The Effects of Naringenin on miRNA-mRNA Profiles in HepaRG Cells
Abstract
:1. Introduction
2. Results
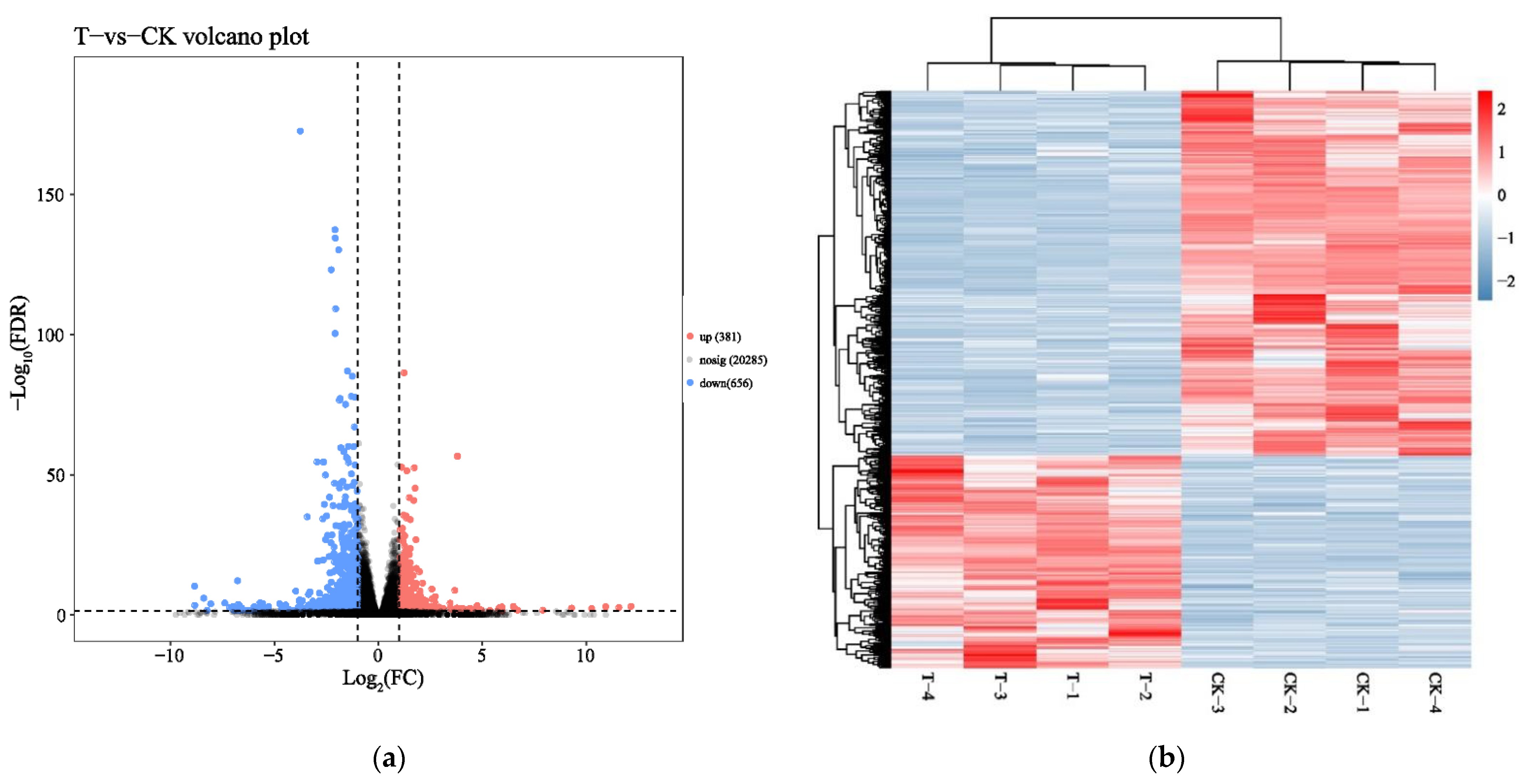
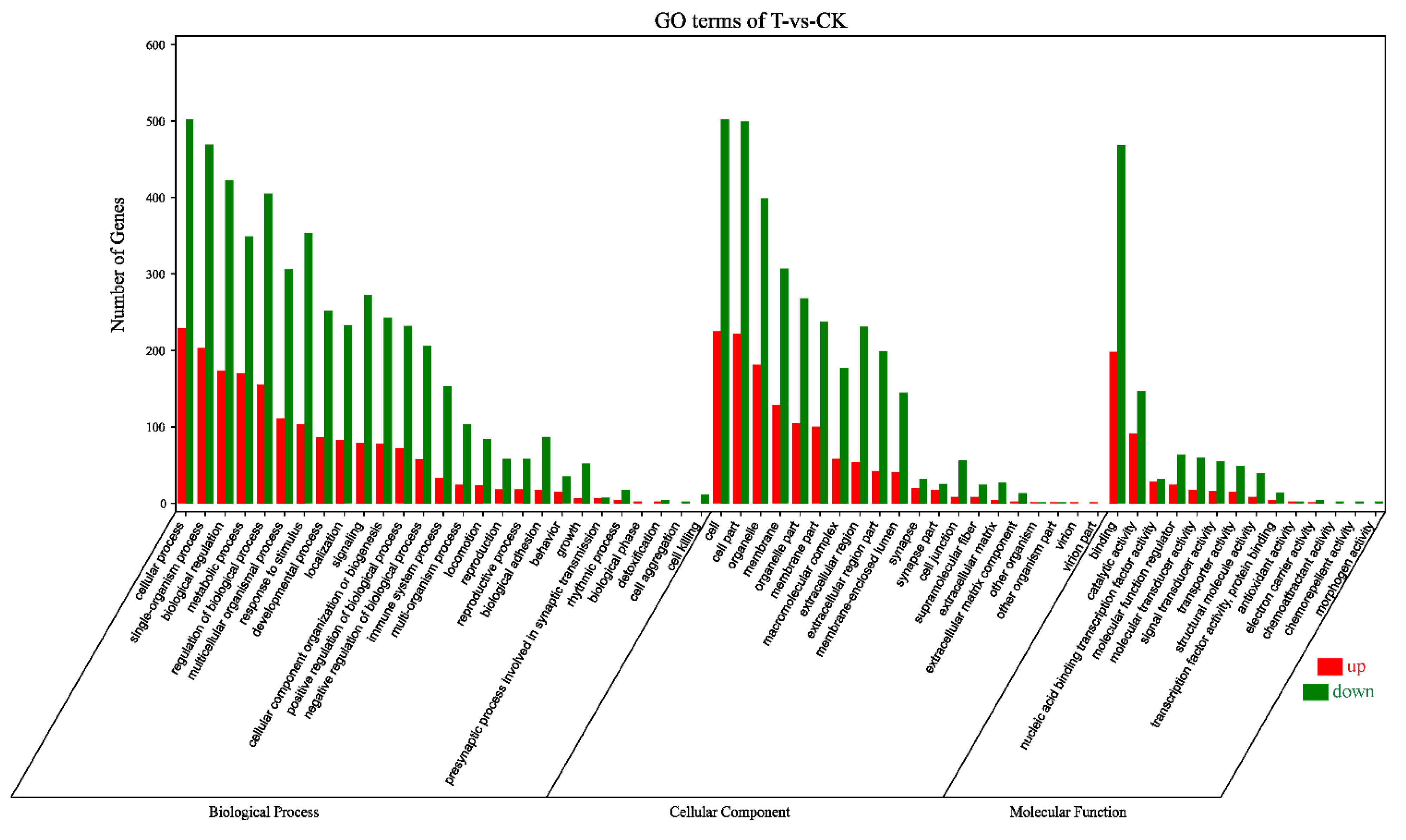
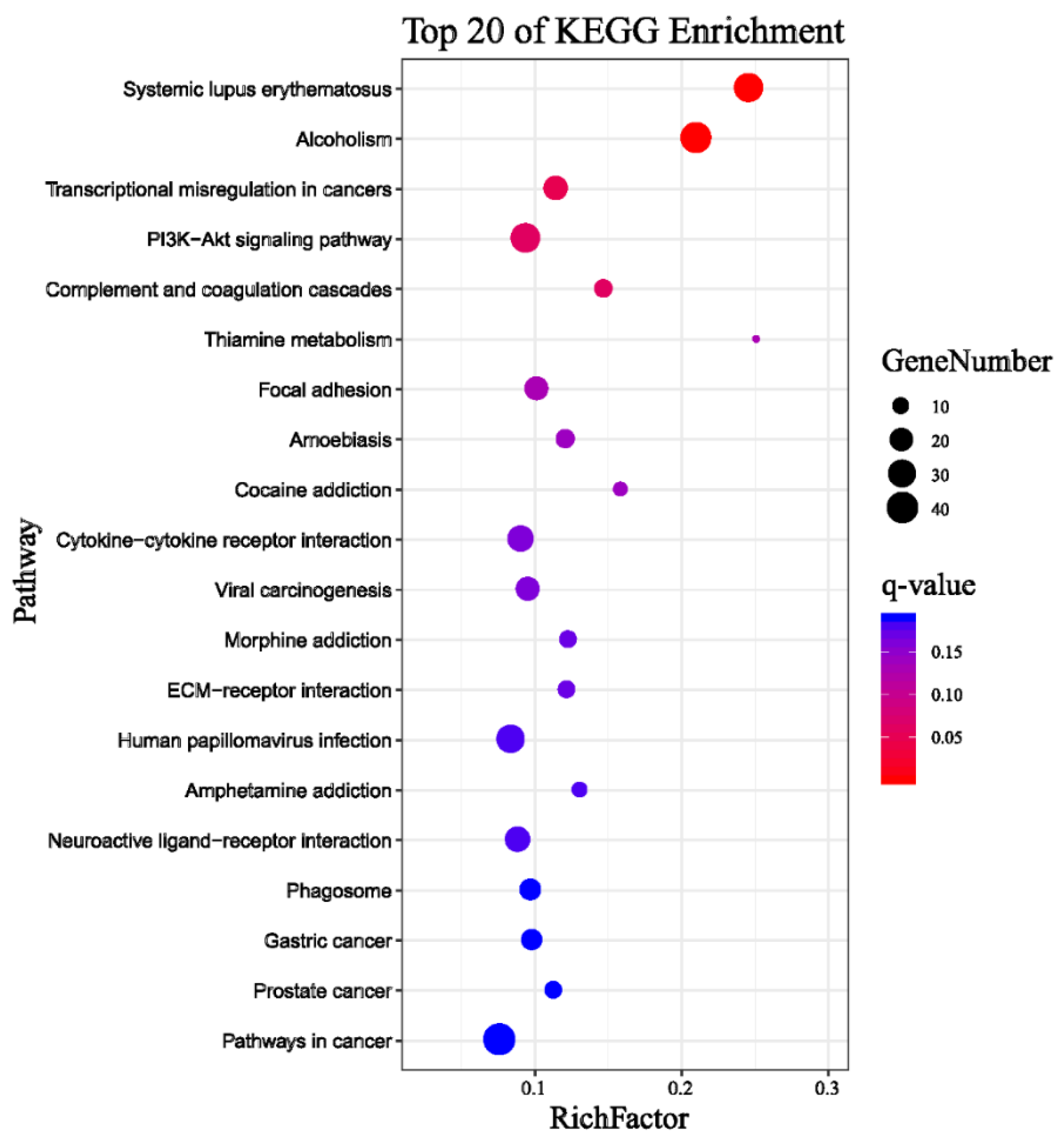
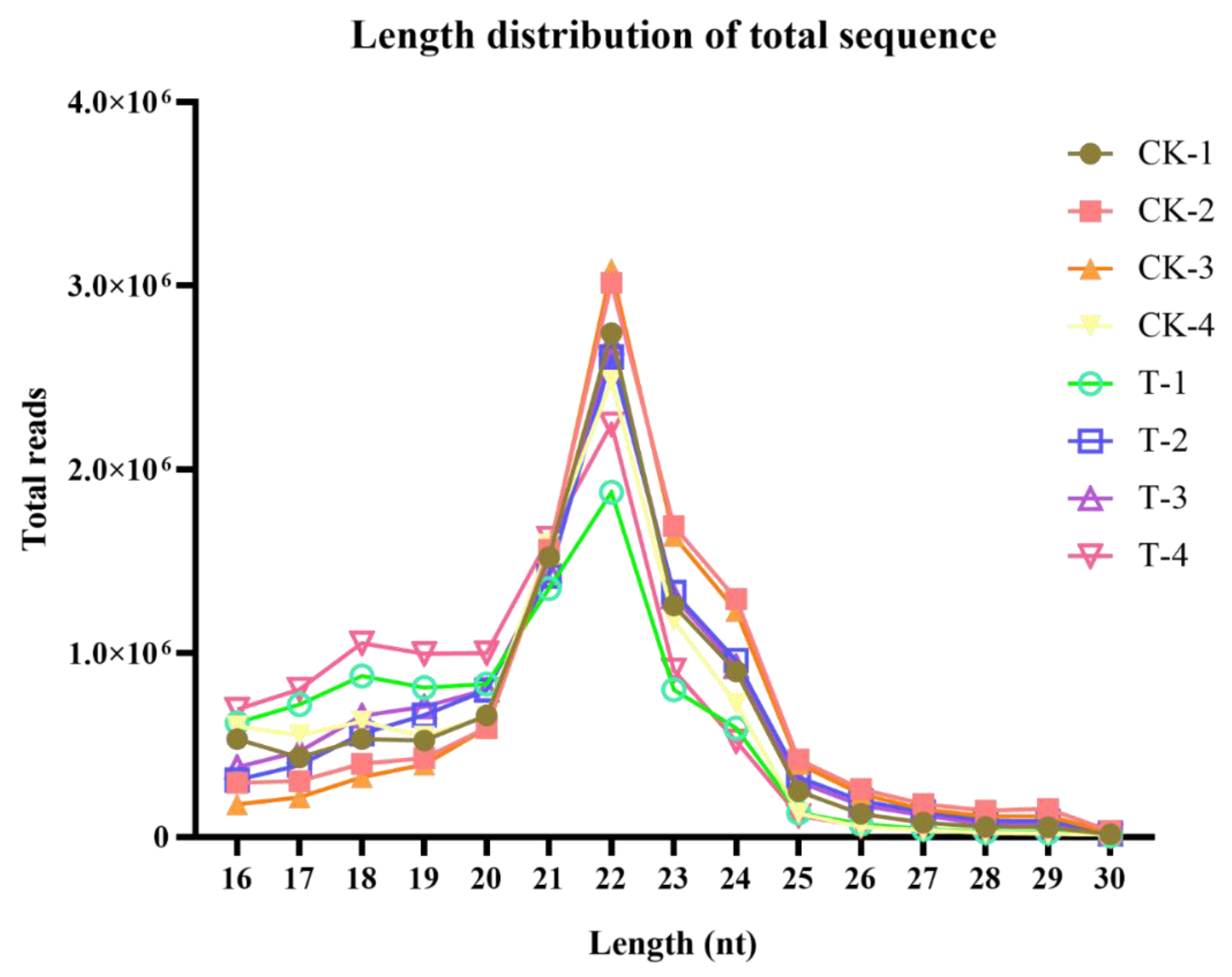
2.1. Analysis of Transcriptome Sequencing in the Response to Naringenin
2.2. Analysis of miRNA Transcript Levels in Response to Naringenin
2.3. Target Prediction and Integration Analysis of mRNA and miRNA Expression Profiles in Response to Naringenin
2.4. Real-Time qPCR Validation of Naringenin Regulation in Liver Metabolism and Potential Regulatory miRNA-mRNA Pairs
3. Discussion and Conclusions
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. HepaRG Cells Culture
4.3. Total RNA Extraction
4.4. RNA Sequencing
4.5. Real-Time qPCR
4.6. Bioinformatic Analysis and SStatistics
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Salehi, B.; Fokou, P.V.T. The Therapeutic Potential of Naringenin: A Review of Clinical Trials. Pharmaceuticals 2019, 12, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, Y.; Peng, W.; Yang, C.; Zou, W.; Liu, M.; Wu, H.; Fan, L.; Li, P.; Zeng, X.; Su, W. Pharmacokinetics and Metabolism of Naringin and Active Metabolite Naringenin in Rats, Dogs, Humans, and the Differences Between Species. Front. Pharmacol. 2020, 11, 364. [Google Scholar] [CrossRef] [Green Version]
- Joshi, R.; Kulkarni, Y.A.; Wairkar, S. Pharmacokinetic, pharmacodynamic and formulations aspects of Naringenin: An update. Life Sci. 2018, 215, 43–56. [Google Scholar] [CrossRef]
- Hernández-Aquino, E.; Muriel, P. Beneficial effects of naringenin in liver diseases: Molecular mechanisms. World J. Gastroenterol. 2018, 24, 1679–1707. [Google Scholar] [CrossRef]
- Wang, Q.; Ou, Y.; Hu, G.; Wen, C.; Yue, S.; Chen, C.; Xu, L.; Xie, J.; Dai, H.; Xiao, H.; et al. Naringenin attenuates non-alcoholic fatty liver disease by down-regulating the NLRP3/NF-κB pathway in mice. Br. J. Pharmacol. 2020, 177, 1806–1821. [Google Scholar] [CrossRef]
- Khan, T.H.; Ganaie, M.A. Naringenin prevents doxorubicin-induced toxicity in kidney tissues by regulating the oxidative and inflammatory insult in Wistar rats. Arch. Physiol. Biochem. 2020, 126, 300–307. [Google Scholar] [CrossRef]
- Wang, J.; Ding, Y.; Zhou, W. Albumin self-modified liposomes for hepatic fibrosis therapy via SPARC-dependent pathways. Int. J. Pharm. 2020, 574, 118940. [Google Scholar] [CrossRef]
