Cloning, Expression, Purification and Crystallization of the PR Domain of Human Retinoblastoma Protein-Binding Zinc Finger Protein 1 (RIZ1)
Abstract
:1. Introduction
2. Results and Discussion
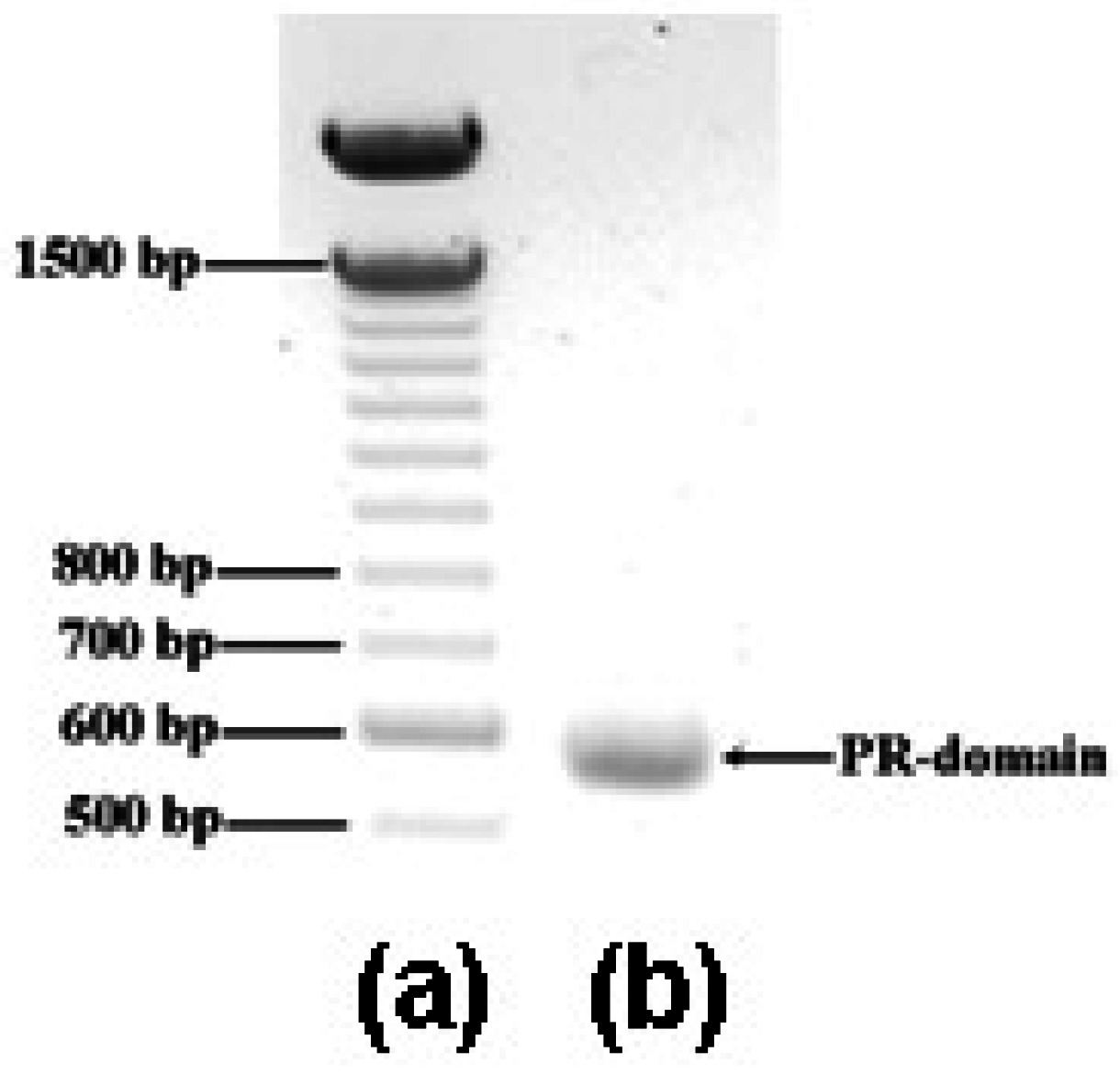
2.1. Cloning and expression of DNA fragment encoding the PR domain
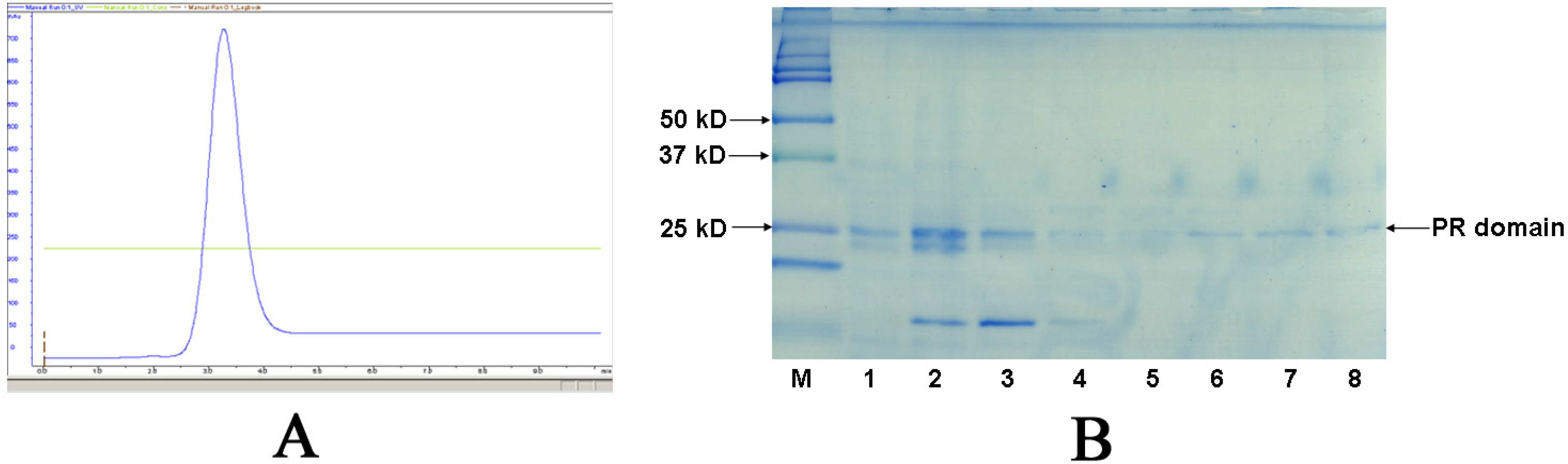
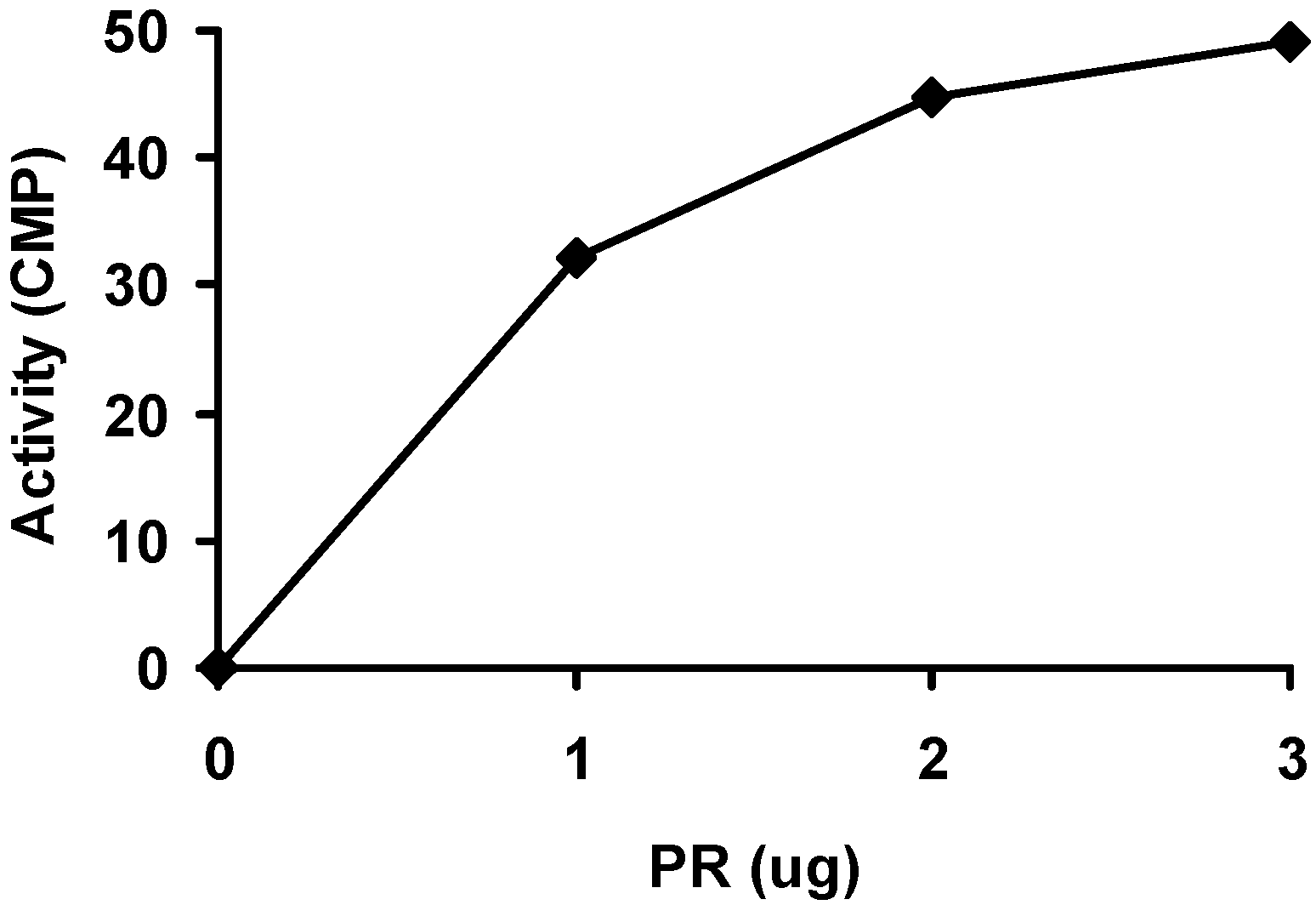
2.2. Purification and activity assay of PR domain
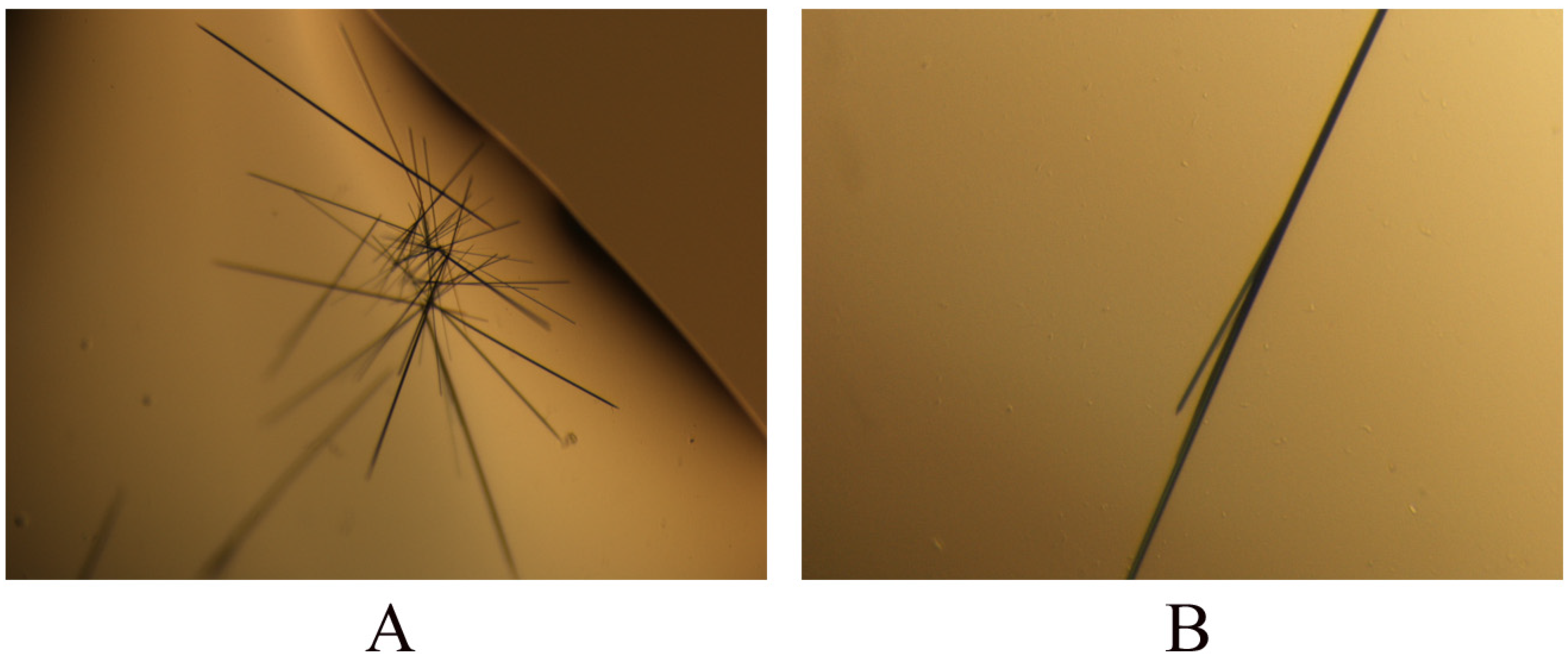
2.3. Crystallization of PR domain
3. Experimental Section
3.1. Cloning and expression of the gene fragment encoding the PR domain
3.2. Purification of the PR domain
3.3. Activity assay
3.4. Crystallization of the PR domain
4. Conclusion
Acknowledgments
References and Notes
- Buyse, IM; Shao, G; Huang, S. The retinoblastoma protein binds to RIZ, a zinc-finger protein that shares an epitope with the adenovirus E1A protein. Proc. Natl. Acad. Sci. USA 1995, 92, 4467–4471. [Google Scholar]
- Buyse, IM; Huang, S. In vitro analysis of the E1A-homologous sequences of RIZ. J. Virol. 1997, 71, 6200–6203. [Google Scholar]
- Buyse, IM; Takahashi, EI; Huang, S. Physical mapping of the retinoblastoma interacting zinc finger gene RIZ to D1S228 on chromosome 1p36. Genomics 1996, 34, 119–121. [Google Scholar]
