E. Retinoic acid induces neurite outgrowth and growth cone turning in invertebrate neurons. Dev. Biol. 2006, 294, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Hunter, K.; Maden, M.; Summerbell, D.; Eriksson, U.; Holder, N. Retinoic acid stimulates neurite outgrowth in the amphibian spinal cord. Proc. Natl. Acad. Sci. USA 1991, 88, 3666–3670. [Google Scholar] [CrossRef] [PubMed]
- Prince, D.J.; Carlone, R.L. Retinoic acid involvement in the reciprocal neurotrophic interactions between newt spinal cord and limb blastemas in vitro. Dev. Brain Res. 2003, 140, 67–73. [Google Scholar] [CrossRef]
- Carter, C.; Clark, A.; Spencer, G.; Carlone, G. Cloning and expression of a retinoic acid receptor β2 subtype from the adult newt: Evidence for an early role in tail and caudal spinal cord regeneration. Dev. Dyn. 2011, 240, 2613–2625. [Google Scholar] [CrossRef] [PubMed]
- Lepp, A.; Carlone, R. RARβ2 expression is induced by the down-regulation of microRNA 133a during caudal spinal cord regeneration in the adult newt. Dev. Dyn. 2014. [Google Scholar] [CrossRef] [PubMed]
- Lepp, A.; Carlone, R. MicroRNA dysregulation in response to RARβ2 inhibition reveals a negative feedback loop between microRNAs 1, 133a, and RARβ2 during tail and spinal cord regeneration in the adult newt. Dev. Dyn. 2015, 244, 1519–1537. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.M.; Gibbs, K.M.; Davila, J.; Campbell, N.; Sung, S.; Todorova, T.I.; Otsuka, S.; Sabaawy, H.E.; Hart, R.P.; Schachner, M. MicroRNA miR-133b is essential for functional recovery after spinal cord injury in adult zebrafish. Eur. J. Neurosci. 2011, 33, 1587–1597. [Google Scholar] [CrossRef] [PubMed]
- Dmetrichuk, J.M.; Carlone, R.L.; Jones, T.R.B.; Vesprini, N.D.; Spencer, G.E. Detection of endogenous retinoids in the molluscan CNS and characterization of the trophic and tropic actions of 9-cis retinoic acid on isolated neurons. J. Neurosci. 2008, 28, 13014–13024. [Google Scholar] [CrossRef] [PubMed]
- Farrar, N.R.; Dmetrichuk, J.M.; Carlone, R.L.; Spencer, G.E. A novel, nongenomic mechanism underlies retinoic acid-induced growth cone turning. J. Neurosci. 2009, 29, 14136–14142. [Google Scholar] [CrossRef] [PubMed]
- Carter, C.J.; Farrar, N.; Carlone, R.L.; Spencer, G.E. Developmental expression of a molluscan RXR and evidence for its novel, nongenomic role in growth cone guidance. Dev. Biol. 2010, 343, 124–137. [Google Scholar] [CrossRef] [PubMed]
- Carter, C.J.; Rand, C.; Mohammad, I.; Lepp, A.; Vesprini, N.; Wiebe, O.; Carlone, R.; Spencer, G.E. Expression of a retinoic acid receptor (RAR)-like protein in the embryonic and adult nervous system of a protostome species. J. Exp. Zool. B. Mol. Dev. Evol. 2015, 324B, 51–67. [Google Scholar] [CrossRef] [PubMed]
- Starega-Roslan, J.; Krol, J.; Koscianska, E.; Kozlowski, P.; Szlachcic, W.J.; Sobczak, K.; Krzyzosiak, W.J. Structural basis of microRNA length variety. Nucleic. Acid Res. 2011, 39, 257–268. [Google Scholar] [CrossRef] [PubMed]
- Lagos-Quintana, M.; Rauhut, R.; Lendeckel, W.; Tuschl, T. Identification of novel genes coding for small expressed RNAs. Science 2001, 294, 853–858. [Google Scholar] [CrossRef] [PubMed]