- Kometsi, L.; Govender, K.; Mofo Mato, E.P.; Hurchund, R.; Owira, P.M.O. By reducing oxidative stress, naringenin mitigates hyperglycaemia-induced upregulation of hepatic nuclear factor erythroid 2-related factor 2 protein. J. Pharm. Pharmacol. 2020, 72, 1394–1404. [Google Scholar] [CrossRef] [PubMed]
- Rossino, M.G.; Casini, G. Nutraceuticals for the Treatment of Diabetic Retinopathy. Nutrients 2019, 11, 771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernández-Aquino, E.; Quezada-Ramírez, M.A.; Silva-Olivares, A.; Casas-Grajales, S.; Ramos-Tovar, E.; Flores-Beltrán, R.E.; Segovia, J.; Shibayama, M.; Muriel, P. Naringenin attenuates the progression of liver fibrosis via inactivation of hepatic stellate cells and profibrogenic pathways. Eur. J. Pharmacol. 2019, 865, 172730. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Zhao, Z.; Cai, Q.; Zhang, Y.; Zhang, P.; Shi, S.; Xie, H.; Peng, X.; Yin, W.; Tao, Y.; et al. miRNA-based biomarkers, therapies, and resistance in Cancer. Int. J. Biol. Sci. 2020, 16, 2628–2647. [Google Scholar] [CrossRef]
- Li, D.; Zhang, J.; Li, J. Role of miRNA sponges in hepatocellular carcinoma. Clin. Chim. Acta Int. J. Clin. Chem. 2020, 500, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Bofill-De Ros, X.; Yang, A.; Gu, S. IsomiRs: Expanding the miRNA repression toolbox beyond the seed. Biochim. Biophys. Acta. Gene Regul. Mech. 2020, 1863, 194373. [Google Scholar] [CrossRef]
- Sun, H.; Zhao, L.; Pan, K.; Zhang, Z.; Zhou, M.; Cao, G. Integrated analysis of mRNA and miRNA expression profiles in pancreatic ductal adenocarcinoma. Oncol. Rep. 2017, 37, 2779–2786. [Google Scholar] [CrossRef] [Green Version]
- Lou, W.; Liu, J.; Ding, B.; Chen, D.; Xu, L.; Ding, J.; Jiang, D.; Zhou, L.; Zheng, S.; Fan, W. Identification of potential miRNA-mRNA regulatory network contributing to pathogenesis of HBV-related HCC. J. Transl. Med. 2019, 17, 7. [Google Scholar] [CrossRef]
- Vaschetto, L.M. miRNA activation is an endogenous gene expression pathway. RNA Biol. 2018, 15, 826–828. [Google Scholar] [CrossRef]
- Zhang, H.; Chen, X.; Yuan, Y. Investigation of the miRNA and mRNA Coexpression Network and Their Prognostic Value in Hepatocellular Carcinoma. BioMed Res. Int. 2020, 2020, 8726567. [Google Scholar] [CrossRef]
