In Silico Design in Homogeneous Catalysis Using Descriptor Modelling
Abstract
:Table of Content
- 1.
- Introduction
- 1.1
- Combinatorial Methods in Homogeneous Catalysis
- 1.2
- Computational Approaches in Catalysis Research
- 2.
- Descriptors and Molecular Modelling in Homogeneous Catalysis
- 2.1
- 3D-Descriptors
- 2.2
- The CoMFA Method
- 2.3
- The Ligand Repulsive Energy Method
- 2.4
- 2D and 1D Descriptors
- 2.5
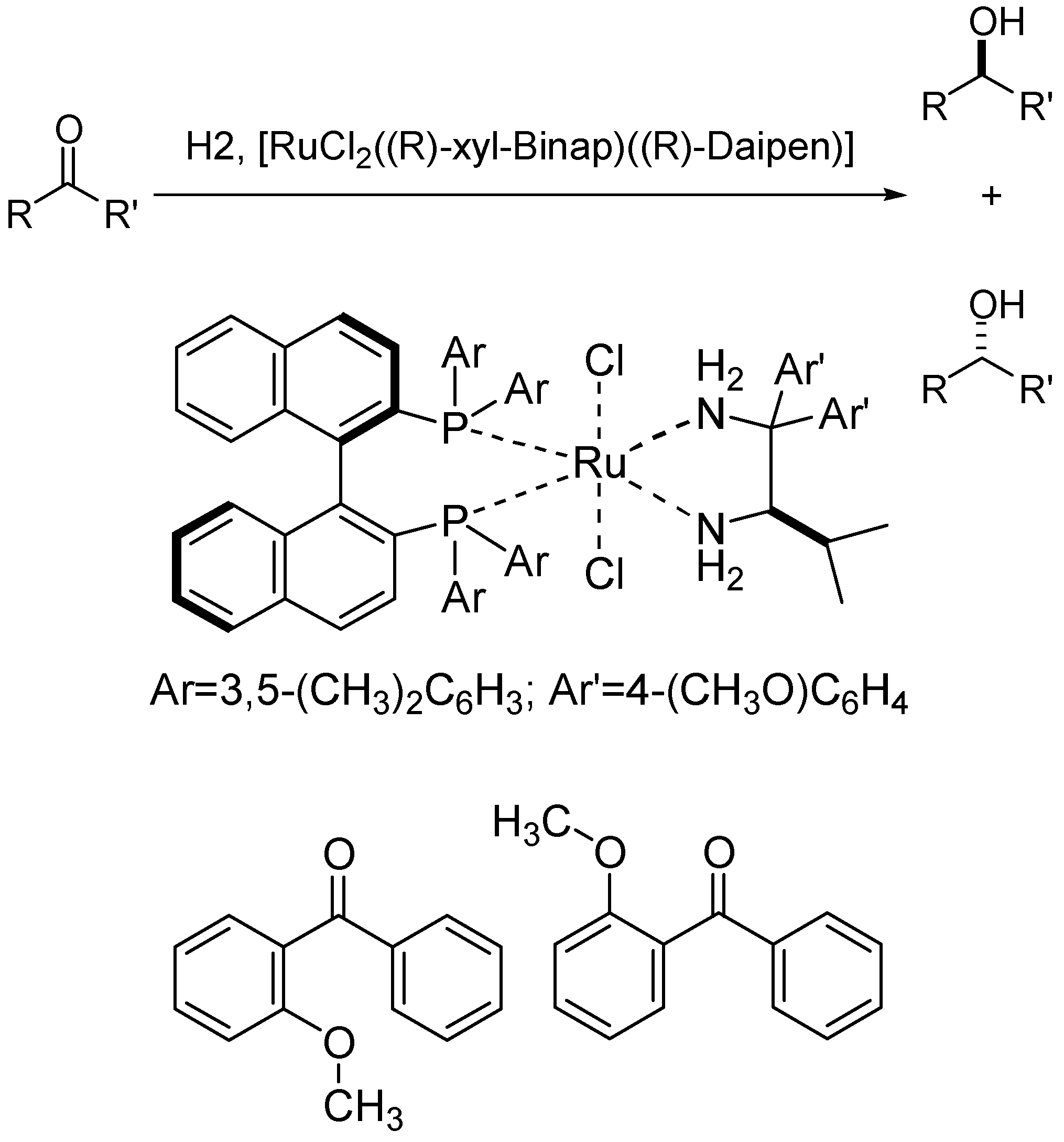
- Modelling the Chemical and Physical Properties of Solvents
- 2.6
- Using Descriptors: Pros and Cons
- 3.
- Modelling and data analysis
- 3.1
- Partial Least-Squares Models
- 3.2
- Artificial Neural Networks and Classification Analysis
- 3.3
- General Methodology in Data Analysis
- 4.
- Conclusions and Outlook
1. Introduction
1.1 Combinatorial Methods in Homogeneous Catalysis
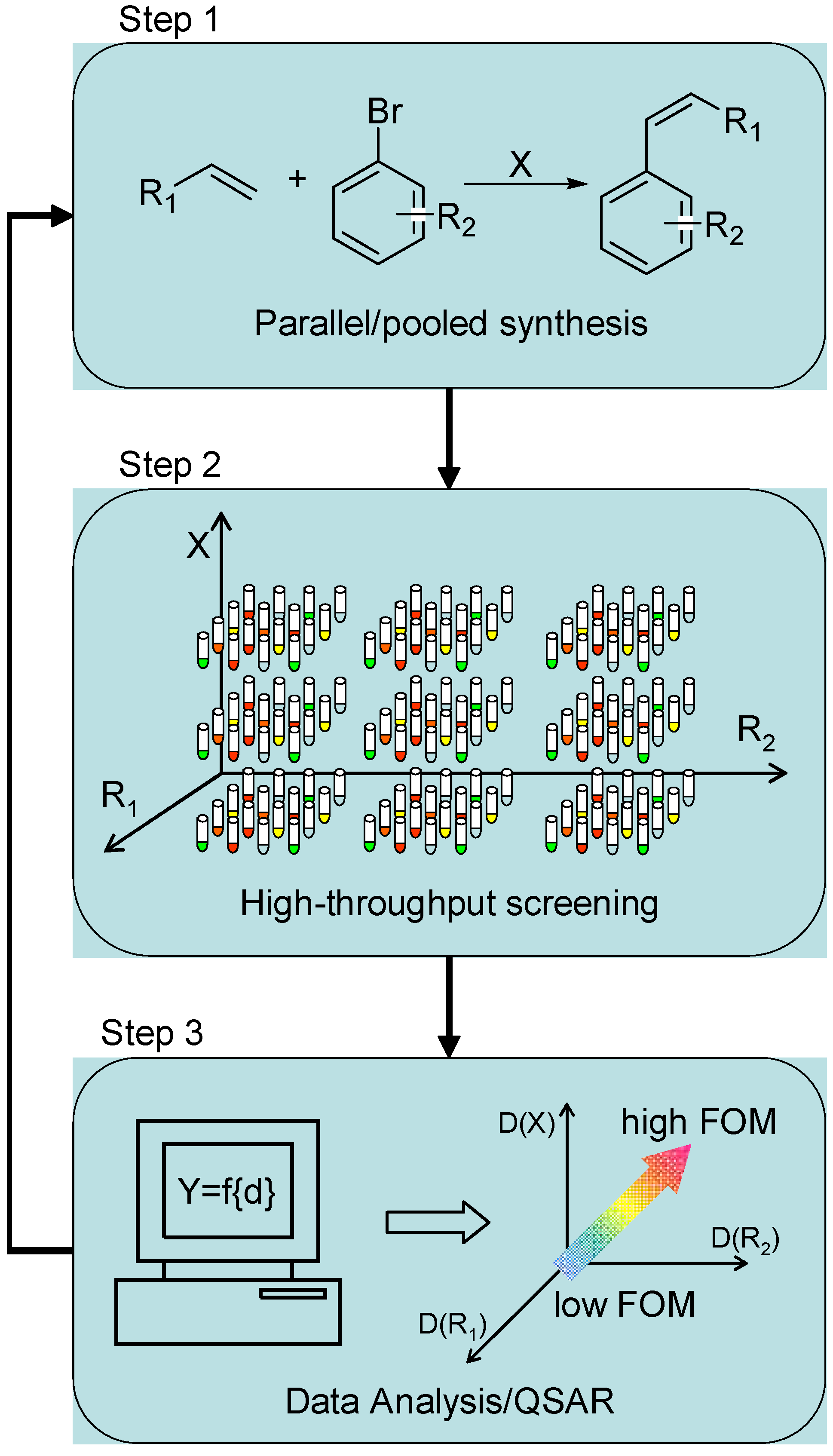
| Entry | Test reaction | Discovered/optimized catalyst |
| 1 |  | 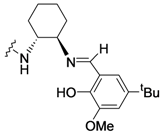 |
| 2a |  | 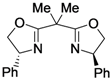 |
| 3 | 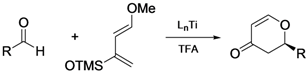 | 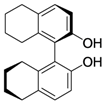 |
| 4 | 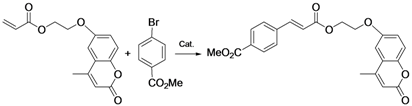 | 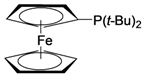 |
| 5b |  |  |
1.2 Computational Approaches in Catalysis Research
2. Descriptors and Molecular Modelling for Homogeneous Catalysis
2.1 3D-Descriptors
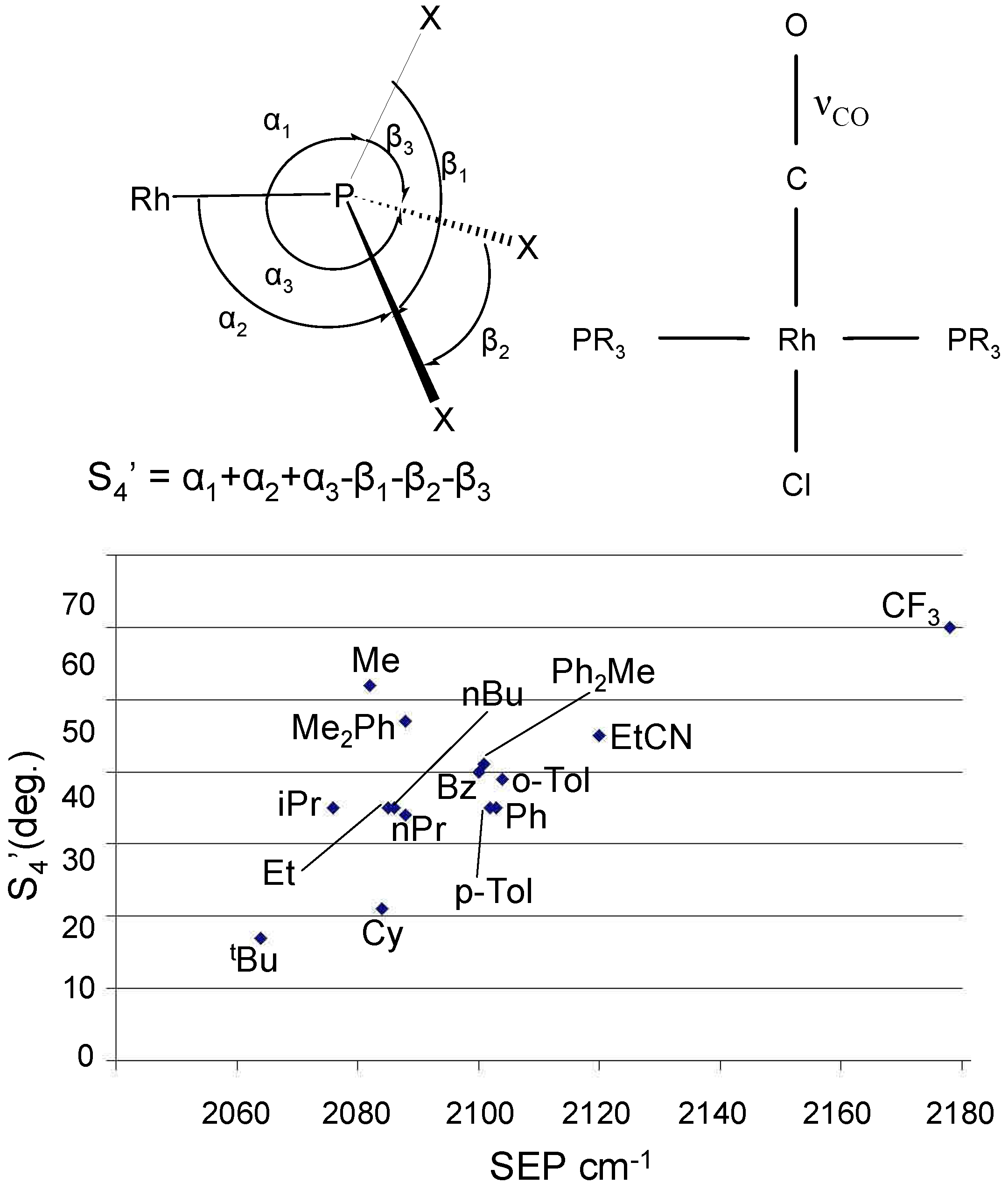
2.2 The CoMFA Method
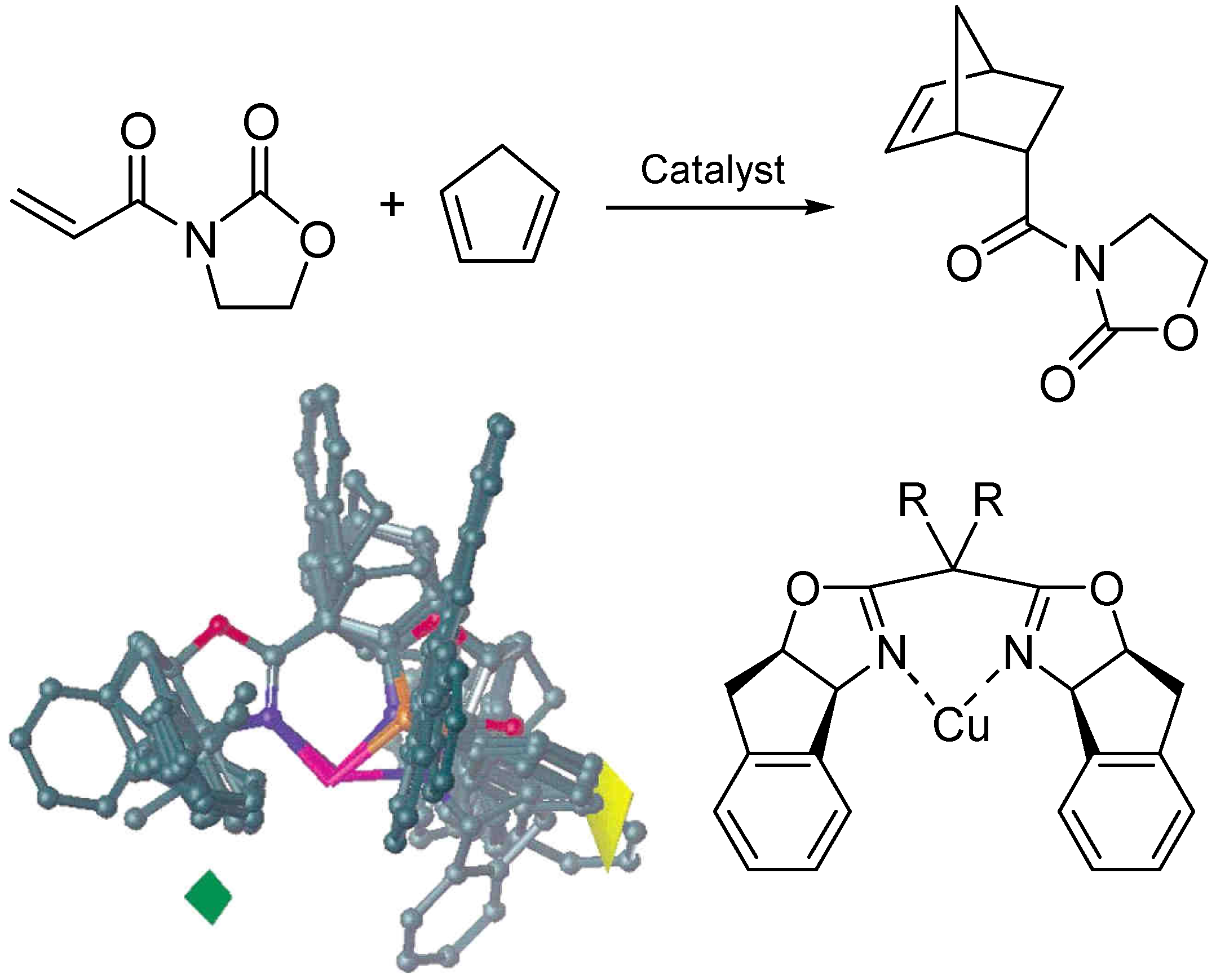
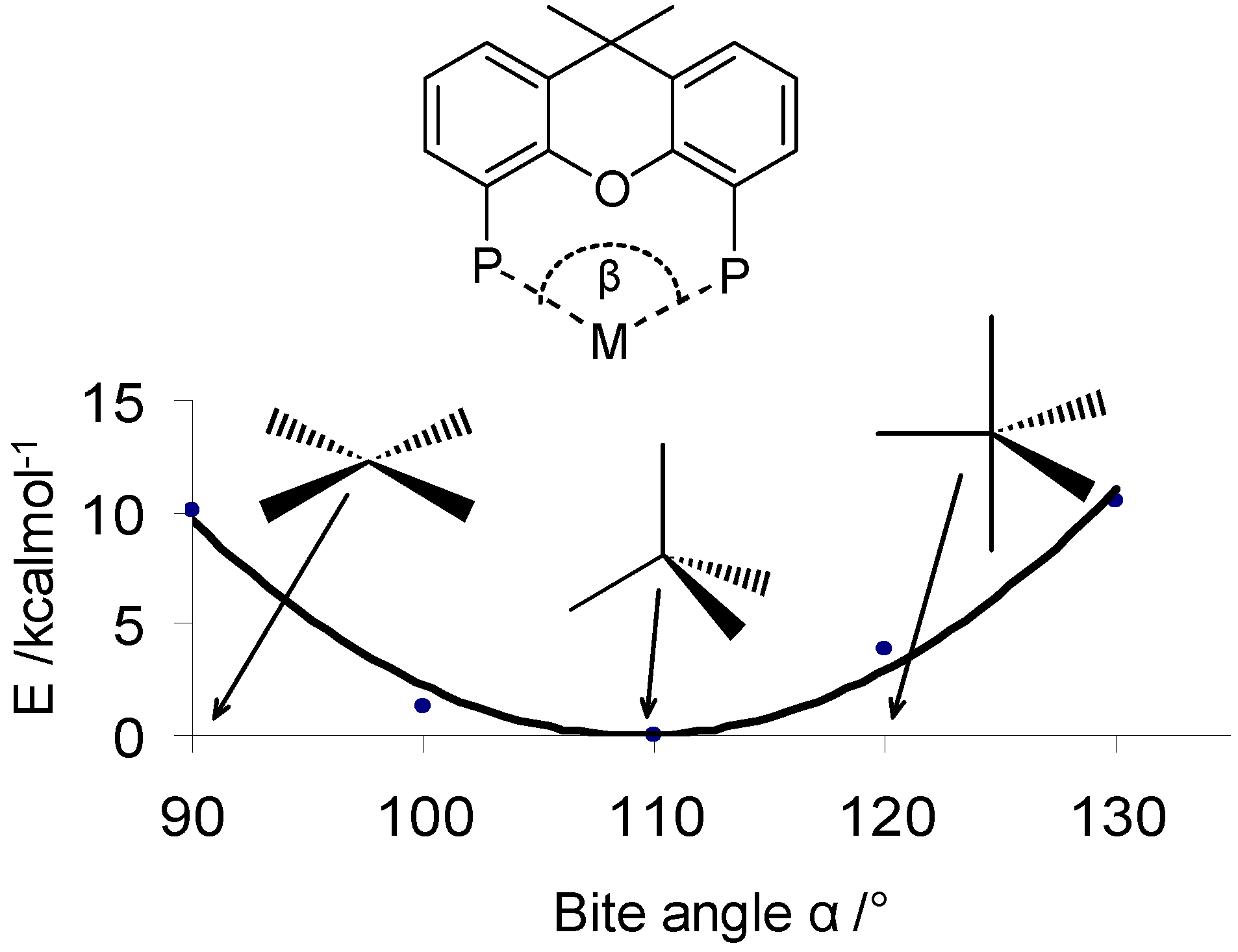
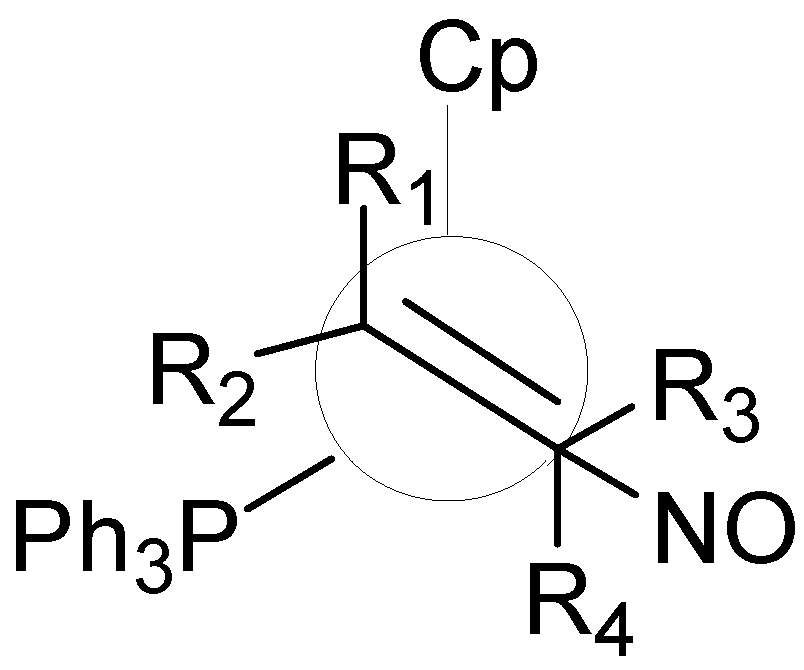
2.3 The Ligand Repulsive Energy Method

2.4 2D and 1D Descriptors
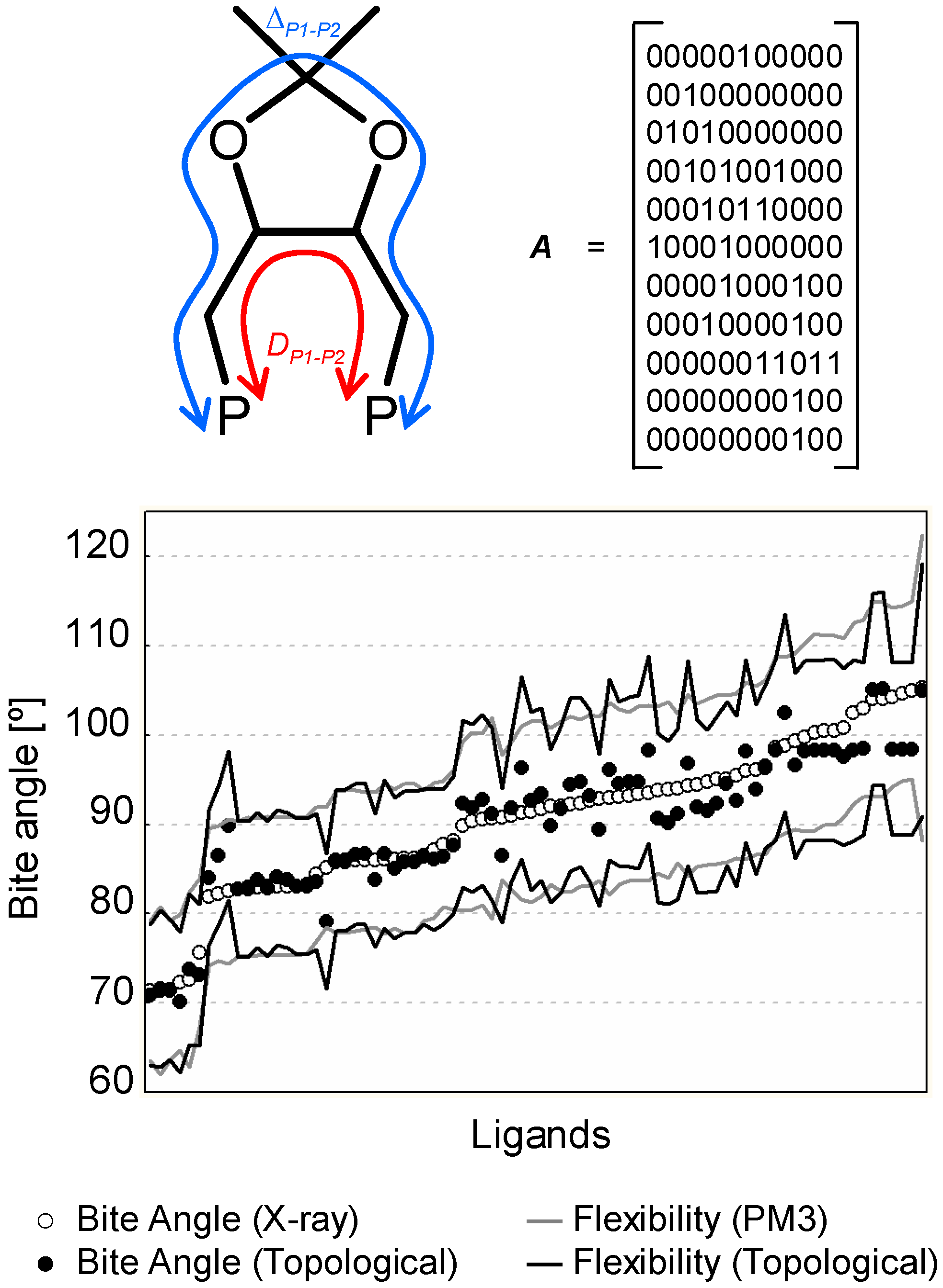
2.5 Modelling the Chemical and Physical Properties of Solvents

