What Do We Know about Barley miRNAs?
Abstract
:1. Introduction
2. Plant miRNAs—Biogenesis and Regulatory Potential
3. miRNAs in Barley Physiology and Stress Responses
4. Target Transcripts of Barley miRNAs
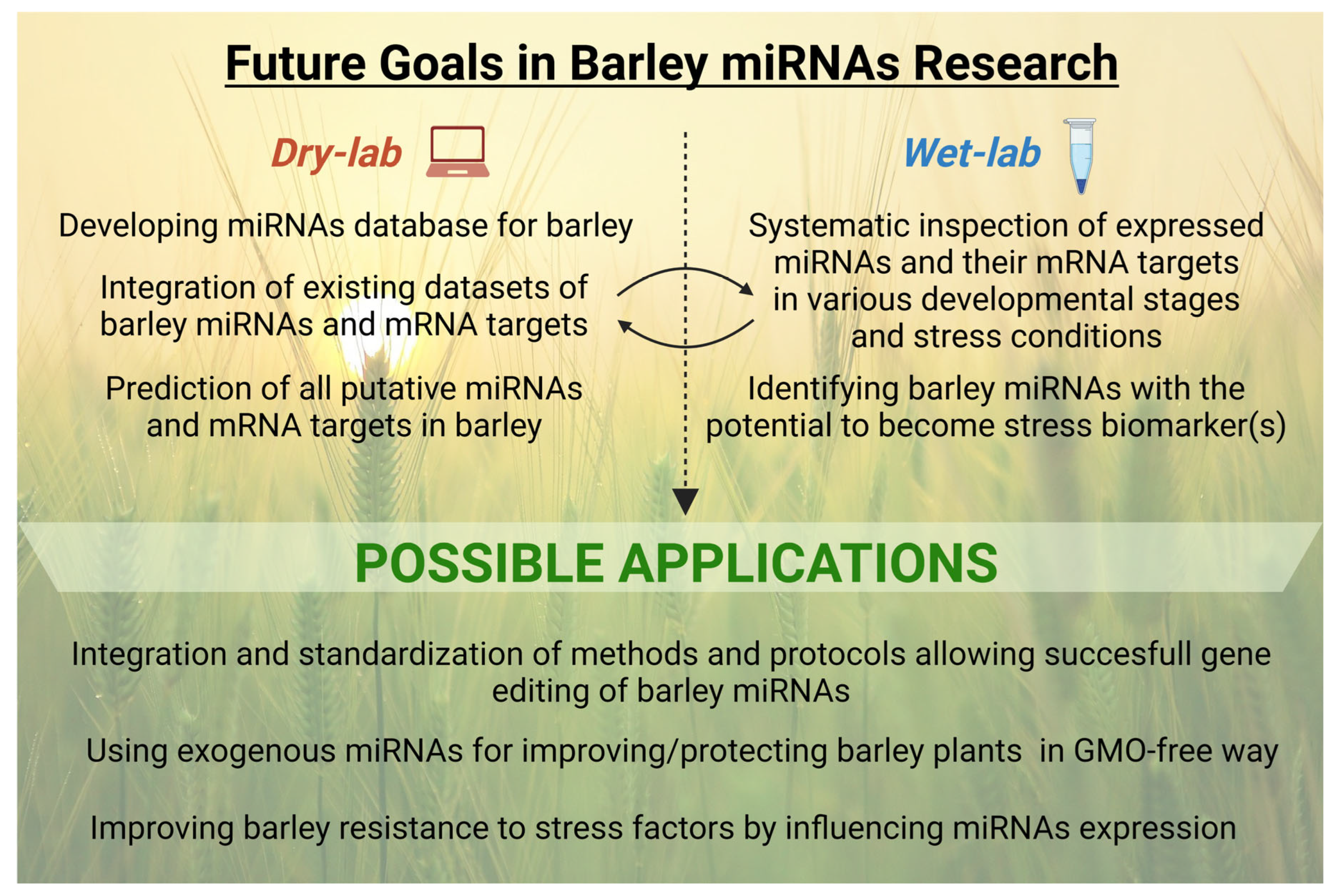
5. Conclusions and Future Directions
- (a)
- Showing co-expression of miRNA and target mRNA in vivo;
- (b)
- Proving interaction between miRNA and a specific site within target mRNA;
- (c)
- Demonstrating miRNA-mediated effects on target protein expression;
- (d)
- Demonstrating miRNA effects on biological function.
- (1)
- Are some of the barley miRNAs tissue/developmental, or stage-specific? Are we able to catalog it in some integrative and user-friendly way? For this purpose, it would be beneficial to have something like a barley miRNA atlas (similar to PmiRExAt, where wheat, rice, maize, and Arabidopsis miRNAs in multiple tissues and developmental stages can be found) [171].
- (2)
- Which barley miRNAs have the potential to become a useful stress biomarker? In other words, do some stress-specific miRNAs exist?
- (3)
- Is barley miRNome rather complete, or not? Compared to rice, wheat, and Arabidopsis, the total number of known barley miRNAs is still lack behind, and bona fide many discoveries waiting for us!
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Lam, J.K.W.; Chow, M.Y.T.; Zhang, Y.; Leung, S.W.S. SiRNA versus MiRNA as Therapeutics for Gene Silencing. Mol. Ther. Nucleic Acids 2015, 4, e252. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Mei, J.; Ren, G. Plant MicroRNAs: Biogenesis, Homeostasis, and Degradation. Front. Plant Sci. 2019, 10, 360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sen, G.L.; Blau, H.M. A Brief History of RNAi: The Silence of the Genes. FASEB J. 2006, 20, 1293–1299. [Google Scholar] [CrossRef] [Green Version]
- Napoli, C.; Lemieux, C.; Jorgensen, R. Introduction of a Chimeric Chalcone Synthase Gene into Petunia Results in Reversible Co-Suppression of Homologous Genes in Trans. Plant Cell 1990, 2, 279–289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romano, N.; Macino, G. Quelling: Transient Inactivation of Gene Expression in Neurospora Crassa by Transformation with Homologous Sequences. Mol. Microbiol. 1992, 6, 3343–3353. [Google Scholar] [CrossRef]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans Heterochronic Gene Lin-4 Encodes Small RNAs with Antisense Complementarity to Lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef] [PubMed]
- Ambros, V.; Bartel, B.; Bartel, D.P.; Burge, C.B.; Carrington, J.C.; Chen, X.; Dreyfuss, G.; Eddy, S.R.; Griffiths-Jones, S.; Marshall, M.; et al. A Uniform System for MicroRNA Annotation. RNA 2003, 9, 277–279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gebert, L.F.R.; MacRae, I.J. Regulation of MicroRNA Function in Animals. Nat. Rev. Mol. Cell Biol. 2019, 20, 21–37. [Google Scholar] [CrossRef] [PubMed]
- Garcia, D. A MiRacle in Plant Development: Role of MicroRNAs in Cell Differentiation and Patterning. Semin. Cell Dev. Biol. 2008, 19, 586–595. [Google Scholar] [CrossRef]
- Millar, A.A. The Function of MiRNAs in Plants. Plants 2020, 9, 198. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, T.; Zhang, F.; Zhang, Y.; Liang, Y. RNA Interference: A Natural Immune System of Plants to Counteract Biotic Stressors. Cells 2019, 8, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Galili, G. Tuning the Orchestra: MiRNAs in Plant Immunity. Trends Plant Sci. 2019, 24, 189–191. [Google Scholar] [CrossRef] [PubMed]
- Cui, C.; Wang, J.-J.; Zhao, J.-H.; Fang, Y.-Y.; He, X.-F.; Guo, H.-S.; Duan, C.-G. A Brassica MiRNA Regulates Plant Growth and Immunity through Distinct Modes of Action. Mol. Plant 2020, 13, 231–245. [Google Scholar] [CrossRef]
- Mengistu, A.A.; Tenkegna, T.A. The Role of MiRNA in Plant–Virus Interaction: A Review. Mol. Biol. Rep. 2021, 48, 2853–2861. [Google Scholar] [CrossRef]
- Pagano, L.; Rossi, R.; Paesano, L.; Marmiroli, N.; Marmiroli, M. MiRNA Regulation and Stress Adaptation in Plants. Environ. Exp. Bot. 2021, 184, 104369. [Google Scholar] [CrossRef]
- Nevo, E. Evolution of Wild Barley and Barley Improvement. In Advance in Barley Sciences; Springer: Dordrecht, The Netherlands, 2013; pp. 1–23. [Google Scholar]
- Pourkheirandish, M.; Komatsuda, T. The Importance of Barley Genetics and Domestication in a Global Perspective. Ann. Bot. 2007, 100, 999–1008. [Google Scholar] [CrossRef] [Green Version]
- Ullrich, S.E. Barley: Production, Improvement, and Uses; John Wiley & Sons: New York, NY, USA, 2010. [Google Scholar]
- Tosh, S.M.; Bordenave, N. Emerging Science on Benefits of Whole Grain Oat and Barley and Their Soluble Dietary Fibers for Heart Health, Glycemic Response, and Gut Microbiota. Nutr. Rev. 2020, 78, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Lahouar, L.; El-Bok, S.; Achour, L. Therapeutic Potential of Young Green Barley Leaves in Prevention and Treatment of Chronic Diseases: An Overview. Am. J. Chin. Med. 2015, 43, 1311–1329. [Google Scholar] [CrossRef]
- Pech, R.; Volná, A.; Hunt, L.; Bartas, M.; Červeň, J.; Pečinka, P.; Špunda, V.; Nezval, J. Regulation of Phenolic Compound Production by Light Varying in Spectral Quality and Total Irradiance. Int. J. Mol. Sci. 2022, 23, 6533. [Google Scholar] [CrossRef]
- Harwood, W.A. An Introduction to Barley: The Crop and the Model. In Barley; Humana Press: New York, NY, UAS, 2019; pp. 1–5. [Google Scholar]
- Sato, K. History and Future Perspectives of Barley Genomics. DNA Res. 2020, 27, dsaa023. [Google Scholar] [CrossRef] [PubMed]
- Saski, C.; Lee, S.-B.; Fjellheim, S.; Guda, C.; Jansen, R.K.; Luo, H.; Tomkins, J.; Rognli, O.A.; Daniell, H.; Clarke, J.L. Complete Chloroplast Genome Sequences of Hordeum vulgare, Sorghum bicolor and Agrostis stolonifera, and Comparative Analyses with Other Grass Genomes. Theor. Appl. Genet. 2007, 115, 571–590. [Google Scholar] [CrossRef] [Green Version]
- Schoch, C.L.; Ciufo, S.; Domrachev, M.; Hotton, C.L.; Kannan, S.; Khovanskaya, R.; Leipe, D.; Mcveigh, R.; O’Neill, K.; Robbertse, B. NCBI Taxonomy: A Comprehensive Update on Curation, Resources and Tools. Database 2020, 2020, baaa062. [Google Scholar] [CrossRef] [PubMed]
- Sato, F.; Tsuchiya, S.; Meltzer, S.J.; Shimizu, K. MicroRNAs and Epigenetics. FEBS J. 2011, 278, 1598–1609. [Google Scholar] [CrossRef] [PubMed]
- Shapulatov, U.; van Hoogdalem, M.; Schreuder, M.; Bouwmeester, H.; Abdurakhmonov, I.Y.; van der Krol, A.R. Functional Intron-Derived MiRNAs and Host-Gene Expression in Plants. Plant Methods 2018, 14, 83. [Google Scholar] [CrossRef]
- Wu, X.; Hornyik, C.; Bayer, M.; Marshall, D.; Waugh, R.; Zhang, R. In Silico Identification and Characterization of Conserved Plant MicroRNAs in Barley. Open Life Sci. 2014, 9, 841–852. [Google Scholar] [CrossRef] [Green Version]