- Schultz, J.; Lorenz, P.; Gross, G.; Ibrahim, S.; Kunz, M. MicroRNA let-7b targets important cell cycle molecules in malignant melanoma cells and interferes with anchorage-independent growth. Cell Res. 2008, 18, 549–557. [Google Scholar] [CrossRef] [PubMed]
- Sun Lee, Y.; Dutta, A. The tumo suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007, 21, 1025–1030. [Google Scholar]
- Guo, Y.; Liu, H.; Zhang, H.; Shang, C.; Song, Y. miR-96 regulates FOXO1-mediated cell apoptosis in bladder cancer. Oncol. Lett. 2012, 4, 561–565. [Google Scholar] [CrossRef] [PubMed]
- Feng, T.; Xu, D.; Tu, C.; Li, W.; Ning, Y.; Ding, J.; Wang, S.; Yuan, L.; Xu, N.; Qian, K.; et al. miR-124 inhibits cell proliferation in breast cancer through downregulation of CDK4. Tumour Biol. 2015, 38, 5987–5997. [Google Scholar] [CrossRef] [PubMed]
- Budd, W.T.; Seashols-Williams, S.J.; Clark, G.C.; Weaver, D.; Calvert, V.; Petricoin, E.; Dragoescu, E.A.; O’Hanlon, K.; Zehner, Z.E. Dual action of miR-125b as a tumor suppressor and oncomiR-22 promotes prostate cancer tumorigenesis. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Feliciano, A.; Castellvi, J.; Artero-Castro, A.; Leal, J.A.; Romagosa, C.; Hernandez-Losa, J.; Peg, V.; Fabra, A.; Vidal, F.; Kondoh, H.; et al. miR-125b acts as a tumor suppressor in breast cancer tumorgenesis via its novel direct targets ENPEP, CK2-α, CCNJ, and MEGF9. PLoS ONE 2013, 8, e76247. [Google Scholar] [CrossRef] [PubMed]
- Moskwa, P.; Buffa, F.M.; Pan, Y.; Panchakshari, R.; Gottipati, P.; Muschel, R.J.; Beech, J.; Kulshrestha, R.; Abdelmohsen, K.; Weinstock, D.M.; et al. miR-182-mediated downregulation of BRCA1 impacts DNA repair and sensitivity to PARP inhibitors. Mol. Cell 2011, 41, 210–220. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Liu, M.; Yan, X.; Zhou, Y.; Wang, W.; Wang, X.; Fu, Z.; Wang, N.; Zhang, S.; Wang, Y.; et al. miR-193a-3p functions as a tumor suppressor in lung cancer by downregulating ERBB4. J. Biol. Chem. 2015, 290, 926–940. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S.M.; Grosshans, H.; Shingara, J.; Byrom, M.; Jarvis, R.; Cheng, A.; Labourier, E.; Reinert, K.L.; Brown, D.; Slack, F.J. RAS is regulated by the let-7 microRNA family. Cell 2005, 120, 635–647. [Google Scholar] [CrossRef] [PubMed]
- Cushing, L.; Crostinean, S.; Xu, W.; Jiang, Z.; Madden, L.; Kuang, P.; Huang, J.; Weisman, A.; Hata, A.; Croce, C.M.; et al. Disruption of miR-29 leads to aberrant differentiation of smooth muscle cells selectively associated with distal lung vasculature. PLoS Genet. 2015, 11, e1005238. [Google Scholar] [CrossRef] [PubMed]
- Iovino, N.; Pane, A.; Gaul, U. miR-184 has multiple roles in Drosophila female germline development. Dev. Cell 2009, 17, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Brennecke, J.; Hipfner, D.R.; Stark, A.; Russell, R.B.; Cohen, S.M. Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 2003, 113, 25–36. [Google Scholar] [CrossRef]
- Liu, M.; Wang, N.; Ye, C.; Zhao, C.; Liu, Y.; Fan, Q.; Zhang, C.Y.; Sang, J.; Zen, K.; Chen, X. miR-96 promotes cell proliferation, migration and invasion by targeting PTPN9 in breast cancer. Sci. Rep. 2016, 6, 37421. [Google Scholar]
- Hong, Y.; Liang, H.; Rehman, U.U.; Wang, Y.; Zhang, W.; Zhou, Y.; Chen, S.; Yu, M.; Cui, S.; Zheng, A.; et al. miR-125b regulates proliferation and apoptosis of nasopharyngeal carcinoma by targeting A20/NF-κB signaling pathway. Cell Death Dis. 2017, 8, e2855. [Google Scholar]