2.6 Using Descriptors: Pros and Cons
3. Modelling and data analysis
3.1 Partial Least-Squares Analysis
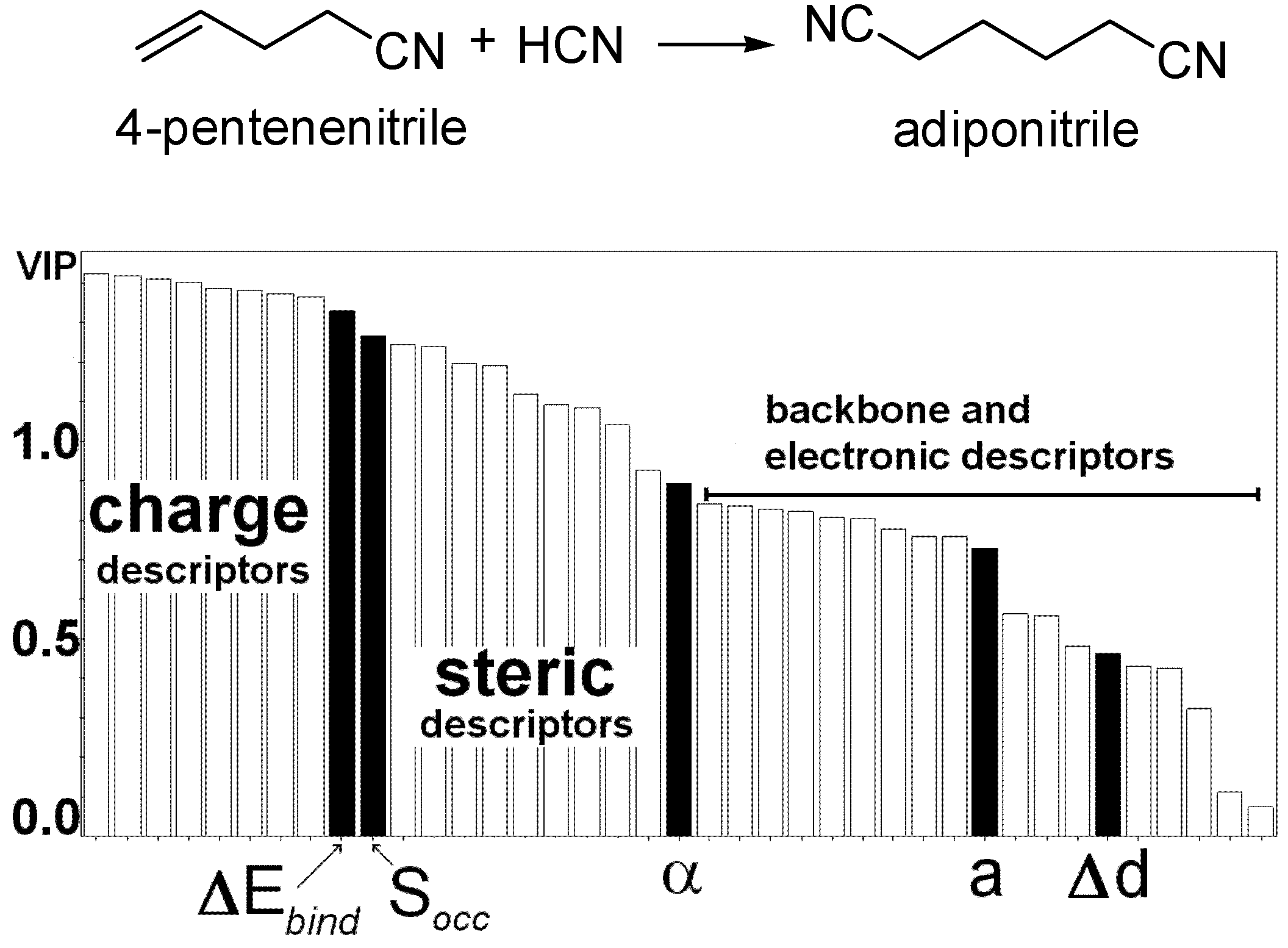
3.2 Artificial Neural Networks and Classification Analysis
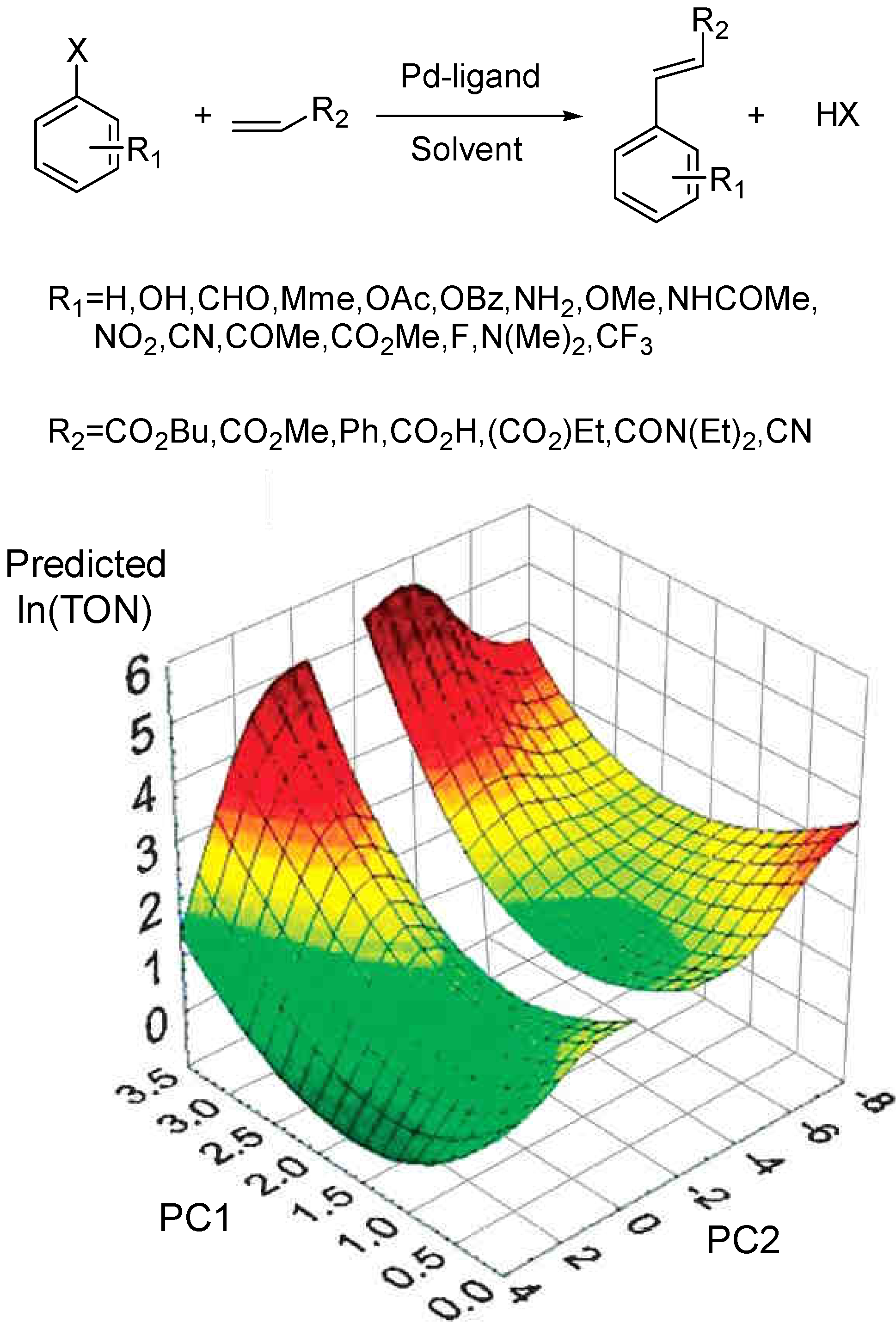
3.3 General Methodology in Data Analysis
4. Conclusions and Outlook
References and Notes
- Dolle, R.E. Comprehensive Survey of Combinatorial Library Synthesis: 1999. J. Comb. Chem. 2000, 2, 383–433. [Google Scholar] [CrossRef]
- Boring, E.; Geletii, Y.V.; Hill, C.L. A homogeneous catalyst for selective O-2 oxidation at ambient temperature. Diversity-based discovery and mechanistic investigation of thioether-oxidation by the Au(III)Cl2NO3(thioether)/O-2 system. J. Am. Chem. Soc. 2001, 123, 1625–1635. [Google Scholar] [CrossRef]
- Schareina, T.; Kempe, R. Combinatorial libraries with p-functionalized aminopyridines: Ligands for the preparation of efficient C(Aryl)-Cl activation catalysts. Angew. Chem. Int. Ed. 2002, 41, 1521–1523. [Google Scholar] [CrossRef]
- Schiedel, M.S.; Briehn, C.A.; Bauerle, P. C-C cross-coupling reactions for the combinatorial synthesis of novel organic materials. J. Organomet. Chem. 2002, 653, 200–208. [Google Scholar] [CrossRef]
- Stambuli, J.P.; Hartwig, J.F. Recent advances in the discovery of organometallic catalysts using high-throughput screening assays. Curr. Opin. Chem. Biol. 2003, 7, 420–426. [Google Scholar] [CrossRef]
- Stambuli, J.P.; Stauffer, S.R.; Shaughnessy, K.H.; Hartwig, J.F. Screening of homogeneous catalysts by fluorescence resonance energy transfer. Identification of catalysts for room-temperature Heck reactions. J. Am. Chem. Soc. 2001, 123, 2677–2678. [Google Scholar] [CrossRef]