- Baldrich, P.; Hsing, Y.-I.C.; San Segundo, B. Genome-Wide Analysis of Polycistronic MicroRNAs in Cultivated and Wild Rice. Genome. Biol. Evol. 2016, 8, 1104–1114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zou, Q.; Mao, Y.; Hu, L.; Wu, Y.; Ji, Z. MiRClassify: An Advanced Web Server for MiRNA Family Classification and Annotation. Comput. Biol. Med. 2014, 45, 157–160. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.; Dou, Y.; Li, S.; Ren, G.; Chevalier, D.; Zhang, C.; Yu, B. DAWDLE Interacts with DICER-LIKE Proteins to Mediate Small RNA Biogenesis. Plant Physiol. 2018, 177, 1142–1151. [Google Scholar] [CrossRef]
- Zhang, L.; Xiang, Y.; Chen, S.; Shi, M.; Jiang, X.; He, Z.; Gao, S. Mechanisms of MicroRNA Biogenesis and Stability Control in Plants. Front. Plant Sci. 2022, 13, 844149. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Yang, Z.; Li, J.; Minakhina, S.; Yang, M.; Padgett, R.W.; Steward, R.; Chen, X. Methylation as a Crucial Step in Plant MicroRNA Biogenesis. Science 2005, 307, 932–935. [Google Scholar] [CrossRef] [Green Version]
- Djami-Tchatchou, A.T.; Sanan-Mishra, N.; Ntushelo, K.; Dubery, I.A. Functional Roles of MicroRNAs in Agronomically Important Plants—Potential as Targets for Crop Improvement and Protection. Front. Plant Sci. 2017, 8, 378. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Yu, B. Recent Advances in the Regulation of Plant MiRNA Biogenesis. RNA Biol. 2021, 18, 2087–2096. [Google Scholar] [CrossRef] [PubMed]
- Medley, J.C.; Panzade, G.; Zinovyeva, A.Y. MicroRNA Strand Selection: Unwinding the Rules. Wiley Interdiscip. Rev. RNA 2021, 12, e1627. [Google Scholar] [CrossRef] [PubMed]
- Meijer, H.A.; Smith, E.M.; Bushell, M. Regulation of MiRNA Strand Selection: Follow the Leader? Biochem. Soc. Trans. 2014, 42, 1135–1140. [Google Scholar] [CrossRef]
- Vimalraj, S.; Selvamurugan, N. MicroRNAs: Synthesis, Gene Regulation and Osteoblast Differentiation. Curr. Issues Mol. Biol. 2013, 15, 7–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forman, J.J.; Coller, H.A. The Code within the Code: MicroRNAs Target Coding Regions. Cell Cycle 2010, 9, 1533–1541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, B.; Wang, H. Translational Inhibition by MicroRNAs in Plants. Prog. Mol. Subcell. Biol. 2010, 50, 41–57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Teng, C.; Xia, R.; Meyers, B.C. PhasiRNAs in Plants: Their Biogenesis, Genic Sources, and Roles in Stress Responses, Development, and Reproduction. Plant Cell 2020, 32, 3059–3080. [Google Scholar] [CrossRef] [PubMed]
- Araki, S.; Le, N.T.; Koizumi, K.; Villar-Briones, A.; Nonomura, K.-I.; Endo, M.; Inoue, H.; Saze, H.; Komiya, R. MiR2118-Dependent U-Rich PhasiRNA Production in Rice Anther Wall Development. Nat. Commun. 2020, 11, 3115. [Google Scholar] [CrossRef]
- Xia, R.; Chen, C.; Pokhrel, S.; Ma, W.; Huang, K.; Patel, P.; Wang, F.; Xu, J.; Liu, Z.; Li, J.; et al. 24-Nt Reproductive PhasiRNAs Are Broadly Present in Angiosperms. Nat. Commun. 2019, 10, 627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, Z.; Khanna, K.; Ruan, S. Expression of MicroRNAs and Its Regulation in Plants. Semin. Cell Dev. Biol. 2010, 21, 790–797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chhabra, R. MiRNA and Methylation: A Multifaceted Liaison. ChemBioChem 2015, 16, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Świda-Barteczka, A.; Krieger-Liszkay, A.; Bilger, W.; Voigt, U.; Hensel, G.; Szweykowska-Kulinska, Z.; Krupinska, K. The Plastid-Nucleus Located DNA/RNA Binding Protein WHIRLY1 Regulates MicroRNA-Levels during Stress in Barley (Hordeum vulgare L.). RNA Biol. 2018, 15, 886–891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Mo, B.; Chen, X. Mechanisms That Impact MicroRNA Stability in Plants. RNA Biol. 2012, 9, 1218–1223. [Google Scholar] [CrossRef] [Green Version]
- Gallego-Bartolomé, J. DNA Methylation in Plants: Mechanisms and Tools for Targeted Manipulation. New Phytol. 2020, 227, 38–44. [Google Scholar] [CrossRef] [Green Version]
- Wu, L.; Zhou, H.; Zhang, Q.; Zhang, J.; Ni, F.; Liu, C.; Qi, Y. DNA Methylation Mediated by a MicroRNA Pathway. Mol. Cell 2010, 38, 465–475. [Google Scholar] [CrossRef] [PubMed]
- Bao, N.; Lye, K.-W.; Barton, M.K. MicroRNA Binding Sites in Arabidopsis Class III HD-ZIP MRNAs Are Required for Methylation of the Template Chromosome. Dev. Cell 2004, 7, 653–662. [Google Scholar] [CrossRef] [PubMed]
- Vasudevan, S. Posttranscriptional Upregulation by MicroRNAs. WIREs RNA 2012, 3, 311–330. [Google Scholar] [CrossRef] [PubMed]
- Lauressergues, D.; Couzigou, J.-M.; Clemente, H.S.; Martinez, Y.; Dunand, C.; Bécard, G.; Combier, J.-P. Primary Transcripts of MicroRNAs Encode Regulatory Peptides. Nature 2015, 520, 90–93. [Google Scholar] [CrossRef] [PubMed]
- Prasad, A.; Sharma, N.; Prasad, M. Noncoding but Coding: Pri-MiRNA into the Action. Trends Plant Sci. 2021, 26, 204–206. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Badola, P.K.; Bhatia, C.; Sharma, D.; Trivedi, P.K. Primary Transcript of MiR858 Encodes Regulatory Peptide and Controls Flavonoid Biosynthesis and Development in Arabidopsis. Nat. Plants 2020, 6, 1262–1274. [Google Scholar] [CrossRef]
- Couzigou, J.-M.; André, O.; Guillotin, B.; Alexandre, M.; Combier, J.-P. Use of MicroRNA-Encoded Peptide MiPEP172c to Stimulate Nodulation in Soybean. New Phytol. 2016, 211, 379–381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Q.; Deng, B.; Gao, J.; Zhao, Z.; Chen, Z.; Song, S.; Wang, L.; Zhao, L.; Xu, W.; Zhang, C.; et al. A MiRNA-Encoded Small Peptide, Vvi-MiPEP171d1, Regulates Adventitious Root Formation. Plant Physiol. 2020, 183, 656–670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, C.E.; Zhao, Y.-T.; Wang, X.-J.; Croft, L.; Wang, Z.-H.; Haerizadeh, F.; Mattick, J.S.; Singh, M.B.; Carroll, B.J.; Bhalla, P.L. MicroRNAs in the Shoot Apical Meristem of Soybean. J. Exp. Bot. 2011, 62, 2495–2506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choudhary, A.; Kumar, A.; Kaur, H.; Kaur, N. MiRNA: The Taskmaster of Plant World. Biologia 2021, 76, 1551–1567. [Google Scholar] [CrossRef]
- Waheed, S.; Zeng, L. The Critical Role of MiRNAs in Regulation of Flowering Time and Flower Development. Genes 2020, 11, 319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Curaba, J.; Spriggs, A.; Taylor, J.; Li, Z.; Helliwell, C. MiRNA Regulation in the Early Development of Barley Seed. BMC Plant Biol. 2012, 12, 120. [Google Scholar] [CrossRef] [Green Version]
- Curaba, J.; Talbot, M.; Li, Z.; Helliwell, C. Over-Expression of MicroRNA171 Affects Phase Transitions and Floral Meristem Determinancy in Barley. BMC Plant Biol. 2013, 13, 6. [Google Scholar] [CrossRef] [Green Version]
- Smoczynska, A.; Szweykowska-Kulinska, Z. MicroRNA-Mediated Regulation of Flower Development in Grasses. Acta Biochim. Pol. 2016, 63, 687–692. [Google Scholar] [CrossRef] [PubMed]