- Zhang, X.; Ma, G.; Liu, J.; Zhang, Y. MicroRNA-182 promotes proliferation and metastasis by targeting FOXF2 in triple-negative breast cancer. Oncol. Lett. 2017, 14, 4805–4811. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Teng, Z.Q.; Santistevan, N.J.; Szulwach, K.E.; Guo, W.; Jin, P.; Zhao, X. Epigenetic regulation of miR-184 by MBD1 governs neural stem cell proliferation and differentiation. Cell Stem Cell 2010, 6, 433–444. [Google Scholar] [CrossRef] [PubMed]
- Selcuklu, S.D.; Donoghue, M.T.; Rehmet, K.; de Souza Gomes, M.; Fort, A.; Kovvuru, P.; Muniyappa, M.K.; Kerin, M.J.; Enright, A.J.; Spillane, C. MicroRNA-9 inhibition of cell proliferation and identification of novel miR-9 targets by transcriptome profliling in breast cancer cells. J. Biol. Chem. 2012, 287, 29516–29528. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhong, H.; Wang, L.; Dong, Y.; Jia, A.; Mo, Q.; Zhang, C. miR-29 induced K562 cell apoptosis by downregulating FoxM1. Med. Sci. Monit. 2015, 21, 3115–3120. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.M.; Park, S.J.; Jung, S.H.; Kim, E.J.; Jogeswar, G.; Ajita, J.; Rhee, Y.; Kim, C.H.; Lim, S.K. miR-182 is a negative regulator of osteoblast proliferation, differentiation, and skeletogenesis through targeting FoxO1. J. Bone Miner. Res. 2012, 27, 1669–1679. [Google Scholar] [CrossRef] [PubMed]
- Nakano, H.; Yamada, Y.; Miyazawa, T.; Yoshida, T. Gain-of-function microRNA screens identify miR-193a regulating proliferation and apoptosis in epithelial ovarian cancer cells. Int. J. Oncol. 2013, 42, 1875–1882. [Google Scholar] [CrossRef] [PubMed]
- Legesse-Miller, A.; Elemento, O.; Pfau, S.J.; Forman, J.J.; Tavazoie, S.; Coller, H.A. let-7 overexpression leads to an increased fraction of cells in G2/M, direct down-regulation of Cdc34, and stabilization of Wee1 kinase in primary fibroblasts. J. Biol. Chem. 2009, 284, 6605–6609. [Google Scholar] [CrossRef] [PubMed]
- Kouri, F.M.; Hurley, L.A.; Daniel, W.L.; Day, E.S.; Hua, Y.; Hao, L.; Peng, C.Y.; Merkel, T.J.; Queisser, M.A.; Ritner, C.; et al. miR-182 integrates apoptosis, growth, and differentiation programs in glioblastoma. Genes. Dev. 2015, 29, 732–745. [Google Scholar] [CrossRef] [PubMed]
- Qi, X. The role of miR-9 during neuron differentiation of mouse retinal stem cells. Artif. Cells Nanomed. Biotechnol. 2016, 44, 1883–1890. [Google Scholar] [CrossRef] [PubMed]
- Xue, A.; Yu, C.; Wang, Y.; Liu, L.; Zhang, K.; Fang, C.; Liu, F.; Bian, G.; Song, B.; Yang, A.; et al. miR-9 and miR-124 synergistically affect regulation of dendritic branching via the AKT/GSK3β pathway by targeting Rap2a. Sci. Rep. 2016, 6, 26781. [Google Scholar] [CrossRef] [PubMed]
- Makeyev, E.V.; Zhang, J.; Carrasco, M.A.; Maniatis, T. The microRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol. Cell. 2007, 27, 435–448. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.C.; Pastrana, E.; Tavazoie, M.; Doetsch, F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat. Neurosci. 2009, 12, 399–408. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.S.; Chopp, M.; Zhang, R.L.; Tao, T.; Wang, X.L.; Kassis, H.; Hozeska-Solgot, A.; Zhang, L.; Chen, C.; Zhang, Z.G. MicroRNA profiling in subventricular zone after stroke: miR-124a regulates proliferation of neural progenitor cells through Notch signaling pathway. PLoS ONE 2011, 6. [Google Scholar] [CrossRef] [PubMed]