- Fagan, P.J.; Hauptman, E.; Shapiro, R.; Casalnuovo, A. Using intelligent/random library screening to design focused libraries for the optimization of homogeneous catalysts: Ullmann ether formation. J. Am. Chem. Soc. 2000, 122, 5043–5051. [Google Scholar] [CrossRef]
- Yue, T.Y.; Nugent, W.A. Enantioselective hydrogenation of 3-alkylidenelactams: High-throughput screening provides a surprising solution. J. Am. Chem. Soc. 2002, 124, 13692–13693. [Google Scholar] [CrossRef]
- Boele, M.D.K.; van Strijdonck, G.P.F.; de Vries, A.H.M.; Kamer, P.C.J.; de Vries, J.G.; van Leeuwen, P.W.N.M. Selective Pd-catalyzed oxidative coupling of anilides with olefins through C-H bond activation at room temperature. J. Am. Chem. Soc. 2002, 124, 1586–1587. [Google Scholar] [CrossRef]
- Long, J.; Hu, J.Y.; Shen, X.Q.; Ji, B.M.; Ding, K.L. Discovery of exceptionally efficient catalysts for solvent-free enantioselective hetero-Diels-Alder reaction. J. Am. Chem. Soc. 2002, 124, 10–11. [Google Scholar] [CrossRef]
- Sigman, M.S.; Jacobsen, E.N. Schiff base catalysts for the asymmetric Strecker reaction identified and optimized from parallel synthetic libraries. J. Am. Chem. Soc. 1998, 120, 4901–4902. [Google Scholar] [CrossRef]
- Burgess, K.; Lim, H.J.; Porte, A.M.; Sulikowski, G.A. New catalysts and conditions for a C-H insertion reaction identified by high throughput catalyst screening. Angew. Chem. Int. Ed. Engl. 1996, 35, 220–222. [Google Scholar] [CrossRef]
- Shaughnessy, K.H.; Kim, P.; Hartwig, J.F. A fluorescence-based assay for high-throughput screening of coupling reactions. Application to Heck chemistry. J. Am. Chem. Soc. 1999, 121, 2123–2132. [Google Scholar] [CrossRef]
- Gao, X.; Kagan, H.B. One-pot multi-substrate screening in asymmetric catalysis. Chirality 1998, 10, 120–124. [Google Scholar]