- Zhang, W.; Zhong, W.; Sun, Q.; Sun, X.; Zhou, Z. Hepatic overproduction of 13-HODE due to ALOX15 upregulation contributes to alcohol-induced liver injury in mice. Sci. Rep. 2017, 7, 8976. [Google Scholar] [CrossRef] [Green Version]
- Sweeney, N.W.; Gomes, C.J.; De Armond, R.; Centuori, S.M. Hypoxia Suppresses High Fat Diet-Induced Steatosis And Development Of Hepatic Adenomas. Hypoxia 2019, 7, 53–63. [Google Scholar] [CrossRef] [Green Version]
- Sinha, R.A.; Bruinstroop, E.; Singh, B.K.; Yen, P.M. Nonalcoholic Fatty Liver Disease and Hypercholesterolemia: Roles of Thyroid Hormones, Metabolites, and Agonists. Thyroid Off. J. Am. Thyroid Assoc. 2019, 29, 1173–1191. [Google Scholar] [CrossRef]
- Pusec, C.M.; De Jesus, A.; Khan, M.W.; Terry, A.R.; Ludvik, A.E.; Xu, K.; Giancola, N.; Pervaiz, H.; Daviau Smith, E.; Ding, X.; et al. Hepatic HKDC1 Expression Contributes to Liver Metabolism. Endocrinology 2019, 160, 313–330. [Google Scholar] [CrossRef] [Green Version]
- Iverson, S.V.; Eriksson, S.; Xu, J.; Prigge, J.R.; Talago, E.A.; Meade, T.A.; Meade, E.S.; Capecchi, M.R.; Arnér, E.S.; Schmidt, E.E. A Txnrd1-dependent metabolic switch alters hepatic lipogenesis, glycogen storage, and detoxification. Free Radic. Biol. Med. 2013, 63, 369–380. [Google Scholar] [CrossRef] [Green Version]
- Lai, R.K.; Xu, I.M.; Chiu, D.K.; Tse, A.P.; Wei, L.L.; Law, C.T.; Lee, D.; Wong, C.M.; Wong, M.P.; Ng, I.O.; et al. NDUFA4L2 Fine-tunes Oxidative Stress in Hepatocellular Carcinoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2016, 22, 3105–3117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Peng, J.; Zhou, Y.; Xie, B.; Wang, J. Silencing RRM2 inhibits multiple myeloma by targeting the Wnt/β-catenin signaling pathway. Mol. Med. Rep. 2019, 20, 2159–2166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Chen, Z.; Huang, Z.; Yu, H.; Li, Y.; Huang, W. Formiminotransferase Cyclodeaminase Suppresses Hepatocellular Carcinoma by Modulating Cell Apoptosis, DNA Damage, and Phosphatidylinositol 3-Kinases (PI3K)/Akt Signaling Pathway. Med Sci. Monit. Int. Med. J. Exp. Clin. Res. 2019, 25, 4474–4484. [Google Scholar] [CrossRef]
- Bowman, T.A.; O’Keeffe, K.R.; D’Aquila, T.; Yan, Q.W.; Griffin, J.D.; Killion, E.A.; Salter, D.M.; Mashek, D.G.; Buhman, K.K.; Greenberg, A.S. Acyl CoA synthetase 5 (ACSL5) ablation in mice increases energy expenditure and insulin sensitivity and delays fat absorption. Mol. Metab. 2016, 5, 210–220. [Google Scholar] [CrossRef]