- Nair, S.K.; Wang, N.; Turuspekov, Y.; Pourkheirandish, M.; Sinsuwongwat, S.; Chen, G.; Sameri, M.; Tagiri, A.; Honda, I.; Watanabe, Y.; et al. Cleistogamous Flowering in Barley Arises from the Suppression of MicroRNA-Guided HvAP2 MRNA Cleavage. Proc. Natl. Acad. Sci. USA 2010, 107, 490–495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anwar, N.; Ohta, M.; Yazawa, T.; Sato, Y.; Li, C.; Tagiri, A.; Sakuma, M.; Nussbaumer, T.; Bregitzer, P.; Pourkheirandish, M.; et al. MiR172 Downregulates the Translation of Cleistogamy 1 in Barley. Ann. Bot. 2018, 122, 251–265. [Google Scholar] [CrossRef]
- Yuan, W.; Suo, J.; Shi, B.; Zhou, C.; Bai, B.; Bian, H.; Zhu, M.; Han, N. The Barley MiR393 Has Multiple Roles in Regulation of Seedling Growth, Stomatal Density, and Drought Stress Tolerance. Plant Physiol. Biochem. 2019, 142, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Tombuloglu, H. Genome-Wide Analysis of the Auxin Response Factors (ARF) Gene Family in Barley (Hordeum vulgare L.). J. Plant Biochem. Biotechnol. 2019, 28, 14–24. [Google Scholar] [CrossRef]
- Shriram, V.; Kumar, V.; Devarumath, R.M.; Khare, T.S.; Wani, S.H. MicroRNAs as Potential Targets for Abiotic Stress Tolerance in Plants. Front. Plant Sci. 2016, 7, 817. [Google Scholar] [CrossRef] [Green Version]
- Barczak-Brzyżek, A.; Brzyżek, G.; Koter, M.; Siedlecka, E.; Gawroński, P.; Filipecki, M. Plastid Retrograde Regulation of MiRNA Expression in Response to Light Stress. BMC Plant Biol. 2022, 22, 150. [Google Scholar] [CrossRef]
- Subburaj, S.; Ha, H.-J.; Jin, Y.-T.; Jeon, Y.; Tu, L.; Kim, J.-B.; Kang, S.-Y.; Lee, G.-J. Identification of γ-Radiation-Responsive MicroRNAs and Their Target Genes in Tradescantia (BNL Clone 4430). J. Plant Biol. 2017, 60, 116–128. [Google Scholar] [CrossRef]
- Visentin, I.; Pagliarani, C.; Deva, E.; Caracci, A.; Turečková, V.; Novák, O.; Lovisolo, C.; Schubert, A.; Cardinale, F. A Novel Strigolactone-MiR156 Module Controls Stomatal Behaviour during Drought Recovery. Plant Cell Environ. 2020, 43, 1613–1624. [Google Scholar] [CrossRef] [PubMed]
- Saminathan, T.; Alvarado, A.; Lopez, C.; Shinde, S.; Gajanayake, B.; Abburi, V.L.; Vajja, V.G.; Jagadeeswaran, G.; Raja Reddy, K.; Nimmakayala, P.; et al. Elevated Carbon Dioxide and Drought Modulate Physiology and Storage-Root Development in Sweet Potato by Regulating MicroRNAs. Funct. Integr. Genom. 2019, 19, 171–190. [Google Scholar] [CrossRef] [PubMed]
- Deng, P.; Wang, L.; Cui, L.; Feng, K.; Liu, F.; Du, X.; Tong, W.; Nie, X.; Ji, W.; Weining, S. Global Identification of MicroRNAs and Their Targets in Barley under Salinity Stress. PLoS ONE 2015, 10, e0137990. [Google Scholar] [CrossRef] [PubMed]
- Kuang, L.; Yu, J.; Shen, Q.; Fu, L.; Wu, L. Identification of MicroRNAs Responding to Aluminium, Cadmium and Salt Stresses in Barley Roots. Plants 2021, 10, 2754. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Sun, G. Micro RNA s Contribute to Enhanced Salt Adaptation of the Autopolyploid Hordeum bulbosum Compared with Its Diploid Ancestor. Plant J. 2017, 91, 57–69. [Google Scholar] [CrossRef] [Green Version]
- Kuang, L.; Shen, Q.; Wu, L.; Yu, J.; Fu, L.; Wu, D.; Zhang, G. Identification of MicroRNAs Responding to Salt Stress in Barley by High-Throughput Sequencing and Degradome Analysis. Environ. Exp. Bot. 2019, 160, 59–70. [Google Scholar] [CrossRef]
- Smoczynska, A.; Pacak, A.M.; Nuc, P.; Swida-Barteczka, A.; Kruszka, K.; Karlowski, W.M.; Jarmolowski, A.; Szweykowska-Kulinska, Z. A Functional Network of Novel Barley MicroRNAs and Their Targets in Response to Drought. Genes 2020, 11, 488. [Google Scholar] [CrossRef] [PubMed]
- Ferdous, J.; Sanchez-Ferrero, J.C.; Langridge, P.; Milne, L.; Chowdhury, J.; Brien, C.; Tricker, P.J. Differential Expression of MicroRNAs and Potential Targets under Drought Stress in Barley. Plant Cell Environ. 2017, 40, 11–24. [Google Scholar] [CrossRef]
- Kantar, M.; Unver, T.; Budak, H. Regulation of Barley MiRNAs upon Dehydration Stress Correlated with Target Gene Expression. Funct. Integr. Genom. 2010, 10, 493–507. [Google Scholar] [CrossRef]
- Hackenberg, M.; Gustafson, P.; Langridge, P.; Shi, B.-J. Differential Expression of MicroRNAs and Other Small RNAs in Barley between Water and Drought Conditions. Plant Biotechnol. J. 2015, 13, 2–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiu, C.-W.; Liu, L.; Feng, X.; Hao, P.-F.; He, X.; Cao, F.; Wu, F. Genome-Wide Identification and Characterization of Drought Stress Responsive MicroRNAs in Tibetan Wild Barley. Int. J. Mol. Sci. 2020, 21, 2795. [Google Scholar] [CrossRef] [Green Version]
- Grabowska, A.; Smoczynska, A.; Bielewicz, D.; Pacak, A.; Jarmolowski, A.; Szweykowska-Kulinska, Z. Barley MicroRNAs as Metabolic Sensors for Soil Nitrogen Availability. Plant Sci. 2020, 299, 110608. [Google Scholar] [CrossRef] [PubMed]
- Ozhuner, E.; Eldem, V.; Ipek, A.; Okay, S.; Sakcali, S.; Zhang, B.; Boke, H.; Unver, T. Boron Stress Responsive MicroRNAs and Their Targets in Barley. PLoS ONE 2013, 8, e59543. [Google Scholar] [CrossRef]
- Hackenberg, M.; Huang, P.-J.; Huang, C.-Y.; Shi, B.-J.; Gustafson, P.; Langridge, P. A Comprehensive Expression Profile of MicroRNAs and Other Classes of Non-Coding Small RNAs in Barley Under Phosphorous-Deficient and -Sufficient Conditions. DNA Res. 2013, 20, 109–125. [Google Scholar] [CrossRef] [Green Version]
- Sega, P.; Kruszka, K.; Bielewicz, D.; Karlowski, W.; Nuc, P.; Szweykowska-Kulinska, Z.; Pacak, A. Pi-Starvation Induced Transcriptional Changes in Barley Revealed by a Comprehensive RNA-Seq and Degradome Analyses. BMC Genom. 2021, 22, 165. [Google Scholar] [CrossRef]
- Bai, B.; Bian, H.; Zeng, Z.; Hou, N.; Shi, B.; Wang, J.; Zhu, M.; Han, N. MiR393-Mediated Auxin Signaling Regulation Is Involved in Root Elongation Inhibition in Response to Toxic Aluminum Stress in Barley. Plant Cell Physiol. 2017, 58, 426–439. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Yu, J.; Shen, Q.; Huang, L.; Wu, D.; Zhang, G. Identification of MicroRNAs in Response to Aluminum Stress in the Roots of Tibetan Wild Barley and Cultivated Barley. BMC Genom. 2018, 19, 560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, J.; Wu, L.; Fu, L.; Shen, Q.; Kuang, L.; Wu, D.; Zhang, G. Genotypic Difference of Cadmium Tolerance and the Associated MicroRNAs in Wild and Cultivated Barley. Plant Growth Regul. 2019, 87, 389–401. [Google Scholar] [CrossRef]
- Chen, F.; He, J.; Jin, G.; Chen, Z.-H.; Dai, F. Identification of Novel MicroRNAs for Cold Deacclimation in Barley. Plant Growth Regul. 2020, 92, 389–400. [Google Scholar] [CrossRef]
- Kruszka, K.; Pacak, A.; Swida-Barteczka, A.; Nuc, P.; Alaba, S.; Wroblewska, Z.; Karlowski, W.; Jarmolowski, A.; Szweykowska-Kulinska, Z. Transcriptionally and Post-Transcriptionally Regulated MicroRNAs in Heat Stress Response in Barley. J. Exp. Bot. 2014, 65, 6123–6135. [Google Scholar] [CrossRef] [Green Version]