- Rappe, A.T.; Skiff, W.M.; Casewit, C.J. Modeling metal-catalyzed olefin polymerization. Chemical Reviews 2000, 100, 1435–1456. [Google Scholar] [CrossRef]
- Willett, P. Chemoinformatics - similarity and diversity in chemical libraries. Curr. Opin. Biotechnol. 2000, 11, 85–88. [Google Scholar] [CrossRef]
- Fergus, S.; Bender, A.; Spring, D.R. Assessment of structural diversity in combinatorial synthesis. Curr. Opin. Chem. Biol. 2005, 9, 304–309. [Google Scholar] [CrossRef]
- Maldonado, A.G.; Doucet, J.P.; Petitjean, M.; Fan, B.T. Molecular similarity and diversity in chemoinformatics: From theory to applications. Molecular Diversity 2006, 10, 39–79. [Google Scholar] [CrossRef]
- Tolman, C.A. Phosphorus ligand exchange equilibriums on zerovalent nickel. Dominant role for steric effects. J. Am. Chem. Soc. 1970, 92, 2956–2965. [Google Scholar] [CrossRef]
- Tolman, C.A. Electron donor-acceptor properties of phosphorus ligands. Substituent additivity. J. Am. Chem. Soc. 1970, 92, 2953–2956. [Google Scholar] [CrossRef]
- Tolman, C.A. Electronic effects of phosphorus ligands on the protonation of NiL4 complexes. Inorg. Chem. 1972, 11, 3128–3129. [Google Scholar] [CrossRef]
- Tolman, C.A. Steric effects of phosphorus ligands in organometallic chemistry and homogeneous catalysis. Chem. Rev. 1977, 77, 313–348. [Google Scholar] [CrossRef]
- Brown, T.L.; Lee, K.J. Ligand Steric Properties. Coord. Chem. Rev. 1993, 128, 89–116. [Google Scholar] [CrossRef]
- Imyanitov, N.S. Cone angle of ligands - Group IV and V compounds. Koordinatsionnaya Khimiya 1985, 11, 1171–1178. [Google Scholar]
- Imyanitov, N.S. Numerical determination of the steric characteristics of ligands. Conical angles of Group IV and V element hydrides and halides. Koordinatsionnaya Khimiya 1985, 11, 1041–1045. [Google Scholar]