- Ohyama, K.; Suzuki, K. Dihydrocapsiate improved age-associated impairments in mice by increasing energy expenditure. Am. J. Physiol. Endocrinol. Metab. 2017, 313, E586–E597. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Liu, S.; Zhang, X.; Lam, S.M.; Hu, X.; Zhou, Y.; Chen, J.; Wang, Y.; Wu, C.; Shui, G.; et al. Requirement of cytosolic phospholipase A2 gamma in lipid droplet formation. Biochim. Et Biophys. Acta. Mol. Cell Biol. Lipids 2017, 1862, 692–705. [Google Scholar] [CrossRef] [PubMed]
- Mayr, J.A.; Feichtinger, R.G.; Tort, F.; Ribes, A.; Sperl, W. Lipoic acid biosynthesis defects. J. Inherit. Metab. Dis. 2014, 37, 553–563. [Google Scholar] [CrossRef] [PubMed]
- Virtanen, K.A. The rediscovery of BAT in adult humans using imaging. Best Pract. Res. Clin. Endocrinol. Metab. 2016, 30, 471–477. [Google Scholar] [CrossRef]
- Acosta-Andrade, C.; Lambertos, A.; Urdiales, J.L.; Sánchez-Jiménez, F.; Peñafiel, R.; Fajardo, I. A novel role for antizyme inhibitor 2 as a regulator of serotonin and histamine biosynthesis and content in mouse mast cells. Amino Acids 2016, 48, 2411–2421. [Google Scholar] [CrossRef]
- Xu, Y.; Qi, X.; Hu, M.; Lin, R.; Hou, Y.; Wang, Z.; Zhou, H.; Zhao, Y.; Luan, Y.; Zhao, S.; et al. Transcriptome Analysis of Adipose Tissue Indicates That the cAMP Signaling Pathway Affects the Feed Efficiency of Pigs. Genes 2018, 9, 336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Z.; Michal, J.J.; Wu, X.L.; Pan, Z.; MacNeil, M.D. The heparan and heparin metabolism pathway is involved in regulation of fatty acid composition. Int. J. Biol. Sci. 2011, 7, 659–663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kübeck, R.; Bonet-Ripoll, C.; Hoffmann, C.; Walker, A.; Müller, V.M.; Schüppel, V.L.; Lagkouvardos, I.; Scholz, B.; Engel, K.H.; Daniel, H.; et al. Dietary fat and gut microbiota interactions determine diet-induced obesity in mice. Mol. Metab. 2016, 5, 1162–1174. [Google Scholar] [CrossRef]
- Ueda, T.; Yokota, T.; Okuzaki, D.; Uno, Y.; Mashimo, T.; Kubota, Y.; Sudo, T.; Ishibashi, T.; Shingai, Y.; Doi, Y.; et al. Endothelial Cell-Selective Adhesion Molecule Contributes to the Development of Definitive Hematopoiesis in the Fetal Liver. Stem Cell Rep. 2019, 13, 992–1005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramani, K.; Lu, S.C. Methionine adenosyltransferases in liver health and diseases. Liver Res. 2017, 1, 103–111. [Google Scholar] [CrossRef]