- Muraosa, Y; Takahashi, K; Yoshizawa, M; Shibahara, S. cDNA cloning of a novel protein containing two zinc-finger domains that may function as a transcription factor for the human heme-oxygenase-1 gene. Eur. J. Biochem. 1996, 235, 471–479. [Google Scholar]
- Juneja, S; Lukeis, R; Tan, L; Cooper, I; Szelag, G; Parkin, JD; Ironside, P; Garson, OM. Cytogenetic analysis of 147 cases of non-Hodgkin's lymphoma: non-random chromosomal abnormalities and histological correlations. Br. J. Haemotol. 1990, 76, 231–237. [Google Scholar]
- Whang-Peng, J; Knutsen, T; Jaffe, ES; Steinberg, SM; Raffeld, M; Zhao, WP; Duffey, P; Condron, K; Yano, T; Longo, DL. Sequential analysis of 43 patients with non-Hodgkin's lymphoma: clinical correlations with cytogenetic, histologic, immunophenotyping, and molecular studies. Blood 1995, 85, 203–216. [Google Scholar]
- Cigudosa, JC; Parsa, NZ; Louie, DC; Filippa, DA; Jhanwar, SC; Johansson, B; Mitelman, F; Chaganti, RS. Cytogenetic analysis of 363 consecutively ascertained diffuse large B-cell lymphomas. Genes Chromosomes Cancer 1999, 25, 123–133. [Google Scholar]
- Dave, BJ. Rearrangements of chromosome band 1p36 in non-Hodgkin's lymphoma. Clin. Cancer Res. 1999, 5, 1401–1409. [Google Scholar]
- Sasaki, O; Meguro, K; Tohmiya, Y; Funato, T; Shibahara, S; Sasaki, T. Altered expression of retinoblastoma protein-interacting zinc finger gene, RIZ, in human leukaemia. Br. J. Haematol. 2002, 119, 940–948. [Google Scholar]