- Alyea, E.C.; Dias, S.A.; Ferguson, G.; Restivo, R.J. Structural Studies of Steric Effects in Phosphine Complexes. Synthesis and Crystal and Molecular Structure of the Dinitrato( tricyclohexylphosphine)mercury (II) Dimer. Inorg. Chem. 1977, 16, 2329–2334. [Google Scholar] [CrossRef]
- Alyea, E.C.; Dias, S.A.; Ferguson, G.; Parvez, M. Structural studies of steric effects in phosphine complexes. Part 6. The synthesis, characterization and molecular structure of the dinitro(trimesitylphosphine)mercury(II)dimer. Inorg. Chim. Acta 1979, 37, 45–52. [Google Scholar] [CrossRef]
- Smith, J.D.; Oliver, J.D. Ligand profiles of tricyclohexylphosphine. Structure of (.pi.-allyl)bis(tricyclohexylphosphine)platinum hexafluorophosphate. Inorg. Chem. 1978, 17, 2585–2589. [Google Scholar] [CrossRef]
- Zakharov, L.N.; Saf'yanov, Y.N.; Domrachev, G.A. A role of non-bonding interactions in the chemistry of organometalic compounds. Inorg. Chim. Acta 1989, 160, 77–82. [Google Scholar] [CrossRef]
- Xing-Fu, L.; Ao-Ling, G. The Nature of seat-ligand fitting in coordination space. V. Steric hindrances and reaction mechanisms — a further discussion on the structure and chemistry of compounds containing three pi-bonded cyclopentadienyl groups. Inorg. Chim. Acta 1987, 134, 143–153. [Google Scholar] [CrossRef]
- Xing-Fu, L.; Eggers, S.; Kopf, J.; Jahn, W.; Fischer, R.D.; Apostolidis, C.; Kanellakopulos, B.; Benetollo, F.; Polo, A.; Bombieri, G. Preparation and characterization of the first triscyclopentadienyl lanthanoid complexes containing two aliphatic nitrile ligands: Crystal and molecular structures of the isomorphous compounds Trans-bis(acetonitrile)tris(η5cyclopentadienyl)lanthanoid(III) (Ln = La, Ce, Pr). A successful confirmation of the solid ‘solid angle sum rule’. Inorg. Chim. Acta 1985, 100, 183–189. [Google Scholar] [CrossRef]