- Xie, Y.; Shi, X.; Sheng, K.; Han, G.; Li, W.; Zhao, Q.; Jiang, B.; Feng, J.; Li, J.; Gu, Y. PI3K/Akt signaling transduction pathway, erythropoiesis and glycolysis in hypoxia (Review). Mol. Med. Rep. 2019, 19, 783–791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoxhaj, G.; Manning, B.D. The PI3K-AKT network at the interface of oncogenic signalling and cancer metabolism. Nat. Rev. Cancer 2020, 20, 74–88. [Google Scholar] [CrossRef]
- Tewari, D.; Patni, P.; Bishayee, A.; Sah, A.N.; Bishayee, A. Natural products targeting the PI3K-Akt-mTOR signaling pathway in cancer: A novel therapeutic strategy. Semin. Cancer Biol. 2019. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhao, Y.; He, F.; Wang, H. Artificial MicroRNA-Mediated Tgfbr2 and Pdgfrb Co-Silencing Ameliorates Carbon Tetrachloride-Induced Hepatic Fibrosis in Mice. Hum. Gene Ther. 2019, 30, 179–196. [Google Scholar] [CrossRef] [PubMed]
- Katoh, M. FGFR inhibitors: Effects on cancer cells, tumor microenvironment and whole-body homeostasis (Review). Int. J. Mol. Med. 2016, 38, 3–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sung, J.S.; Kang, C.W. ITGB4-mediated metabolic reprogramming of cancer-associated fibroblasts. Oncogene 2020, 39, 664–676. [Google Scholar] [CrossRef] [PubMed]
- Satapati, S.; Kucejova, B.; Duarte, J.A.; Fletcher, J.A.; Reynolds, L.; Sunny, N.E.; He, T.; Nair, L.A.; Livingston, K.A.; Fu, X.; et al. Mitochondrial metabolism mediates oxidative stress and inflammation in fatty liver. J. Clin. Investig. 2015, 125, 4447–4462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hatting, M.; Tavares, C.D.J.; Sharabi, K.; Rines, A.K.; Puigserver, P. Insulin regulation of gluconeogenesis. Ann. N. Y. Acad. Sci. 2018, 1411, 21–35. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, Y.; Satoh, A.; Yabe, S.; Furusawa, M.; Tokushige, N.; Tezuka, H.; Mikami, M.; Iwata, W.; Shingyouchi, A.; Matsuzaka, T.; et al. Hepatic CREB3L3 controls whole-body energy homeostasis and improves obesity and diabetes. Endocrinology 2014, 155, 4706–4719. [Google Scholar] [CrossRef] [Green Version]