- Schreiber, A.W.; Shi, B.-J.; Huang, C.-Y.; Langridge, P.; Baumann, U. Discovery of Barley MiRNAs through Deep Sequencing of Short Reads. BMC Genom. 2011, 12, 129. [Google Scholar] [CrossRef] [Green Version]
- Lv, S.; Nie, X.; Wang, L.; Du, X.; Biradar, S.S.; Jia, X.; Weining, S. Identification and Characterization of MicroRNAs from Barley (Hordeum vulgare L.) by High-Throughput Sequencing. Int. J. Mol. Sci. 2012, 13, 2973–2984. [Google Scholar] [CrossRef] [Green Version]
- Kruszka, K.; Pacak, A.; Swida-Barteczka, A.; Stefaniak, A.K.; Kaja, E.; Sierocka, I.; Karlowski, W.; Jarmolowski, A.; Szweykowska-Kulinska, Z. Developmentally Regulated Expression and Complex Processing of Barley Pri-MicroRNAs. BMC Genom. 2013, 14, 34. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Cheng, X.; Liu, D.; Xu, W.; Wise, R.; Shen, Q.-H. The MiR9863 Family Regulates Distinct Mla Alleles in Barley to Attenuate NLR Receptor-Triggered Disease Resistance and Cell-Death Signaling. PLoS Genet. 2014, 10, e1004755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kis, A.; Tholt, G.; Ivanics, M.; Várallyay, É.; Jenes, B.; Havelda, Z. Polycistronic Artificial MiRNA-Mediated Resistance to Wheat Dwarf Virus in Barley Is Highly Efficient at Low Temperature. Mol. Plant Pathol. 2016, 17, 427–437. [Google Scholar] [CrossRef]
- Deng, P.; Bian, J.; Yue, H.; Feng, K.; Wang, M.; Du, X.; Weining, S.; Nie, X. Characterization of MicroRNAs and Their Targets in Wild Barley (Hordeum vulgare Subsp. Spontaneum) Using Deep Sequencing. Genome 2016, 59, 339–348. [Google Scholar] [CrossRef] [Green Version]
- Pacak, A.M.; Kruszka, K.; Świda-Barteczka, A.; Nuc, P.; Karlowski, W.; Jarmołowski, A.; Szweykowska-Kulińska, Z. Developmental Changes in Barley MicroRNA Expression Profiles Coupled with MiRNA Targets Analysis. Acta Biochim. Pol. 2016, 63, 799–809. [Google Scholar] [CrossRef] [Green Version]
- Bai, B.; Shi, B.; Hou, N.; Cao, Y.; Meng, Y.; Bian, H.; Zhu, M.; Han, N. MicroRNAs Participate in Gene Expression Regulation and Phytohormone Cross-Talk in Barley Embryo during Seed Development and Germination. BMC Plant Biol. 2017, 17, 150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, O.; Palmer, S.A.; Clapham, A.J.; Rose, P.; Liu, Y.; Wang, J.; Allaby, R.G. Small RNA Activity in Archeological Barley Shows Novel Germination Inhibition in Response to Environment. Mol. Biol. Evol. 2017, 34, 2555–2562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tripathi, R.K.; Bregitzer, P.; Singh, J. Genome-Wide Analysis of the SPL/MiR156 Module and Its Interaction with the AP2/MiR172 Unit in Barley. Sci. Rep. 2018, 8, 7085. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plaksenkova, I.; Kokina, I.; Petrova, A.; Jermaļonoka, M.; Gerbreders, V.; Krasovska, M. The Impact of Zinc Oxide Nanoparticles on Cytotoxicity, Genotoxicity, and MiRNA Expression in Barley (Hordeum vulgare L.) Seedlings. Sci. World J. 2020, 2020, 6649746. [Google Scholar] [CrossRef]
- Ye, Z.; Zeng, J.; Long, L.; Ye, L.; Zhang, G. Identification of MicroRNAs in Response to Low Potassium Stress in the Shoots of Tibetan Wild Barley and Cultivated. Curr. Plant Biol. 2021, 25, 100193. [Google Scholar] [CrossRef]
- Puchta, M.; Groszyk, J.; Małecka, M.; Koter, M.D.; Niedzielski, M.; Rakoczy-Trojanowska, M.; Boczkowska, M. Barley Seeds MiRNome Stability during Long-Term Storage and Aging. Int. J. Mol. Sci. 2021, 22, 4315. [Google Scholar] [CrossRef]
- Yao, X.; Wang, Y.; Yao, Y.; Bai, Y.; Wu, K.; Qiao, Y. Identification MicroRNAs and Target Genes in Tibetan Hulless Barley to BLS Infection. Agron. J. 2021, 113, 2273–2292. [Google Scholar] [CrossRef]
- Wang, N.-H.; Zhou, X.-Y.; Shi, S.-H.; Zhang, S.; Chen, Z.-H.; Ali, M.A.; Ahmed, I.M.; Wang, Y.; Wu, F. An MiR156-Regulated Nucleobase-Ascorbate Transporter 2 Confers Cadmium Tolerance via Enhanced Anti-Oxidative Capacity in Barley. J. Adv. Res. 2022, in press. [Google Scholar] [CrossRef]
- Liao, P.; Li, S.; Cui, X.; Zheng, Y. A Comprehensive Review of Web-Based Resources of Non-Coding RNAs for Plant Science Research. Int. J. Biol. Sci. 2018, 14, 819–832. [Google Scholar] [CrossRef]
- Yi, X.; Zhang, Z.; Ling, Y.; Xu, W.; Su, Z. PNRD: A Plant Non-Coding RNA Database. Nucleic Acids Res. 2015, 43, D982–D989. [Google Scholar] [CrossRef] [Green Version]
- Guo, Z.; Kuang, Z.; Zhao, Y.; Deng, Y.; He, H.; Wan, M.; Tao, Y.; Wang, D.; Wei, J.; Li, L. PmiREN2.0: From Data Annotation to Functional Exploration of Plant MicroRNAs. Nucleic Acids Res. 2022, 50, D1475–D1482. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. MiRBase: From MicroRNA Sequences to Function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef] [PubMed]
- Lunardon, A.; Johnson, N.R.; Hagerott, E.; Phifer, T.; Polydore, S.; Coruh, C.; Axtell, M.J. Integrated Annotations and Analyses of Small RNA–Producing Loci from 47 Diverse Plants. Genome Res. 2020, 30, 497–513. [Google Scholar] [CrossRef] [PubMed]
- Szcześniak, M.W.; Makalowska, I. MiRNEST 2.0: A Database of Plant and Animal MicroRNAs. Nucleic Acids Res. 2014, 42, D74–D77. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Liu, X.; Zhang, S.; Liang, S.; Luan, W.; Ma, X. TarDB: An Online Database for Plant MiRNA Targets and MiRNA-Triggered Phased SiRNAs. BMC Genom. 2021, 22, 348. [Google Scholar] [CrossRef] [PubMed]
- Dou, X.; Zhou, Z.; Zhao, L. Identification and Expression Analysis of MiRNAs in Germination and Seedling Growth of Tibetan Hulless Barley. Genomics 2021, 113, 3735–3749. [Google Scholar] [CrossRef]
- Xing, S.; Salinas, M.; Höhmann, S.; Berndtgen, R.; Huijser, P. MiR156-Targeted and Nontargeted SBP-Box Transcription Factors Act in Concert to Secure Male Fertility in Arabidopsis. Plant Cell 2010, 22, 3935–3950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, L.; Zheng, F.; Wang, J.; Zhang, C.; Xiao, F.; Ye, J.; Li, C.; Ye, Z.; Zhang, J. MiR156a-Targeted SBP-Box Transcription Factor SlSPL13 Regulates Inflorescence Morphogenesis by Directly Activating SFT in Tomato. Plant Biotechnol. J. 2020, 18, 1670–1682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Cheng, X.; Liu, P.; Sun, J. MiR156-Targeted SBP-Box Transcription Factors Interact with DWARF53 to Regulate TEOSINTE BRANCHED1 and BARREN STALK1 Expression in Bread Wheat. Plant Physiol. 2017, 174, 1931–1948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Millar, A.A.; Lohe, A.; Wong, G. Biology and Function of MiR159 in Plants. Plants 2019, 8, 255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Csukasi, F.; Donaire, L.; Casañal, A.; Martínez-Priego, L.; Botella, M.A.; Medina-Escobar, N.; Llave, C.; Valpuesta, V. Two Strawberry MiR159 Family Members Display Developmental-Specific Expression Patterns in the Fruit Receptacle and Cooperatively Regulate Fa-GAMYB. New Phytol. 2012, 195, 47–57. [Google Scholar] [CrossRef] [PubMed]