- Immirzi, A.; Musco, A. A method to measure the size of phosphorus ligands in coordination complexes. Inorg. Chim. Acta 1977, 25, L41–L42. [Google Scholar] [CrossRef]
- White, D.; Taverner, B.C.; Leach, P.G.L.; Coville, N.J. Quantification of Substituent and Ligand Size by the Use of Solid Angles. J. Comp. Chem. 1993, 14, 1042–1049. [Google Scholar] [CrossRef]
- White, D.; Tavener, B.C.; Leach, P.G.L.; Coville, N.J. Solid Angles . 1. The Radial Profile. J. Organomet. Chem. 1994, 478, 205–211. [Google Scholar] [CrossRef]
- White, D.; Coville, N.J. Quantification of steric effects in organometallic chemistry. In Advances in Organometallic Chemistry; Vol 36, 1994; pp. 95–158. [Google Scholar]
- White, D.; Taverner, B.C.; Coville, N.J.; Wade, P.W. Solid Angles .3. The Role of Conformers in Solid Angle Calculations. J. Organomet. Chem. 1995, 495, 41–51. [Google Scholar] [CrossRef]
- White, D.P.; Anthony, J.C.; Oyefeso, A.O. Computational measurement of steric effects: the size of organic substituents computed by ligand repulsive energies. J. Org. Chem. 1999, 64, 7707–7716. [Google Scholar] [CrossRef]
- Taverner, B.C.; Smith, J.M.; White, D.P.; Coville, N.J. Quantification of ligand-ligand interactions using solid angles. South African Journal of Chemistry-Suid-Afrikaanse Tydskrif Vir Chemie 1997, 50, 59–66. [Google Scholar]
- Steric®. Steric is available in binary for Linux (1.2.8), IRIX (5.2) and 32-bit extended DOS (gjgpp), as well as in source code for other Unix platforms. See http://www.ccl.net/cca/software/SOURCES/C/steric/index.shtml.