- Feng, Y.X.; Jin, D.X.; Sokol, E.S.; Reinhardt, F.; Miller, D.H.; Gupta, P.B. Cancer-specific PERK signaling drives invasion and metastasis through CREB3L1. Nat. Commun. 2017, 8, 1079. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, S.; Vargas, J. Signaling via the NFκB system. Wiley Interdiscip. Rev. Syst. Biology Med. 2016, 8, 227–241. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Chen, P. Evaluation of Naringenin as a Promising Treatment Option for COPD Based on Literature Review and Network Pharmacology. Biomolecules 2020, 10, 1644. [Google Scholar] [CrossRef] [PubMed]
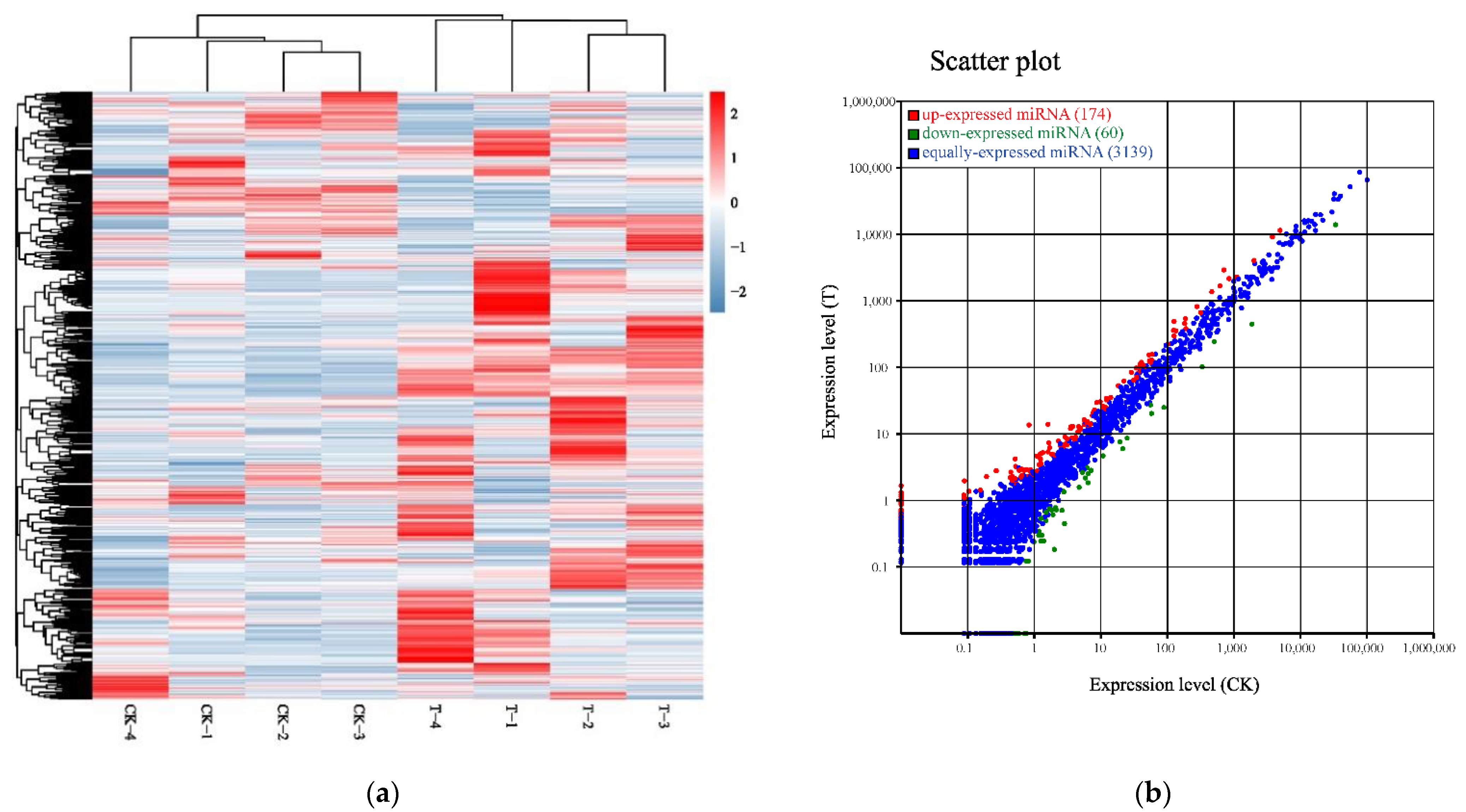
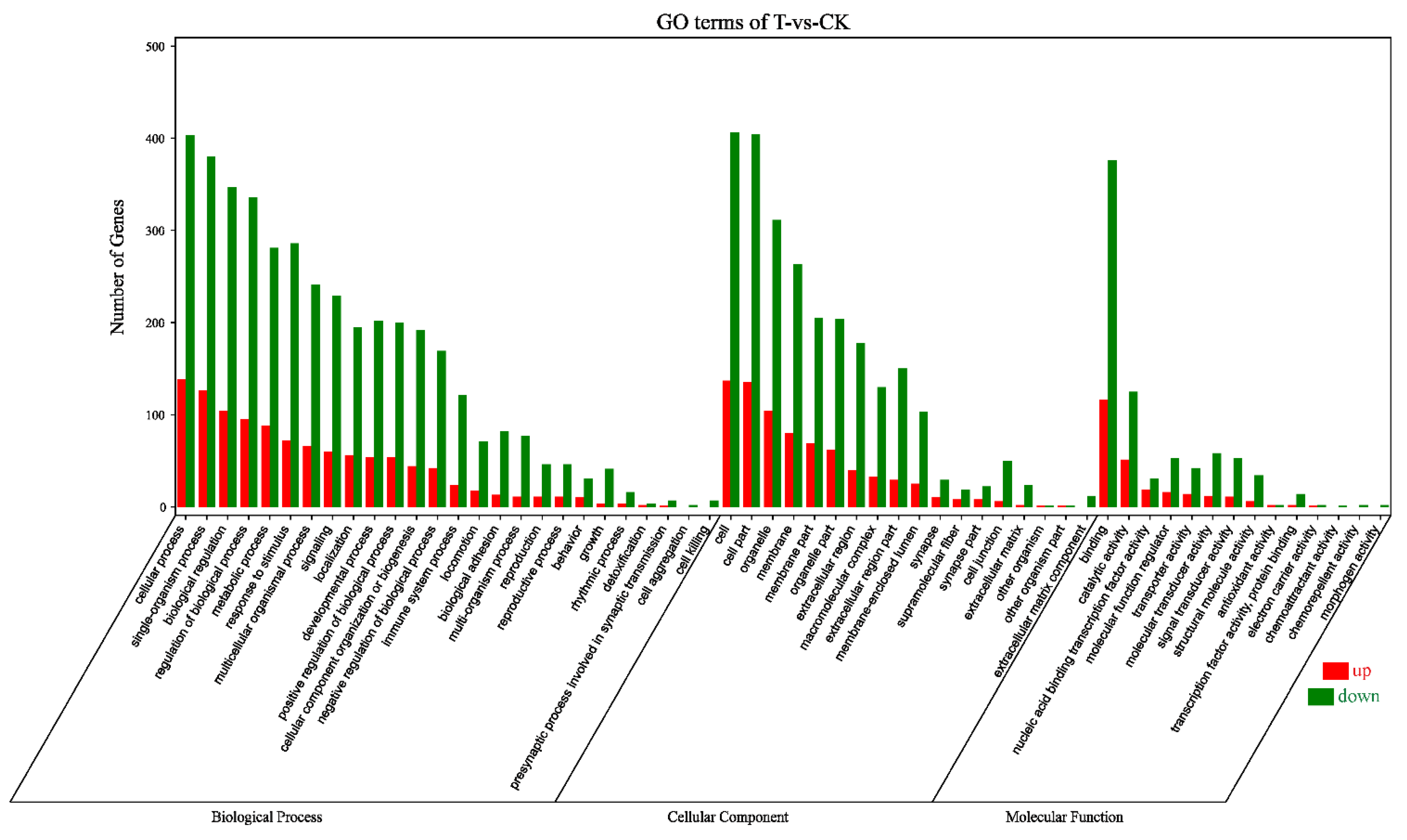



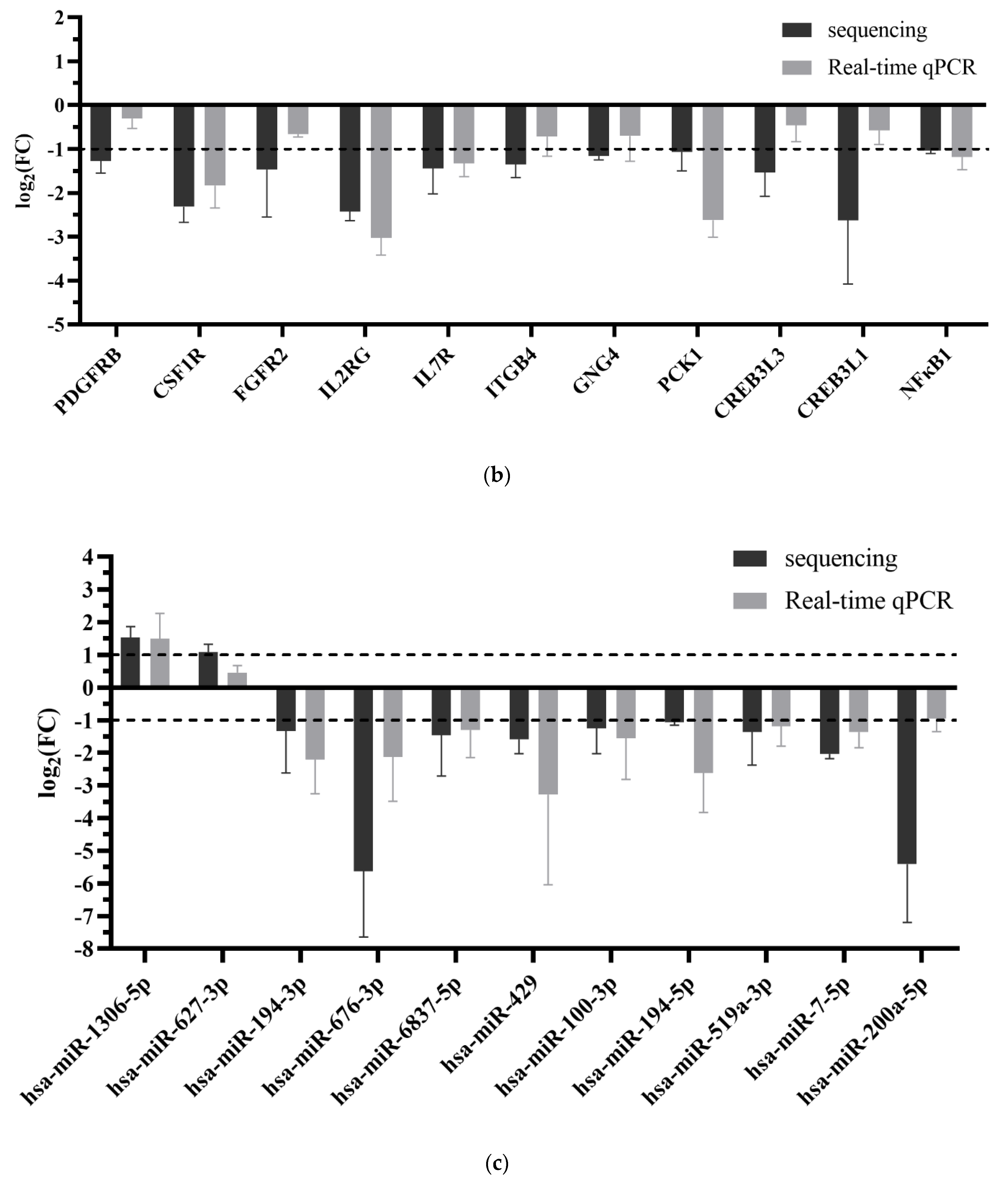
| Sample | Raw Data | Clean Data (%) | Raw Data (bp) | Clean Data (bp) | After Filtering Q20 (%) | After Filtering Q30 (%) | After Filtering N (%) | After Filtering GC (%) |
|---|---|---|---|---|---|---|---|---|
| CK-1 | 47,663,932 | 47,577,906 (99.82%) | 7,149,589,800 | 7,114,281,738 | 6,992,811,543 (98.29%) | 6,756,607,154 (94.97%) | 19,427 (0.00%) | 3,804,122,670 (53.47%) |
| CK-2 | 42,079,942 | 42,026,040 (99.87%) | 6,311,991,300 | 6,283,554,811 | 6,189,856,925 (98.51%) | 6,000,864,261 (95.50%) | 8392 (0.00%) | 3,360,455,066 (53.48%) |
| CK-3 | 41,188,030 | 41,131,120 (99.86%) | 6,178,204,500 | 6,147,638,474 | 6,050,714,205 (98.42%) | 5,857,735,081 (95.28%) | 8775 (0.00%) | 3,273,834,153 (53.25%) |
| CK-4 | 43,096,530 | 43,039,450 (99.87%) | 6,464,479,500 | 6,433,433,599 | 6,333,636,124 (98.45%) | 6,134,820,735 (95.36%) | 8660 (0.00%) | 3,439,680,095 (53.47%) |
| T-1 | 51,658,770 | 51,539,258 (99.77%) | 7,748,815,500 | 7,706,270,162 | 7,562,514,956 (98.13%) | 7,292,630,398 (94.63%) | 26,068 (0.00%) | 4,098,720,423 (53.19%) |
| T-2 | 48,606,134 | 48,501,812 (99.79%) | 7,290,920,100 | 7,250,461,713 | 7,113,064,471 (98.10%) | 6,85,1340,567 (94.50%) | 23,956 (0.00%) | 3,887,698,839 (53.62%) |
| T-3 | 62,602,866 | 62,433,856 (99.73%) | 9,390,429,900 | 9,333,102,578 | 9,155,158,477 (98.09%) | 8,825,370,611 (94.56%) | 45,571 (0.00%) | 4,878,494,560 (52.27%) |
| T-4 | 41,346,744 | 41,292,738 (99.87%) | 6,202,011,600 | 6,166,582,431 | 6,070,483,986 (98.44%) | 5,876,108,215 (95.29%) | 8644 (0.00%) | 3,277,271,342 (53.15%) |
| Sample | Clean Reads | High Quality | Smaller than 18 Nt | Polya | Low Cutoff | Clean Tags |
|---|---|---|---|---|---|---|
| CK-1 | 13,123,917 | 13,076,410 | 2,570,571 | 540 | 328,987 | 10,051,993 |