- Chawla, G.; Deosthale, P.; Childress, S.; Wu, Y.C.; Sokol, N.S. A let-7-to-miR-125 microRNA switch regulates neuronal integrity and lifespan in Drosophila. PLoS Genet. 2016, 12, e1006247. [Google Scholar] [CrossRef] [PubMed]
- Shibata, M.; Nakao, H.; Kiyonari, H.; Abe, T.; Aizawa, S. MicroRNA-9 regulates neurogenesis in mouse telencephalon by targeting multiple transcription factors. J. Neurosci. 2011, 31, 3407–3422. [Google Scholar] [CrossRef] [PubMed]
- Baudet, M.L.; Zivraj, K.H.; Abreu-Goodger, C.; Muldal, A.; Armisen, J.; Blenkiron, C.; Goldstein, L.D.; Miska, E.A.; Holt, C.E. miR-124 acts through CoREST to control onset of Sema3A sensitivity in navigating retinal growth cones. Nat. Neurosci. 2011, 15, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Quiroz, J.F.D.; Tsai, E.; Coyle, M.; Sehm, T.; Echeverri, K. Precise control of miR-125b is required to create a regeneration-permissive environment after spinal cord injury. Dis. Model. Mech. 2014. [Google Scholar] [CrossRef]
- Lu, C.S.; Zhai, B.; Mauss, A.; Landgraf, M.; Gygi, S.; Van Vactor, D. MicroRNA-8 promotes robust motor axon targeting by coordinate regulation of cell adhesion molecules during synapse development. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014, 369. [Google Scholar] [CrossRef] [PubMed]
- Hou, Q.; Ruan, H.; Gilbert, J.; Wang, G.; Ma, Q.; Yao, W.D.; Man, H.Y. MicroRNA miR124 is required for the expression of homeostatic synaptic plasticity. Nat. Commun. 2015, 6, 10045. [Google Scholar]
- Rajasethupathy, P.; Fiumara, F.; Sheridan, R.; Betel, D.; Puthanveettil, S.V.; Russo, J.J.; Sander, C.; Tuschi, T.; Kandel, E. Characterization of small RNAs in Aplysia reveals a role for miR-124 in constraining synaptic plasticity through CREB. Neuron 2009, 63, 803–817. [Google Scholar] [CrossRef] [PubMed]
- Maden, M. Retinoic acid in the development, regeneration and maintenance of the nervous system. Nat. Rev. Neurosci. 2007, 8, 755–765. [Google Scholar] [CrossRef] [PubMed]
- Sempere, L.F.; Freemantle, S.; Pitha-Rowe, I.; Moss, E.; Dmitrovsky, E.; Ambros, V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004, 5, R13. [Google Scholar] [CrossRef] [PubMed]
- Annibali, D.; Gioia, U.; Savino, M.; Laneve, P.; Caffarelli, E.; Nasi, S. A new module in neural differentiation control: Two microRNAs upregulated by retinoic acid, miR-9 and -103, target the differentiation inhibitor ID2. PLoS ONE 2012, 7, e40269. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.Y.; Chung, K.H.; Deo, M.; Thompson, R.; Turner, D.L. MicroRNA miR-124 regulated neurite outgrowth during neuronal differentiation. Exp. Cell Res. 2008, 314, 2618–2633. [Google Scholar] [CrossRef] [PubMed]
- Smirnova, L.; Grafe, A.; Seiler, A.; Schumacher, S.; Nitsch, R.; Wilczyn, F.G. Regulation of miRNA expression during neural cell specification. Eur. J. Neurosci. 2005, 21, 1469–1477. [Google Scholar] [CrossRef] [PubMed]
- Griffiths-Jones, S. miR-Base: The microRNA Sequence Database. MicroRNA Protocols 2006, 342, 129–138. [Google Scholar]