- Cooney, K.D.; Cundari, T.R.; Hoffman, N.W.; Pittard, K.A.; Temple, M.D.; Zhao, Y. A priori assessment of the stereoelectronic profile of phosphines and phosphites. J. Am. Chem. Soc. 2003, 125, 4318–4324. [Google Scholar] [CrossRef]
- Kamer, P.C.J.; Reek, J.N.H.; van Leeuwen, P.W.N.M. Designing ligands with the right bite. Chemtech 1998, 28, 27–33. [Google Scholar]
- Kamer, P.C.J.; van Leeuwen, P.W.N.M.; Reek, J.N.H. Wide bite angle diphosphines: Xantphos ligands in transition metal complexes and catalysis. Acc. Chem. Res. 2001, 34, 895–904. [Google Scholar] [CrossRef]
- van der Veen, L.A.; Kamer, P.C.J.; van Leeuwen, P.W.N.M. Unraveling the bite angle effect - New ligands for selective hydroformylation of internal alkenes. Cattech 2002, 6, 116–120. [Google Scholar] [CrossRef]
- Van Haaren, R.J.; Van Leeuwen, P.W.N.M.; Van Strijdonck, G.P.F.; Oevering, H.; Reek, J.N.H.; Kamer, P.C.J. Bite angle effects on the regioselection of allylic alkylation. Abs. Pap. Am. Chem. Soc. 2000, 220, U522–U522. [Google Scholar]
- van Leeuwen, P.W.N.M.; Kamer, P.C.J.; Reek, J.N.H. The bite angle makes the catalyst. Pure Appl. Chem. 1999, 71, 1443–1452. [Google Scholar] [CrossRef]
- van Leeuwen, P.W.N.M.; Kamer, P.C.J.; Reek, J.N.H.; Dierkes, P. Ligand bite angle effects in metal-catalyzed C-C bond formation. Chem. Rev. 2000, 100, 2741–2769. [Google Scholar] [CrossRef]
- van Leeuwen, P.W.N.M.; Kamer, P.C.J.; van der Veen, L.A.; Reek, J.N.H. Bite angle effects in hydroformylation catalysis. Chin. J. Chem. 2001, 19, 1–8. [Google Scholar]
- Dierkes, P.; van Leeuwen, P.W.N.M. The bite angle makes the difference: a practical ligand parameter for diphosphine ligands. J. Chem. Soc., Dalton Trans. 1999, 1519–1529. [Google Scholar] [CrossRef]
- Freixa, Z.; van Leeuwen, P.W.N.M. Bite angle effects in diphosphine metal catalysts: steric or electronic? Dalton Trans. 2003, 1890–1901. [Google Scholar] [CrossRef]
- Kranenburg, M.; Kamer, P.C.J.; van Leeuwen, P.W.N.M. The effect of the bite angle of diphosphane ligands on activity and selectivity in palladium-catalyzed allylic alkylation. Eur. J. Inorg. Chem. 1998, 25–27. [Google Scholar] [CrossRef]
- Lenero, K.A.; Kranenburg, M.; Guari, Y.; Kamer, P.C.J.; van Leeuwen, P.W.N.M.; Sabo-Etienne, S.; Chaudret, B. Ruthenium dihydrogen complexes with wide bite angle diphosphines. Inorg. Chem. 2003, 42, 2859–2866. [Google Scholar] [CrossRef]
- Aires-de-Sousa, J.; Gasteiger, J. Prediction of enantiomeric excess in a combinatorial library of catalytic enantioselective reactions. J. Comb. Chem. 2005, 7, 298–301. [Google Scholar] [CrossRef]
- Aires-de-Sousa, J.; Gasteiger, J. New description of molecular chirality and its application to the prediction of the preferred enantiomer in stereoselective reactions. J. Chem. Inf. Comp. Sci. 2001, 41, 369–375. [Google Scholar]
- A demo version of the 2D-to-3D conversion program Corina is accessible at no charge from http://www.molecular-networks.com/online_demos/corina_demo.html.
- Cramer, R.D.I.; Patterson, D.E.; Bunce, D.J. Comparative Molecular Field Analysis (CoMFA). 1. Effect of Shape on Binding of Steroids to Carrier Proteins. J. Am. Chem. Soc. 1988, 110, 5959–5967. [Google Scholar] [CrossRef]
- Lipkowitz, K.B.; Schefzick, S.; Avnir, D. Enhancement of enantiomeric excess by ligand distortion. J. Am. Chem. Soc. 2001, 123, 6710–6711. [Google Scholar] [CrossRef]
- Lipkowitz, K.B.; Schefzick, S. Ligand distortion modes leading to increased chirality content of Katsuki-Jacobsen catalysts. Chirality 2002, 14, 677–682. [Google Scholar] [CrossRef]
- Lipkowitz, K.B.; Kozlowski, M.C. Understanding stereoinduction in catalysis via computer: New tools for asymmetric synthesis. Synlett 2003, 1547–1565. [Google Scholar] [CrossRef]
- Lipkowitz, K.B.; Pradhan, M. Computational studies of chiral catalysts: A comparative molecular field analysis of an asymmetric Diels-Alder reaction with catalysts containing bisoxazoline or phosphinooxazoline ligands. J. Org. Chem. 2003, 68, 4648–4656. [Google Scholar] [CrossRef]
- Bubel, R.J.; Douglass, W.; White, D.P. Molecular mechanics-based measures of steric effects: Customized code to compute ligand repulsive energies. J. Comp. Chem. 2000, 21, 239–246. [Google Scholar] [CrossRef]
- Gillespie, A.M.; White, D.P. Understanding the steric control of stereoselective olefin binding in cyclopentadienyl complexes of rhenium: An application of de novo ligand design. Organometallics 2001, 20, 5149–5155. [Google Scholar] [CrossRef]
- Gillespie, A.M.; Morello, G.R.; White, D.P. De novo ligand design: Understanding stereoselective olefin binding to η(5)-C5H5)Re(NO)(PPh3) (+) with molecular mechanics, semiempirical quantum mechanics, and density functional theory. Organometallics 2002, 21, 3913–3921. [Google Scholar] [CrossRef]
- Burello, E.; Rothenberg, G. Topological mapping of bidentate ligands: A fast approach for screening homogeneous catalysts. Adv. Synth. Catal. 2005, 347, 1969–1977. [Google Scholar] [CrossRef]
- Chavali, S.; Lin, B.; Miller, D.C.; Camarda, K.V. Environmentally-benign transition metal catalyst design using optimization techniques. Comp. Chem. Eng. 2004, 28, 605–611. [Google Scholar] [CrossRef]
- Lin, B.; Chavali, S.; Camarda, K.; Miller, D.C. Computer-aided molecular design using Tabu search. Comp. Chem. Eng. 2005, 29, 337–347. [Google Scholar] [CrossRef]
- Schmid, R. Effect of Solvent on Chemical Reactions and Reactivity. In Handbook of Solvents; Wypych, G., Ed.; 2001; ChemTech Publishing: Toronto. [Google Scholar]