| (100%) | (99.6380%) | (19.6581%) | (0.0041%) | (2.5159%) | (76.8712%) | |
| CK-2 | 13,744,858 | 13,687,348 | 1,981,919 | 807 | 443,665 | 11,100,847 |
| (100%) | (99.5816%) | (14.4799%) | (0.0059%) | (3.2414%) | (81.1030%) | |
| CK-3 | 12,609,501 | 12,563,626 | 1,535,545 | 766 | 306,124 | 10,571,146 |
| (100%) | (99.6362%) | (12.2221%) | (0.0061%) | (2.4366%) | (84.1409%) | |
| CK-4 | 13,923,071 | 13,865,587 | 3,070,687 | 377 | 386,734 | 10,259,540 |
| (100%) | (99.5871%) | (22.1461%) | (0.0027%) | (2.7892%) | (73.9928%) | |
| T-1 | 12,156,840 | 12,100,462 | 2,354,606 | 464 | 465,005 | 9,161,791 |
| (100%) | (99.5362%) | (19.4588%) | (0.0038%) | (3.8429%) | (75.7144%) | |
| T-2 | 12,620,973 | 12,574,364 | 1,472,821 | 741 | 337,320 | 10,592,975 |
| (99.6307%) | (11.7129%) | (0.0059%) | (2.6826%) | (84.2426%) | ||
| T-3 | 12,806,071 | 12,758,280 | 1,603,461 | 1012 | 307,185 | 10,700,322 |
| (100%) | (99.6268%) | (12.5680%) | (0.0079%) | (2.4077%) | (83.8696%) | |
| T-4 | 14,891,138 | 14,831,250 | 2,514,654 | 556 | 317,307 | 11,831,747 |
| (100%) | (99.5978%) | (16.9551%) | (0.0037%) | (2.1394%) | (79.7758%) |
| Gene | Forward Sequence (5′→3′) | Reverse Sequence (5′→3′) |
|---|---|---|
| PDGFRB | TGCAGACATCGAGTCCTCCAAC | GCTTAGCACTGGAGACTCGTTG |
| CSF1R | GCTGCCTTACAACGAGAAGTGG | CATCCTCCTTGCCCAGACCAAA |
| FGFR2 | GTGCCGAATGAAGAACACGACC | GGCGTGTTGTTATCCTCACCAG |
| IL2RG | CACTCTGTGGAAGTGCTCAGCA | GAGCCAACAGAGATAACCACGG |
| IL7R | ATCGCAGCACTCACTGACCTGT | TCAGGCACTTTACCTCCACGAG |
| ITGB4 | AGGATGACGACGAGAAGCAGCT | ACCGAGAACTCAGGCTGCTCAA |
| GNG4 | CTCCAGATTCAGCCTCCGTTTTG | TGCCATAGGTCTGGAAGAGGTG |
| PCK1 | CATTGCCTGGATGAAGTTTGACG | GGGTTGGTCTTCACTGAAGTCC |
| CREB3L3 | GAAGCCTCTGTGACCATAGACC | GGAGGTCTTTCACGGTGAGATTG |
| CREB3L1 | GCCTTGTGCTTTGTTCTGGTGC | CCGTCATCGTAGAATAGGAGGC |
| NFκB1 | GCAGCACTACTTCTTGACCACC | TCTGCTCCTGAGCATTGACGTC |
| ALOX15 | ACCTTCCTGCTCGCCTAGTGTT | GGCTACAGAGAATGACGTTGGC |
| CA9 | GTGCCTATGAGCAGTTGCTGTC | AAGTAGCGGCTGAAGTCAGAGG |
| TH | GCTGGACAAGTGTCATCACCTG | CCTGTACTGGAAGGCGATCTCA |
| HKDC1 | ATCGCCGACTTCCTGGACTACA | GCCTTGAAACCTTTGGTCCACC |
| NDUFA4L2 | CTGGGACAGAAAGAACAACCCG | CAGCCTGGCTTAGAAGTCTGGC |
| RRM2 | CTGGCTCAAGAAACGAGGACTG | CTCTCCTCCGATGGTTTGTGTAC |
| ACSL5 | GCTTATGAGCCCACTCCTGATG | GGAAGAATCCAACTCTGGCTCC |
| PLA2G4C | GGAAGACTGGTCAGAACTCACC | GCATTAGCAACAGCCCTTCTCC |