- Liu, L; Shao, G; Steele-Perkins, G; Huang, S. The retinoblastoma interacting zinc finger gene RIZ produces a PR domain-lacking product through an internal promoter. J. Biol. Chem. 1997, 272, 2984–2991. [Google Scholar]
- Huang, S; Shao, G; Liu, L. The PR domain of the Rb-binding zinc finger protein RIZ1 is a protein binding interface and is related to the SET domain functioning in chromatin-mediated gene expression. J. Biol. Chem. 1998, 273, 15933–15939. [Google Scholar]
- Jiang, G; Huang, S. Adenovirus expressing RIZ1 in tumor suppressor gene therapy of microsatellite-unstable colorectal cancers. Cancer Res. 2001, 61, 1796–1798. [Google Scholar]
- Du, Y; Carling, T; Fang, W; Piao, Z; Sheu, JC; Huang, S. Hypermethylation in human cancers of the RIZ1 tumor suppressor gene, a member of a histone/protein methyltransferase superfamily. Cancer Res. 2001, 61, 8094–8099. [Google Scholar]
- Deng, Q; Huang, S. PRDM5 is silenced in human cancers and has growth suppressive activities. Oncogene 2004, 17, 4903–4910. [Google Scholar]
- Mori, N; Morosetti, R; Spira, S; Lee, S; Ben-Yehuda, D; Schiller, G; Landolfi, R; Mizoguchi, H; Koeffler, HP. Chromosome band 1p36 contains a putative tumor suppressor gene important in the evolution of chronic myelocytic leukemia. Blood 1998, 92, 3405–3409. [Google Scholar]