- Drago, R.S.; Hirsch, M.S.; Ferris, D.C.; Chronister, C.W. A Unified Scale of Solvent Polarities for Specific and Nonspecific Interactions. J. Chem. Soc., Perkin Trans. 2 1994, 219–230. [Google Scholar]
- Mu, L.; Drago, R.S.; Richardson, D.E. A model based QSPR analysis of the unified non-specific solvent polarity scale. J. Chem. Soc. Perkin Trans. 2 1998, 159–167. [Google Scholar]
- Kamlet, M.J.; Abboud, J.L.M.; Taft, R.W.; Abraham, M.H. Linear Solvation Energy Relationships. 23. A Comprehensive Collection of the Solvatochromic Parameters, p*, a, and ß, and Some Methods for Simplifying the Generalized Solvatochromic Equation. J. Org. Chem. 1983, 48, 2877–2887. [Google Scholar] [CrossRef]
- Gutmann, V. Donor-Acceptor Approach to Molecular Interactions; Plenum Press: New York, 1978. [Google Scholar]
- Kamlet, M.J.; Taft, R.W. The Solvatochromic Comparison Method. 1. The ß-Scale of Solvent Hydrogen-Bond Acceptor (HBA) Basicities. J. Am. Chem. Soc. 1976, 98, 377–383. [Google Scholar] [CrossRef]
- Kamlet, M.J.; Abboud, J.L.M.; Taft, R.W. The Solvatochromic Comparison Method. 6. The p* Scale of Solvent Polarities. J. Am. Chem. Soc. 1977, 99, 6027–6038. [Google Scholar] [CrossRef]
- Famini, G.R.; Tran, D.; Wilson, L.Y. Application of TLSER molecular descriptors to rate constants. Abs. Pap. Am. Chem. Soc. 1999, 218, U505–U505. [Google Scholar]
- Famini, G.R.; Wilson, L.Y. Using theoretical descriptors in linear free energy relationships: characterizing several polarity, acid and basicity scales. J. Phys. Org. Chem. 1999, 12, 645–653. [Google Scholar] [CrossRef]
- Murray, J.S.; Politzer, P.; Famini, G.R. Theoretical alternatives to linear solvation energy relationships. J. Mol. Struct. - Theochem 1998, 454, 299–306. [Google Scholar] [CrossRef]
- Bonchev, D. Overall connectivities/topological complexities: A new powerful tool for QSPR/QSAR. J. Chem. Inf. Comp. Sci. 2000, 40, 934–941. [Google Scholar]
- Sinha, M.; Achenie, L.E.K.; Ostrovsky, G.M. Environmentally benign solvent design by global optimization. Comp. Chem. Eng. 1999, 23, 1381–1394. [Google Scholar] [CrossRef]
- Gani, R.; Jimenez-Gonzalez, C.; Constable, D.J.C. Method for selection of solvents for promotion of organic reactions. Comp. Chem. Eng. 2005, 29, 1661–1676. [Google Scholar] [CrossRef]
- For information on the Dragon software package see http://www.talete.mi.it/dragon_net.htm.
- Fernandez, A.L.; Reyes, C.; Prock, A.; Giering, W.P. The stereoelectronic parameters of phosphites. The quantitative analysis of ligand effects (QALE). J. Chem. Soc., Perkin Trans. 2 2000, 1033–1041. [Google Scholar]
- For a good general introduction to chemometric methods see http://www.chemometrics.se.
- van der Linden, J.B.; Ras, E.J.; Hooijschuur, S.M.; Klaus, G.M.; Luchters, N.T.; Dani, P.; Verspui, G.; Smith, A.A.; Damen, E.W.P.; McKay, B.; Hoogenraad, M. Asymmetric catalytic ketone hydrogenation: Relating substrate structure and product enantiomeric excess using QSPR. QSAR Comb. Sci. 2005, 24, 94–98. [Google Scholar] [CrossRef]
- Burello, E.; Marion, P.; Galland, J.C.; Chamard, A.; Rothenberg, G. Ligand Descriptor analysis in nickel-catalysed hydrocyanation: A combined experimental and theoretical study. Adv. Synth. Catal. 2005, 347, 803–810. [Google Scholar] [CrossRef]
- Carlson, R.; Gautun, H. Combinatorial libraries and the development of organic synthetic methods. PLS modelling to discriminate between successful and failed reaction systems. Chemom. Intell. lab. Syst. 2005, 78, 113–124. [Google Scholar] [CrossRef]
- Burello, E.; Farrusseng, D.; Rothenberg, G. Combinatorial explosion in homogeneous catalysis: Screening 60,000 cross-coupling reactions. Adv. Synth. Catal. 2004, 346, 1844–1853. [Google Scholar] [CrossRef]
- Cundari, T.R.; Deng, J.; Pop, H.F.; Sarbu, C. Structural analysis of transition metal beta-X substituent interactions. Toward the use of soft computing methods for catalyst modeling. J. Chem. Inf. Comp. Sci. 2000, 40, 1052–1061. [Google Scholar]
- Cundari, T.R.; Russo, M. Database mining using soft computing techniques. An integrated neural network-fuzzy logic-genetic algorithm approach. J. Chem. Inf. Comp. Sci. 2001, 41, 281–287. [Google Scholar]
- Cundari, T.R.; Sarbu, C.; Pop, H.F. Robust fuzzy principal component analysis (FPCA). A comparative study concerning interaction of carbon-hydrogen bonds with molybdenum-oxo bonds. J. Chem. Inf. Comp. Sci. 2002, 42, 1363–1369. [Google Scholar]
- Bocker, A.; Schneider, G.; Teekentrup, A. Status of HTS data mining approaches. QSAR Comb. Sci. 2004, 23, 207–213. [Google Scholar] [CrossRef]
- Walters, W.P.; Goldman, B.B. Feature selection in quantitative structure-activity relationships. Curr. Opin. Drug. Discov. Develop. 2005, 8, 329–333. [Google Scholar]
- Hageman, J.A.; Westerhuis, J.A.; Fruhauf, H.W.; Rothenberg, G. Design and assembly of virtual homogeneous catalyst libraries - Towards in silico catalyst optimisation. Adv. Synth. Catal. 2006, 348, 361–369. [Google Scholar] [CrossRef]
© 2006 by MDPI (http://www.mdpi.org). Reproduction is permitted for noncommercial purposes.
Share and Cite
Burello, E.; Rothenberg, G. In Silico Design in Homogeneous Catalysis Using Descriptor Modelling. Int. J. Mol. Sci. 2006, 7, 375-404. https://doi.org/10.3390/i7090375
Burello E, Rothenberg G. In Silico Design in Homogeneous Catalysis Using Descriptor Modelling. International Journal of Molecular Sciences. 2006; 7(9):375-404. https://doi.org/10.3390/i7090375
Chicago/Turabian StyleBurello, Enrico, and Gadi Rothenberg. 2006. "In Silico Design in Homogeneous Catalysis Using Descriptor Modelling" International Journal of Molecular Sciences 7, no. 9: 375-404. https://doi.org/10.3390/i7090375