- da Silva, E.M.; Silva, G.F.F.E.; Bidoia, D.B.; da Silva Azevedo, M.; de Jesus, F.A.; Pino, L.E.; Peres, L.E.P.; Carrera, E.; López-Díaz, I.; Nogueira, F.T.S. Micro RNA 159-Targeted Sl GAMYB Transcription Factors Are Required for Fruit Set in Tomato. Plant J. 2017, 92, 95–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yadav, A.; Kumar, S.; Verma, R.; Lata, C.; Sanyal, I.; Rai, S.P. MicroRNA 166: An Evolutionarily Conserved Stress Biomarker in Land Plants Targeting HD-ZIP Family. Physiol. Mol. Biol. Plants 2021, 27, 2471–2485. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Fang, R.; Deng, R.; Li, J. The OsmiRNA166b-OsHox32 Pair Regulates Mechanical Strength of Rice Plants by Modulating Cell Wall Biosynthesis. Plant Biotechnol. J. 2021, 19, 1468–1480. [Google Scholar] [CrossRef] [PubMed]
- Javed, M.; Solanki, M.; Sinha, A.; Shukla, L.I. Position Based Nucleotide Analysis of MiR168 Family in Higher Plants and Its Targets in Mammalian Transcripts. MicroRNA 2017, 6, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Um, T.; Choi, J.; Park, T.; Chung, P.J.; Jung, S.E.; Shim, J.S.; Kim, Y.S.; Choi, I.-Y.; Park, S.C.; Oh, S.-J. Rice MicroRNA171f/SCL6 Module Enhances Drought Tolerance by Regulation of Flavonoid Biosynthesis Genes. Plant Direct 2022, 6, e374. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Zhou, J.; Gao, L.; Tang, Y. Plant MiR397 and Its Functions. Funct. Plant Biol. 2020, 48, 361–370. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.-H.; Pei, H. Over-Expression of MiR397 Improves Plant Tolerance to Cold Stress in Arabidopsis Thaliana. J. Plant Biol. 2014, 57, 209–217. [Google Scholar] [CrossRef]
- Pegler, J.L.; Oultram, J.M.; Grof, C.P.; Eamens, A.L. Molecular Manipulation of the MiR399/PHO2 Expression Module Alters the Salt Stress Response of Arabidopsis Thaliana. Plants 2020, 10, 73. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Ahn, H.J.; Chiou, T.-J.; Ahn, J.H. The Role of the MiR399-PHO2 Module in the Regulation of Flowering Time in Response to Different Ambient Temperatures in Arabidopsis Thaliana. Mol. Cells 2011, 32, 83–88. [Google Scholar] [CrossRef]
- Yan, Y.; Wang, H.; Hamera, S.; Chen, X.; Fang, R. MiR444a Has Multiple Functions in the Rice Nitrate-Signaling Pathway. Plant J. 2014, 78, 44–55. [Google Scholar] [CrossRef]
- Wang, H.; Jiao, X.; Kong, X.; Hamera, S.; Wu, Y.; Chen, X.; Fang, R.; Yan, Y. A Signaling Cascade from MiR444 to RDR1 in Rice Antiviral RNA Silencing Pathway. Plant Physiol. 2016, 170, 2365–2377. [Google Scholar] [CrossRef] [Green Version]
- Jiao, X.; Wang, H.; Yan, J.; Kong, X.; Liu, Y.; Chu, J.; Chen, X.; Fang, R.; Yan, Y. Promotion of BR Biosynthesis by MiR444 Is Required for Ammonium-Triggered Inhibition of Root Growth. Plant Physiol. 2020, 182, 1454–1466. [Google Scholar] [CrossRef]
- Sun, L.; Sun, G.; Shi, C.; Sun, D. Transcriptome Analysis Reveals New MicroRNAs-Mediated Pathway Involved in Anther Development in Male Sterile Wheat. BMC Genom. 2018, 19, 333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bano, N.; Fakhrah, S.; Nayak, S.P.; Bag, S.K.; Mohanty, C.S. Identification of MiRNA and Their Target Genes in Cestrum nocturnum L. and Cestrum diurnum L. in Stress Responses. Physiol. Mol. Biol. Plants 2022, 28, 31–49. [Google Scholar] [CrossRef]
- Li, T.; Ma, L.; Geng, Y.; Hao, C.; Chen, X.; Zhang, X. Small RNA and Degradome Sequencing Reveal Complex Roles of MiRNAs and Their Targets in Developing Wheat Grains. PLoS ONE 2015, 10, e0139658. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, X.; Yuan, L.; Liu, Y.; Shen, T.; Zhang, Y. Comparative Small RNA Profiling and Functional Exploration on Wheat with High-and Low-Cadmium Accumulation. Front. Genet. 2021, 12, 635599. [Google Scholar] [CrossRef]
- Samad, A.F.A.; Sajad, M.; Nazaruddin, N.; Fauzi, I.A.; Murad, A.M.A.; Zainal, Z.; Ismail, I. MicroRNA and Transcription Factor: Key Players in Plant Regulatory Network. Front. Plant Sci. 2017, 8, 565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Bel, M.; Diels, T.; Vancaester, E.; Kreft, L.; Botzki, A.; Van de Peer, Y.; Coppens, F.; Vandepoele, K. PLAZA 4.0: An Integrative Resource for Functional, Evolutionary and Comparative Plant Genomics. Nucleic Acids Res. 2018, 46, D1190–D1196. [Google Scholar] [CrossRef]
- Van Bel, M.; Silvestri, F.; Weitz, E.M.; Kreft, L.; Botzki, A.; Coppens, F.; Vandepoele, K. PLAZA 5.0: Extending the Scope and Power of Comparative and Functional Genomics in Plants. Nucleic Acids Res. 2022, 50, D1468–D1474. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.-M.; Kuang, S.; Xiong, X.; Gao, T.; Liu, C.; Guo, A.-Y. Transcription Factor and MicroRNA Co-Regulatory Loops: Important Regulatory Motifs in Biological Processes and Diseases. Brief. Bioinform. 2015, 16, 45–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selbach, M.; Schwanhäusser, B.; Thierfelder, N.; Fang, Z.; Khanin, R.; Rajewsky, N. Widespread Changes in Protein Synthesis Induced by MicroRNAs. Nature 2008, 455, 58–63. [Google Scholar] [CrossRef]
- Baldrich, P.; Beric, A.; Meyers, B.C. Despacito: The Slow Evolutionary Changes in Plant MicroRNAs. Curr. Opin. Plant Biol. 2018, 42, 16–22. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, L.; Chen, D.; Wu, X.; Huang, D.; Chen, L.; Li, L.; Deng, X.; Xu, Q. Genome-Wide Comparison of MicroRNAs and Their Targeted Transcripts among Leaf, Flower and Fruit of Sweet Orange. BMC Genom. 2014, 15, 695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baldrich, P.; Campo, S.; Wu, M.-T.; Liu, T.-T.; Hsing, Y.-I.C.; Segundo, B.S. MicroRNA-Mediated Regulation of Gene Expression in the Response of Rice Plants to Fungal Elicitors. RNA Biol. 2015, 12, 847–863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banks, I.R.; Zhang, Y.; Wiggins, B.E.; Heck, G.R.; Ivashuta, S. RNA Decoys. Plant Signal. Behav. 2012, 7, 1188–1193. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Liu, C.; Gu, L.; Mo, B.; Cao, X.; Chen, X. TarHunter, a Tool for Predicting Conserved MicroRNA Targets and Target Mimics in Plants. Bioinformatics 2018, 34, 1574–1576. [Google Scholar] [CrossRef] [Green Version]
- Unver, T.; Tombuloglu, H. Barley Long Non-Coding RNAs (LncRNA) Responsive to Excess Boron. Genomics 2020, 112, 1947–1955. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Cheng, Y.; Feng, K.; Ruan, M.; Ye, Q.; Wang, R.; Li, Z.; Zhou, G.; Yao, Z.; Yang, Y.; et al. Genome-Wide Identification and Expression Profiling of Tomato Hsp20 Gene Family in Response to Biotic and Abiotic Stresses. Front. Plant Sci. 2016, 7, 1215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved Seed Pairing, Often Flanked by Adenosines, Indicates That Thousands of Human Genes Are MicroRNA Targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ni, W.-J.; Leng, X.-M. Dynamic MiRNA–MRNA Paradigms: New Faces of MiRNAs. Biochem. Biophys. Rep. 2015, 4, 337–341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gruber, A.R.; Lorenz, R.; Bernhart, S.H.; Neuböck, R.; Hofacker, I.L. The Vienna RNA Websuite. Nucleic Acids Res. 2008, 36, W70–W74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kerpedjiev, P.; Hammer, S.; Hofacker, I.L. Forna (Force-Directed RNA): Simple and Effective Online RNA Secondary Structure Diagrams. Bioinformatics 2015, 31, 3377–3379. [Google Scholar] [CrossRef] [Green Version]