- Deo, M.; Yu, J.Y.; Chung, K.H.; Tippens, M.; Turner, D.L. Detection of mammalian microRNA expression by in situ hybridization with RNA oligonucleotides. Dev. Dyn. 2006, 235, 2538–2548. [Google Scholar] [CrossRef] [PubMed]
- Krichevsky, A.M.; Sonntag, K.C.; Isacson, O.; Kosik, K.S. Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells 2006, 24, 857–864. [Google Scholar] [CrossRef] [PubMed]
- Croll, R.P.; Voronezhskaya, E.E. Early elements in gastropod neurogenesis. Dev. Biol 1996, 173, 344–347. [Google Scholar] [CrossRef] [PubMed]
- Nagy, T.; Elekes, K. Embryogenesis of the central nervous system of the pong snail Lymnaea stagnalis L. An ultrastructural study. J. Neurocytol. 2000, 29, 43–60. [Google Scholar] [CrossRef] [PubMed]
- Bak, M.; Silahtaroglu, A.; Moller, M.; Christensen, M.; Rath, M.F.; Skryabin, B.; Tommerup, N.; Kauppinen, S. MicroRNA expression in the adult mouse central nervous system. RNA 2008, 14, 432–444. [Google Scholar] [CrossRef] [PubMed]
- Wienholds, E.; Kloosterman, W.P.; Miska, E.; Alvarez-Saavedra, E.; Berezikov, E.; de Bruijn, E.; Horvitz, H.R.; Kauppinen, S.; Plasterk, R.H. MicroRNA expression in zebrafish embryonic development. Science 2005, 309, 310–311. [Google Scholar] [CrossRef] [PubMed]
- Syed, N.I.; Winlow, W. Morphology and electrophysiology of neurons innervating the ciliated locomotor epithelium in Lymnaea stagnalis (L.). Comp. Biochem. Physiol. A Physiol. 1989, 93, 633–644. [Google Scholar] [CrossRef]
- Magoski, N.S.; Syed, N.I.; Bulloch, A.G. A neuronal network from the mollusc Lymnaea stagnalis. Brain Res. 1994, 645, 201–214. [Google Scholar] [CrossRef]
- Gearhart, M.D.; Erikson, J.R.; Walsh, A.; Echeverri, K. Identification of conserved and novel microRNAs during tail regeneration in the Mexican Axolotl. Int. J. Mol. Sci. 2015, 16, 22046–22061. [Google Scholar] [CrossRef] [PubMed]
- Sasidharan, V.; Marepally, S.; Elliots, S.A.; Baid, S.; Lakshmanan, V.; Nayyar, N.; Bansal, D.; Sanchez Alvarado, A.; Kumar Vemula, P.; Palakodeti, D. The miR-124 family of microRNAs is crucial for regeneration of the brain and visual system in the planarian Schmidtea mediterranea. Development 2017, 144, 3211–3223. [Google Scholar] [CrossRef] [PubMed]
- Mishima, T.; Mizuguchi, Y.; Kawahigashi, Y.; Takizawa, T.; Takizawa, T. RT-PCR-based analysis of microRNA (miR-1 and -124) expression in mouse CNS. Brain Res. 2007, 1131, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Wen, Z.; Lynn, R.C.; Baudet, M.L.; Holt, C.E.; Sasaki, Y.; Bassell, G.J.; Zheng, J.Q. Regulation of chemotropic guidance of nerve growth cones by microRNA. Mol. Brain 2011, 4. [Google Scholar] [CrossRef] [PubMed]
- Hancock, M.L.; Preitner, N.; Quan, J.; Flanagan, J.G. MicroRNA-132 is enriched in developing axons, locally regulated Rasa1 mRNA, and promotes axon extension. J. Neurosci. 2014, 34, 66–78. [Google Scholar] [CrossRef] [PubMed]
- Bellon, A.; Iyer, A.; Bridi, S.; Lee, F.C.Y.; Ovando-Vazquez, C.; Corradi, E.; Longhi, S.; Roccuzzo, M.; Strohbuecker, S.; Naik, S.; et al. miR-182 regulated Slit2-mediated axon guidance by modulating the local translation of a specific mRNA. Cell Rep. 2017, 18, 1171–1186. [Google Scholar] [CrossRef] [PubMed]