- He, L; Yu, JX; Liu, L; Buyse, IM; Wang, MS; Yang, QC; Nakagawara, A; Brodeur, GM; Shi, YE; Huang, S. RIZ1, but not the alternative RIZ2 product of the same gene, is underexpressed in breast cancer, and forced RIZ1 expression causes G2-M cell cycle arrest and/or apoptosis. Cancer Res. 1998, 58, 4238–4244. [Google Scholar]
- Jiang, G; Liu, L; Buyse, IM; Simon, D; Huang, S. Decreased RIZ1 expression but not RIZ2 in hepatoma and suppression of hepatoma tumorigenicity by RIZ1. Int. J. Cancer. 1999, 83, 541–546. [Google Scholar]
- Chadwick, RB; Jiang, GL; Bennington, GA; Yuan, B; Johnson, CK; Stevens, MW; Niemann, TH; Peltomaki, P; Huang, S; de la Chapelle, A. Candidate tumor suppressor RIZ is frequently involved in colorectal carcinogenesis. Proc. Natl. Acad. Sci. USA 2000, 97, 2662–2667. [Google Scholar]
- Tam, W; Gomez, MF; Chadburn, A; Knowles, DM; Houldsworth, J. Lack of A563G (I188V) missense mutation in RIZ/ PRDM2 in human diffuse large B-cell lymphomas. Genes Chromosomes Cancer 2007, 46, 416–418. [Google Scholar]
- Sasaki, O; Meguro, K; Tohmiya, Y; Funato, T; Shibahara, S; Sasaki, T. Nucleotide alteration of retinoblastoma protein-interacting zinc finger gene, RIZ, in human leukemia. Tohoku J. Exp. Med. 2002, 196, 193–201. [Google Scholar]
- Canote, R; Du, Y; Carling, T; Tian, F; Peng, Z; Huang, S. The tumor suppressor gene RIZ in cancer gene therapy (review). Oncol. Rep. 2002, 9, 57–60. [Google Scholar]