- Jones-Rhoades, M.W.; Bartel, D.P. Computational Identification of Plant MicroRNAs and Their Targets, Including a Stress-Induced MiRNA. Mol. Cell 2004, 14, 787–799. [Google Scholar] [CrossRef]
- Sun, Y.-H.; Lu, S.; Shi, R.; Chiang, V.L. Computational Prediction of Plant MiRNA Targets. In RNAi and Plant Gene Function Analysis; Humana Press: Totowa, NJ, USA, 2011; pp. 175–186. [Google Scholar]
- Dai, X.; Zhuang, Z.; Zhao, P.X. Computational Analysis of MiRNA Targets in Plants: Current Status and Challenges. Brief. Bioinform. 2011, 12, 115–121. [Google Scholar] [CrossRef]
- Fahlgren, N.; Carrington, J.C. MiRNA Target Prediction in Plants. In Plant MicroRNAs; Humana Press: Totowa, NJ, USA, 2010; pp. 51–57. [Google Scholar]
- Pandey, P.; Srivastava, P.K.; Pandey, S.P. Prediction of Plant MiRNA Targets. In Plant MicroRNAs; Humana Press: Totowa, NJ, USA, 2019; pp. 99–107. [Google Scholar]
- Rhoades, M.W.; Reinhart, B.J.; Lim, L.P.; Burge, C.B.; Bartel, B.; Bartel, D.P. Prediction of Plant MicroRNA Targets. Cell 2002, 110, 513–520. [Google Scholar] [CrossRef]
- Llave, C.; Xie, Z.; Kasschau, K.D.; Carrington, J.C. Cleavage of Scarecrow-like MRNA Targets Directed by a Class of Arabidopsis MiRNA. Science 2002, 297, 2053–2056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riolo, G.; Cantara, S.; Marzocchi, C.; Ricci, C. MiRNA Targets: From Prediction Tools to Experimental Validation. Methods Protoc. 2020, 4, 1. [Google Scholar] [CrossRef] [PubMed]
- Patel, P.; Yadav, K.; Ganapathi, T.R.; Penna, S. Plant MiRNAome: Cross Talk in Abiotic Stressful Times. In Genetic Enhancement of Crops for Tolerance to Abiotic Stress: Mechanisms and Approaches, Vol. I; Rajpal, V.R., Sehgal, D., Kumar, A., Raina, S.N., Eds.; Sustainable Development and Biodiversity; Springer International Publishing: Cham, Switzesland, 2019; pp. 25–52. ISBN 978-3-319-91956-0. [Google Scholar]
- Zhang, B.; Wang, Q. MicroRNA-Based Biotechnology for Plant Improvement. J. Cell. Physiol. 2015, 230, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B. MicroRNA: A New Target for Improving Plant Tolerance to Abiotic Stress. J. Exp. Bot. 2015, 66, 1749–1761. [Google Scholar] [CrossRef] [PubMed]
- Ferdous, J.; Whitford, R.; Nguyen, M.; Brien, C.; Langridge, P.; Tricker, P.J. Drought-Inducible Expression of Hv-MiR827 Enhances Drought Tolerance in Transgenic Barley. Funct. Integr. Genom. 2017, 17, 279–292. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Yang, T.; Guo, Z.; Wang, Q.; Chai, M.; Wu, M.; Li, X.; Li, W.; Li, G.; Tang, J.; et al. Maize MicroRNA166 Inactivation Confers Plant Development and Abiotic Stress Resistance. Int. J. Mol. Sci. 2020, 21, 9506. [Google Scholar] [CrossRef] [PubMed]
- Chung, P.J.; Chung, H.; Oh, N.; Choi, J.; Bang, S.W.; Jung, S.E.; Jung, H.; Shim, J.S.; Kim, J.-K. Efficiency of Recombinant CRISPR/RCas9-Mediated MiRNA Gene Editing in Rice. Int. J. Mol. Sci. 2020, 21, 9606. [Google Scholar] [CrossRef]
- Hua, K.; Tao, X.; Zhu, J.-K. Expanding the Base Editing Scope in Rice by Using Cas9 Variants. Plant Biotechnol. J. 2019, 17, 499–504. [Google Scholar] [CrossRef] [Green Version]
- Ó’Maoiléidigh, D.S.; van Driel, A.D.; van Singh, A.; Sang, Q.; LeBec, N.; Vincent, C.; de Olalla, E.B.G.; Vayssières, A.; Branchat, M.R.; Severing, E.; et al. Systematic Analyses of the MIR172 Family Members of Arabidopsis Define Their Distinct Roles in Regulation of APETALA2 during Floral Transition. PLoS Biol. 2021, 19, e3001043. [Google Scholar] [CrossRef] [PubMed]
- Deng, F.; Zeng, F.; Shen, Q.; Abbas, A.; Cheng, J.; Jiang, W.; Chen, G.; Shah, A.N.; Holford, P.; Tanveer, M.; et al. Molecular Evolution and Functional Modification of Plant MiRNAs with CRISPR. Trends Plant Sci. 2022, 27, 890–907. [Google Scholar] [CrossRef] [PubMed]
- Dalakouras, A.; Wassenegger, M.; Dadami, E.; Ganopoulos, I.; Pappas, M.L.; Papadopoulou, K. Genetically Modified Organism-Free RNA Interference: Exogenous Application of RNA Molecules in Plants. Plant Physiol. 2020, 182, 38–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mujtaba, M.; Wang, D.; Carvalho, L.B.; Oliveira, J.L.; Espirito Santo Pereira, A.D.; Sharif, R.; Jogaiah, S.; Paidi, M.K.; Wang, L.; Ali, Q.; et al. Nanocarrier-Mediated Delivery of MiRNA, RNAi, and CRISPR-Cas for Plant Protection: Current Trends and Future Directions. ACS Agric. Sci. Technol. 2021, 1, 417–435. [Google Scholar] [CrossRef]
- Ražná, K.; Rataj, V.; Macák, M.; Galambošová, J. MicroRNA-Based Markers as a Tool to Monitor the Barley (Hordeum vulgare L.) Response to Soil Compaction. Acta Fytotech. Zootech. 2020, 23, 139–146. [Google Scholar] [CrossRef]
- Gurjar, A.K.S.; Panwar, A.S.; Gupta, R.; Mantri, S.S. PmiRExAt: Plant MiRNA Expression Atlas Database and Web Applications. Database 2016, 2016, baw060. [Google Scholar] [CrossRef]





| Title of the Study and Reference | Barley Cultivars Inspected | Year of Publication | Most Important Findings |
|---|---|---|---|
| Regulation of barley miRNAs upon dehydration stress correlated with target gene expression [79] | Hordeum vulgare | 2010 | A total of 28 potential miRNAs were identified using bioinformatic approaches (BLASTn of known plant miRNAs and barley expressed sequence tags (ESTs), and RNA folding algorithms). |
| Discovery of barley miRNAs through deep sequencing of short reads [91] | Hordeum vulgare cultivars Golden Promise and Pallas | 2011 | The first large-scale study of miRNAs in Hordeum Vulgare, 100 miRNAs were identified (only 56 of them had orthologs in wheat, rice, or Brachypodium) and 3 candidates were validated in vitro using a Northern blot assay. |
| Identification and Characterization of MicroRNAs from Barley (Hordeum vulgare L.) by High-Throughput Sequencing [92] | Hordeum vulgare L. | 2012 | 126 conserved miRNAs (belonging to 58 families), and 133 novel miRNAs (50 families) were identified in this study. |
| miRNA regulation in the early development of barley seed [61] | Hordeum vulgare | 2012 | 84 known miRNAs and 7 new miRNAs together with 96 putative miRNA target genes were identified during the early development of barley seeds (first 15 days post anthesis). |
| Developmentally regulated expression and complex processing of barley pri-microRNAs [93] | Hordeum vulgare cultivar Rolap | 2013 | miRNA genes in barley often contain introns which may play important role in miRNA processing. |
| A Comprehensive Expression Profile of MicroRNAs and Other Classes of Non-Coding Small RNAs in Barley Under Phosphorous-Deficient and -Sufficient Conditions [84] | Hordeum vulgare L., cultivar Pallas | 2013 | 221 conserved miRNAs and 12 novel miRNAs were identified, many of them were phosphorus condition-specific. A total of 47 miRNAs were significantly differentially expressed between the two phosphorus treatments. |