- Walker, S.E.; Nottrodt, R.; Maddalena, L.; Carter, C.; Spencer, G.E.; Carlone, R.L. Retinoid X receptor α downregulation is required for tail and caudal spinal cord regeneration in the adult newt. Neural Regen. Res. 2018, 13, 1036–1045. [Google Scholar] [PubMed]
- Wong, R.G.; Hadley, R.D.; Kater, S.B.; Hauser, G.C. Neurite outgrowth in molluscan organ and cell cultures: The role of conditioning factor(s). J. Neurosci. 1981, 1, 1008–1021. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Tsourkas, A. Quantification of miRNA abundance in single cells using locked nucleic acid-FISH and enzyme-labeled fluorescence. In Molecular Imaging; Shah, K., Ed.; Humana Press: Totowa, NJ, USA, 2011; Volume 680, pp. 77–88. [Google Scholar]




| Biological Role | Mature miRNA | Target | Reference |
| Tumorigenesis | let-7 | Cdk4 | [23] |
| HMGA2 | [24] | ||
| miR-96 | FOXO1 | [25] | |
| miR-124 | Cdk4 | [26] | |
| miR-125 | ErbB2 | [27] | |
| ENPEP, CK2-α, CCNJ, MEGF9 | [28] | ||
| miR-182 | BRCA1 | [29] | |
| miR-193 | ERBB4 | [30] | |
| Differentiation | miR-7 | RAS | [31] |
| miR-29b | FBXO2 | [32] | |
| miR-184 | Saxophone | [33] | |
| Proliferation | Bantam | Hid | [34] |
| miR-96 | PTPN9 | [35] | |
| miR-125 | A20 | [36] | |
| miR-182 | FOXF2 | [37] | |
| miR-184 | Numbl | [38] | |
| Apoptosis | miR-9 | MTHFD2 | [39] |
| miR-29 | FoxM1 | [40] | |
| miR-96 | FOXO1 | [25] | |
| miR-182 | FOXO1 | [41] | |
| miR-193 | MCL1 | [42] | |
| Cell cycle regulation | let-7 | Cdc34 | [43] |
| Bantam | hid | [34] | |
| miR-182 | c-Met | [44] | |
| Nervous System-Specific Role | Mature miRNA | Target | Reference |
| Neuronal differentiation | miR-9 | PTBP1 | [45] |
| Rap2a | [46] | ||
| miR-124 | PTBP1 | [47] | |
| Rap2a | [46] | ||
| Sox9 | [48] | ||
| JAG1 | [49] | ||
| Neuronal lifespan | let-7 | Chinmo | [50] |
| miR-125 | Chinmo | [50] | |
| Neurite guidance | miR-9 | Gsh2, Foxg1 | [51] |
| miR-124 | coREST | [52] | |
| miR-125 | Sema4d | [53] | |
| Synaptogenesis | miR-8 | Fasciclin III, Neuroglian | [54] |
| miR-124 | GluA2 | [55] | |
| CREB | [56] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Walker, S.E.; Spencer, G.E.; Necakov, A.; Carlone, R.L. Identification and Characterization of microRNAs during Retinoic Acid-Induced Regeneration of a Molluscan Central Nervous System. Int. J. Mol. Sci. 2018, 19, 2741. https://doi.org/10.3390/ijms19092741
Walker SE, Spencer GE, Necakov A, Carlone RL. Identification and Characterization of microRNAs during Retinoic Acid-Induced Regeneration of a Molluscan Central Nervous System. International Journal of Molecular Sciences. 2018; 19(9):2741. https://doi.org/10.3390/ijms19092741
Chicago/Turabian StyleWalker, Sarah E., Gaynor E. Spencer, Aleksandar Necakov, and Robert L. Carlone. 2018. "Identification and Characterization of microRNAs during Retinoic Acid-Induced Regeneration of a Molluscan Central Nervous System" International Journal of Molecular Sciences 19, no. 9: 2741. https://doi.org/10.3390/ijms19092741
APA StyleWalker, S. E., Spencer, G. E., Necakov, A., & Carlone, R. L. (2018). Identification and Characterization of microRNAs during Retinoic Acid-Induced Regeneration of a Molluscan Central Nervous System. International Journal of Molecular Sciences, 19(9), 2741. https://doi.org/10.3390/ijms19092741