| LIPT2 | GTCTGGCTAGACGATCGCAAGA | GCACGATGTGCTCAAACCACGT |
| UGDH | TGTGATGGTGCCCATGCTGTTG | GTCCATCGAAGATAAAGGCTGGC |
| FTCD | GGAGAACCTCTTCATCCTGGAG | ATGATCCGCTCCTTAGGGCTGA |
| ABAT | GCCTCTGATGAAGACGGAAGTC | CATTCGGTTGCCGTCCACATCA |
| AZIN2 | CTTCACTGTGGCAGTCAGCATC | TCCCATACACGCCCTCATCAAG |
| HS6ST3 | ACTGGACGGAGCTCACCAACTG | TCGCTCAGGTAACGTGACACTG |
| B4GALT6 | CTCATTCCTTTCCGTAATCGCCA | GCCCACATTGAAAAGCATCGCAC |
| GUSB | CTGTCACCAAGAGCCAGTTCCT | GGTTGAAGTCCTTCACCAGCAG |
| DCT | CTCAGACCAACTTGGCTACAGC | CAACCAAAGCCACCAGTGTTCC |
| ALAS2 | GCCTCAAAGGATGTGTCCGTCT | TACTGGTGCCTGAGATGTTGCG |
| MAT1A | GCCAAGTCTCTGGTGAAAGCAG | CTGTCTTCTGAGAGGTTCCGTAG |
| β-Actin | TGAATGATGAGCCTTCGTGC | CTGGTCTCAAGTCAGTGTAC |
| U6 | CTCGCTTCGGCAGCACA | AACGCTTCACGAATTTGCGT |
| miRNA Name | miRNA Sequence (5′→3′) | Forward Sequence (5′→3′) |
|---|---|---|
| hsa-miR-1306-5p | CCACCTCCCCTGCAAACGTCCA | GCCACCTCCCCTGC |
| hsa-miR-627-3p | TCTTTTCTTTGAGACTCACT | CGCAGTCTTTTCTTTGAGACTC |
| hsa-miR-194-3p | CCAGTGGGGCTGCTGTTATCTG | CAGTGGGGCTGCTGT |
| hsa-miR-676-3p | CTGTCCTAAGGTTGTTGAGTT | CAGCTGTCCTAAGGTTGTTG |
| hsa-miR-6837-5p | ACCAGGGCCAGCAGGGAATGT | ACCAGGGCCAGCAG |
| hsa-miR-429 | TAATACTGTCTGGTAAAACCGT | CGCAGTAATACTGTCTGGT |
| hsa-miR-100-3p | CAAGCTTGTATCTATAGGTATG | CGCAGCAAGCTTGTATC |
| hsa-miR-194-5p | CGGGTAGAGAGGGCAGTGGGAGG | CGGGTAGAGAGGGCAGT |
| hsa-miR-519a-3p | AAAGTGCATCCTTTTAGAGTGT | GCAGAAAGTGCATCCTTTTAGAG |
| hsa-miR-7-5p | TGGAAGACTAGTGATTTTGTTGTT | CGCAGTGGAAGACTAGTGA |
| hsa-miR-200a-5p | CATCTTACCGGACAGTGCTGGA | AGCATCTTACCGGACAGT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, W.; Shi, R.; Guan, M.; Chen, P.; Wu, H.; Su, W.; Wang, Y.; Li, P. The Effects of Naringenin on miRNA-mRNA Profiles in HepaRG Cells. Int. J. Mol. Sci. 2021, 22, 2292. https://doi.org/10.3390/ijms22052292
Fan W, Shi R, Guan M, Chen P, Wu H, Su W, Wang Y, Li P. The Effects of Naringenin on miRNA-mRNA Profiles in HepaRG Cells. International Journal of Molecular Sciences. 2021; 22(5):2292. https://doi.org/10.3390/ijms22052292
Chicago/Turabian StyleFan, Weiyang, Rui Shi, Minyi Guan, Pan Chen, Hao Wu, Weiwei Su, Yonggang Wang, and Peibo Li. 2021. "The Effects of Naringenin on miRNA-mRNA Profiles in HepaRG Cells" International Journal of Molecular Sciences 22, no. 5: 2292. https://doi.org/10.3390/ijms22052292