| Boron Stress Responsive MicroRNAs and Their Targets in Barley [83] | Hordeum vulgare L. cultivar Sahara | 2013 | 31 known and 3 new miRNAs were identified in barley, and 25 of them were found to respond to boron treatment. |
| Transcriptionally and post-transcriptionally regulated microRNAs in heat stress response in barley [90] | Hordeum vulgare cultivar Rolap | 2014 | Four heat stress up-regulated barley miRNAs were found (miR160a, miR166a, miR167h, and miR5175a). |
| Differential expression of microRNAs and other small RNAs in barley between water and drought conditions [80] | Hordeum vulgare cultivar Golden Promise | 2014 | Three novel miRNAs, designated as hvu-miRX33, hvu-miRX34, and hvu-miRX35 were identified. hvu-miRX34 had no homologous miRNA in wheat. |
| The miR9863 Family Regulates Distinct Mla Alleles in Barley to Attenuate NLR Receptor-Triggered Disease Resistance and Cell-Death Signaling [94] | Hordeum vulgare L. | 2014 | The key role of the miR9863 family in the immune response to the pathogen (powdery mildew fungus, Blumeria graminis f. sp. hordei) was proposed |
| Polycistronic artificial miRNA-mediated resistance to Wheat dwarf virus in barley is highly efficient at low temperature [95] | Artificially transformed Hordeum vulgare cultivar Golden Promise | 2015 | Polycistronic artificial miRNA in plasmid vector was successfully transformed into barley embryos and mediated resistance to Wheat dwarf virus. |
| Global Identification of MicroRNAs and Their Targets in Barley under Salinity Stress [73] | Hordeum vulgare cultivar Morex | 2015 | Authors identified 152 miRNAs (142 conserved and 10 novel ones), and 44 miRNAs (39 conserved and 5 novel ones) were found to be salinity-responsive. |
| Characterization of microRNAs and their targets in wild barley (Hordeum vulgare subsp. spontaneum) using deep sequencing [96] | Hordeum vulgare subsp. spontaneum | 2016 | A total of 70 known miRNAs and 18 novel miRNA candidates were identified and many of them were predicted to target mRNAs encoding transcription factors. |
| Developmental changes in barley microRNA expression profiles coupled with miRNA target analysis [97] | Hordeum vulgare cultivar Rolap | 2016 | miRNA transcriptomes of five barley developmental stages were inspected. Overall, miR168-3p and miR1432-5p levels increased while the 5′U-miR156-5p level decreased during barley development. |
| miR393-Mediated Auxin Signaling Regulation is Involved in Root Elongation Inhibition in Response to Toxic Aluminum Stress in Barley [86] | Hordeum vulgare cultivar Golden Promise | 2017 | Barley miR393 was functionally characterized. It regulates root sensitivity to aluminum through the alteration of auxin signaling. |
| Differential expression of microRNAs and potential targets under drought stress in barley [78] | Hordeum vulgare L. cultivars Commander, Fleet, Hindmarsh, and breeding line WI4304 | 2017 | miRNA regulation under drought stress in barley is genotype-specific. |
| microRNAs participate in gene expression regulation and phytohormone cross-talk in barley embryo during seed development and germination [98] | Hordeum vulgare cultivar Golden Promise | 2017 | A total of 1324 known miRNAs and 448 novel miRNA candidates were identified. miR393-mediated auxin response regulation significantly affected grain development. |
| Small RNA Activity in Archeological Barley Shows Novel Germination Inhibition in Response to Environment [99] | Ancient Hordeum vulgare | 2017 | Sequencing of miRNAs obtained from archeological barley samples (600–900 years BP) revealed their local adaptation to an agrarian environment around the river Nile. |
| Genome-wide analysis of the SPL/miR156 module and its interaction with the AP2/miR172 unit in barley [100] | Hordeum vulgare L. | 2018 | The study identified 17 barley SPL genes, and 7 of them contain a putative miR156 target site. |
| Identification of microRNAs in response to aluminum stress in the roots of Tibetan wild barley and cultivated barley [87] | Hordeum vulgare Al-sensitive Golden Promise and Tibetan wild barley (Al-tolerant XZ29) | 2018 | 50 miRNAs responsive to aluminum stress were detected, and some of them were found to be exclusively expressed in Al-tolerant XZ29. |
| Identification of microRNAs responding to salt stress in barley by high-throughput sequencing and degradome analysis [76] | Tibetan wild barley accession XZ16; Hordeum vulgare cultivar Golden Promise | 2019 | miR393a, miR156d, and miR172b (regulating HvAFB2/HvTIR1, UGTs, and HvAP2) are responsible for salt tolerance in barley roots. |
| Genotypic difference of cadmium tolerance and the associated microRNAs in wild and cultivated barley [88] | Hordeum vulgare cultivar Golden Promise and wild barley WB-1 | 2019 | 216 conserved miRNAs (in 59 miRNA families) and 87 novel miRNAs were identified. Authors suggest that miRNAs may play critical roles underlying the genotypic difference of cadmium tolerance in barley. |
| Genome-Wide Identification and Characterization of Drought Stress Responsive microRNAs in Tibetan Wild Barley [81] | Tibetan wild barley Hordeum vulgare L. ssp. Spontaneum | 2020 | 69 conserved miRNAs and 1574 novel miRNAs were identified, some of them were differentially expressed in drought conditions. |
| Barley microRNAs as metabolic sensors for soil nitrogen availability [82] | Hordeum vulgare cultivar Golden Promise | 2020 | Authors identified 13 barley miRNAs that are nitrogen excess responsive with the possible function of metabolic sensors for soil nitrogen availability. |
| The Impact of Zinc Oxide Nanoparticles on Cytotoxicity, Genotoxicity, and miRNA Expression in Barley (Hordeum vulgare L.) Seedlings [101] | Hordeum vulgare L. var. Abava | 2020 | ZnO nanoparticles significantly changed the expression of barley miR156a, miR159a, and miR159c in a dosage-dependent manner. |
| Identification of microRNAs in response to low potassium stress in the shoots of Tibetan wild barley and cultivated [102] | A Tibetan wild barley accession (XZ153) and a cultivar (ZD9) differing in low K tolerance | 2021 | A total of 1088 miRNAs were identified in the two barley genotypes under low potassium conditions. 65 of them were significantly differentially expressed. |
| Barley Seeds miRNome Stability during Long-Term Storage and Aging [103] | Hordeum vulgare cultivar Damazy | 2021 | miRNome of barley seeds harvested in 1972 was inspected. 61 known and 81 novel miRNA were identified pointing to the fact that miRNAs in dry seeds are extremely stable. |
| Identification microRNAs and target genes in Tibetan hulless barley to BLS infection [104] | Hordeum vulgare L. variety nudum Hook. f. | 2021 | A total of 36 conserved and 56 novel miRNAs were identified, some of them were differentially expressed between BLS (barley leaf stripe fungal disease)-sensitive and BLS-tolerant barley genotypes. |