- Derunes, C; Briknarová, K; Geng, L; Li, S; Gessner, CR; Hewitt, K; Wu, S; Huang, S; Woods, VI, Jr; Ely, KR. Characterization of the PR domain of RIZ1 histone methyltransferase. Biochem. Biophys. Res. Commun. 2005, 333, 925–934. [Google Scholar]
- Kim, KC; Huang, S. Histone methyltransferases in tumor suppression. Cancer Biol. Ther. 2003, 2, 491–499. [Google Scholar]
- Briknarová, K; Zhou, X; Satterthwait, A; Hoyt, DW; Ely, KR; Huang, S. Structural studies of the SET domain from RIZ1 tumor suppressor. Biochem. Biophys. Res. Commun. 2008, 366, 807–813. [Google Scholar]
Share and Cite
Sun, W.; Geyer, C.R.; Yang, J. Cloning, Expression, Purification and Crystallization of the PR Domain of Human Retinoblastoma Protein-Binding Zinc Finger Protein 1 (RIZ1). Int. J. Mol. Sci. 2008, 9, 943-950. https://doi.org/10.3390/ijms9060943
Sun W, Geyer CR, Yang J. Cloning, Expression, Purification and Crystallization of the PR Domain of Human Retinoblastoma Protein-Binding Zinc Finger Protein 1 (RIZ1). International Journal of Molecular Sciences. 2008; 9(6):943-950. https://doi.org/10.3390/ijms9060943
Chicago/Turabian StyleSun, Wanpeng, C. Ronald Geyer, and Jian Yang. 2008. "Cloning, Expression, Purification and Crystallization of the PR Domain of Human Retinoblastoma Protein-Binding Zinc Finger Protein 1 (RIZ1)" International Journal of Molecular Sciences 9, no. 6: 943-950. https://doi.org/10.3390/ijms9060943