| Pi-starvation induced transcriptional changes in barley revealed by a comprehensive RNA-Seq and degradome analyses [85] | Hordeum vulgare L. | 2021 | Authors suggest that barley adapts to inorganic phosphate (Pi)-starvation also via differential expression of several miRNAs. |
| Identification of microRNAs Responding to Aluminium, Cadmium and Salt Stresses in Barley Roots [74] | Hordeum vulgare cultivar Golden Promise | 2021 | 525 miRNAs (198 known and 327 novel miRNAs) were identified through high-throughput sequencing. 31 miRNAs were differentially expressed under inspected stresses. |
| An miR156-regulated nucleobase-ascorbate transporter 2 confers cadmium tolerance via enhanced anti-oxidative capacity in barley [105] | Hordeum vulgare genotypes Zhenong8 (ZN8) (Cd-tolerant genotype) and W6nk2 (Cd-sensitive genotype) | 2022 | miR156g-3p_3 targets a novel nucleobase-ascorbate transporter gene (HvNAT2). HvNAT2 evolved from the Zygnematales in Streptophyte algae and positively regulates cadmium tolerance → genetic engineering of NAT in plants may have potential in the remediation of soil/water cadmium pollution |
| Regulation of Phenolic Compound Production by Light Varying in Spectral Quality and Total Irradiance [21] | Hordeum vulgare L. cultivar Bojos | 2022 | Several barley miRNAs were differentially expressed in response to the spectral quality of incident light. |
| Database Name | Direct Link | The Overall Count of Barley miRNAs | Notes |
|---|---|---|---|
| Plant Non-coding RNA Database (PNRD) | http://structuralbiology.cau.edu.cn/PNRD/index.php | 71 | 58 of them were experimentally validated |
| Plant MicroRNA Encyclopedia (PmiREN) | https://www.pmiren.com/ | 178 | Divided into 94 miRNA families |
| miRBase | https://www.mirbase.org/summary.shtml?org=hvu | 69 | / |
| Plant small RNA genes | https://plantsmallrnagenes.science.psu.edu/ | 49 | Contain also 118 entities similar to miRNAs |
| miRNEST | http://rhesus.amu.edu.pl/mirnest/copy/browse.php | 398 | An integrative miRNA resource |
| miRNA | mRNA Target(s) in Hordeum vulgare | Known Biological Function(s) of miRNA in Plant Species and Further Notes | References |
|---|---|---|---|
| miR156a | SBP-box gene family member | Inflorescence morphogenesis regulation in tomato (Solanum lycopersicum) plants; male fertility regulation in thale cress (Arabidopsis thaliana) plants | [114,115,116] |
| miR156b | |||
| miR159a | MYB family transcription factor;lectin-like receptor kinase | Ensure normal growth via regulation of GAMYB genes | [117,118,119] |
| miR159b | MYB family transcription factor; | ||
| miR166a | START domain-containing protein; MATE domain-containing protein; class III HD-Zip protein 8 | Shoot apical meristem and vascular differentiation, leaf and root development; evolutionarily conserved stress biomarker in land plants—drought, salinity, temperature, biotic stress | [120,121] |
| miR166b | |||
| miR166c | |||
| miR168-5p | receptor-like protein kinase 5 precursor | Function in plants is unclear but targets many important mammalian transcripts (123 in total), including the gene for Low-density lipoprotein receptor adaptor protein 1 (LDLRAP1, also known as ARH)) | [122] |
| miR171-3p | scarecrow transcription factor family protein | Regulation of germination and seedling growth in Tibetan hull-less barley (Hordeum vulgare L. var. nudum); drought tolerance by regulation of flavonoid biosynthesis genes in rice | [113,123] |
| miR397a | laccase precursor protein; transporter family protein; | Plant development; circadian regulation and plant flowering; cold response in thale cress (Arabidopsis thaliana) | [124,125] |
| miR399 | rp1; ubiquitin-conjugating enzyme protein; pentatricopeptide | Salt stress response and flowering regulation in thale cress (Arabidopsis thaliana) | [126,127] |
| miR444a | FAD-binding domain of DNA photolyase domain-containing protein; DnaK family protein; alpha-taxilin; MADS-box family gene with MIKCc type-box; pentatricopeptide; WD domain, G-beta repeat domain-containing protein | Regulation of nitrate signaling pathway in nitrate-dependent root growth, nitrate accumulation, and phosphate-starvation responses in rice (Oryza sativa); antiviral pathway in rice; regulation of brassinosteroids synthesis in rice | [128,129,130] |
| miR444b | MADS-box family gene with MIKCc type-box; methyltransferase; zinc finger, C3HC4 type domain-containing protein | ||
| miR1120 | An enzyme of the cupin superfamily protein; retrotransposon protein; tesmin/TSO1-like CXC domain-containing protein; WD domain, G-beta repeat domain-containing protein; CCR4-NOT transcription factor; glycosyltransferase family 43 protein; amine oxidase-related; Divergent PAP2 family domain-containing protein | Early anther development in wheat (Triticum aestivum). miR1120 in barley has many diverse mRNA targets, however, it is questionable, if this miR1120 is a true miRNA (originating from hairpin RNA precursor), as the miR1120 gene region in barley displays almost 80% sequence similarity to the short transposon element DNA/TcMar-Stowaway | [93,131] |
| miR1436 | pseudogene | Various stress responses in Cestrum nocturnum L. and Cestrum diurnum L. | [132] |
| miR5048a | cysteine-rich receptor-like protein kinase precursor | Wheat (Triticum aestivum) grains development regulation | [133] |
| miR5049c | modifier of rudimentary protein; auxin-induced protein 5NG4; Spc97/Spc98 family protein; protein kinase domain-containing protein; OsWAK receptor-like protein kinase | Hormone, stress (heat, drought, salinity, and excess boron), and light responsiveness in barley (Hordeum vulgare L.) | [67] |
| miR5049f | resistance protein; transcription factor-related; WD domain, G-beta repeat domain-containing protein; TBC domain-containing protein; | Regulation of salt adaptation in Hordeum bulbosum | [75] |
| miR6197 | DUF26 kinase; exosome complex exonuclease rrp4 | Boron stress response regulation in barley (Hordeum vulgare) | [83] |
| miR6201 | C4-dicarboxylate transporter/malic acid transport protein | Cadmium stress response regulation in wheat (Triticum aestivum) | [134] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Volná, A.; Bartas, M.; Pečinka, P.; Špunda, V.; Červeň, J. What Do We Know about Barley miRNAs? Int. J. Mol. Sci. 2022, 23, 14755. https://doi.org/10.3390/ijms232314755
Volná A, Bartas M, Pečinka P, Špunda V, Červeň J. What Do We Know about Barley miRNAs? International Journal of Molecular Sciences. 2022; 23(23):14755. https://doi.org/10.3390/ijms232314755
Chicago/Turabian StyleVolná, Adriana, Martin Bartas, Petr Pečinka, Vladimír Špunda, and Jiří Červeň. 2022. "What Do We Know about Barley miRNAs?" International Journal of Molecular Sciences 23, no. 23: 14755. https://doi.org/10.3390/ijms232314755








