Examination of a Culturable Microbial Population from the Gastrointestinal Tract of the Wood-Eating Loricariid Catfish Panaque nigrolineatus
Abstract
:1. Introduction
2. Experimental Section
2.1. Fish Maintenance and Sacrifice
2.2. Fluorescence Microscopy
2.3. Enrichment Cultures
2.4. Extraction of Microbial DNA
2.5. DNA Sequencing and Analysis
2.6. DNA Stable-Isotope Probing (SIP)
3. Results and Discussion

| Class | Phylotype Detected (Source*) | Closest GenBank Species (Accession No.) | % Sequence Similarity |
|---|---|---|---|
| Actinobacteria | |||
| KC000009 (FG) | Propionibacterium acnes (GU814270) | ||
| KC000071 (W) | “ | ||
| KC000084 (WD) | Dermacoccus nishinomiyaensis (NR044872) | 99 | |
| KC000082 (W) | Cellulomonas chitinilytica (JQ659654) | 99 | |
| KC000097 (WD) | “ | ||
| Bacilli | |||
| KC000067 (W) | Paenibacillus illinoisensis (D85397) | 99 | |
| KC000081 (W) | Paenibacillus turicensis (JN378529) | 97 | |
| KC000085 (WD) | Bacillus circulans (AB680477) | 99 | |
| Bacteroides | |||
| KC000017 (FG) | Bacteroides xylanolyticus (FR850058) | 99 | |
| KC000040 (MG) | “ | ||
| Clostridia | |||
| KC000010 (FG) | Eubacterium contortum (EU980608) | 99 | |
| KC000011(FG) | Clostridium xylanolyticum (AB601097) | 99 | |
| KC000027 (FG) | Clostridium intestinale (AM158323) | 99 | |
| KC000090 (WD) | “ | ||
| KC000058 (HG) | Clostridium hathewayi (AY552788) | 99 | |
| KC000018 (FG) | Anaerosporobacter mobilis (AY534872) | 99 | |
| KC000038 (MG) | “ | ||
| KC000049 (HG) | “ | ||
| Flavobacteria | |||
| KC000048 (HG) | Elizabethkingia anophelis (EF426430) | 99 | |
| KC000079 (W) | Cloacibacterium normanense (FJ544401) | 99 | |
| α Proteobacteria | |||
| KC000087 (WD) | Pleomorphomonas koreensis (AB127971) | 99 | |
| γ Proteobacteria | |||
| KC000008 (FG), | Enterobacter soli (GU814270) | 99 | |
| KC000070 (W), | “ | ||
| KC000028 (MG) | “ | ||
| KC000030 (MG) | Enterobacter ludwigii (JN644550) | 99 | |
| KC000013 (FG) | Aeromonas allosaccharophila (GU205192) | 98 | |
| KC000016 (FG), | Aeromonas enteropelogenes (FJ940838) | 99 | |
| KC000031 (MG) | “ | ||
| KC000064 (HG) | “ | ||
| KC000068 (W) | “ | ||
| KC000032 (MG) | Aeromonas sharmana (NR_043470) | 99 | |
| KC000073 (W) | “ | ||
| KC000035 (MG) | Shewanella putrefaciens (AJ000213) | 99 | |
| KC000051 (HG) | Acinetobacter junii (JN644576) | 99 | |
| KC000098 (WD) | Moraxella osloensis (AB643595) | 99 |
| Class | Phylotype Detected (Accession No.) and Source* | Closest GenBank Species (Accession No.) | % Sequence Similarity |
|---|---|---|---|
| Actinobacteria | |||
| SIP9D (JN169776) HG | Cellulosimicrobium cellulans (AB116667) | 99 | |
| SIP9G (JN169780) MG | Curtobacterium flaccumfaciens (EU977762) | 99 | |
| Bacilli | |||
| SIP9A (JN169768) FG | Bacillus cereus strain (HM068888) | 99 | |
| Bacteroides | |||
| SIP1A (JN169765) FG | Flavobacterium aquatile (AM230485) | 95 | |
| Clostridia | |||
| SIP4D (JN169773) MG, HG | Clostridium saccharolyticum (FJ957875) | 97 | |
| SIP11D (JN169778) HG | Clostridium intestinale (AY781385) | 99 | |
| Negativicutes | |||
| SIP3C (JN169769) MG | Sporomusa aerivorans TMAO3 (NR028991) | 96 | |
| α Proteobacteria | |||
| SIP9C (JN169771) MG, HG | Azospirillum brasilense GR2(FR667907) | 99 | |
| SIP8D (JN169775) HG | Azospirillum brasilense (AB480699) | 97 | |
| SIP12D (JN169779) HG | Bosea thiooxidans (AJ250797) | 99 | |
| β Proteobacteria | |||
| SIP7C (JN169770) MG | Achromobacter denitrificans (FJ810080) | 99 | |
| γ Proteobacteria | |||
| SIP3A (JN169766) FG, HG | Pseudoxanthomonas Mexicana (AB375392) | 99 | |
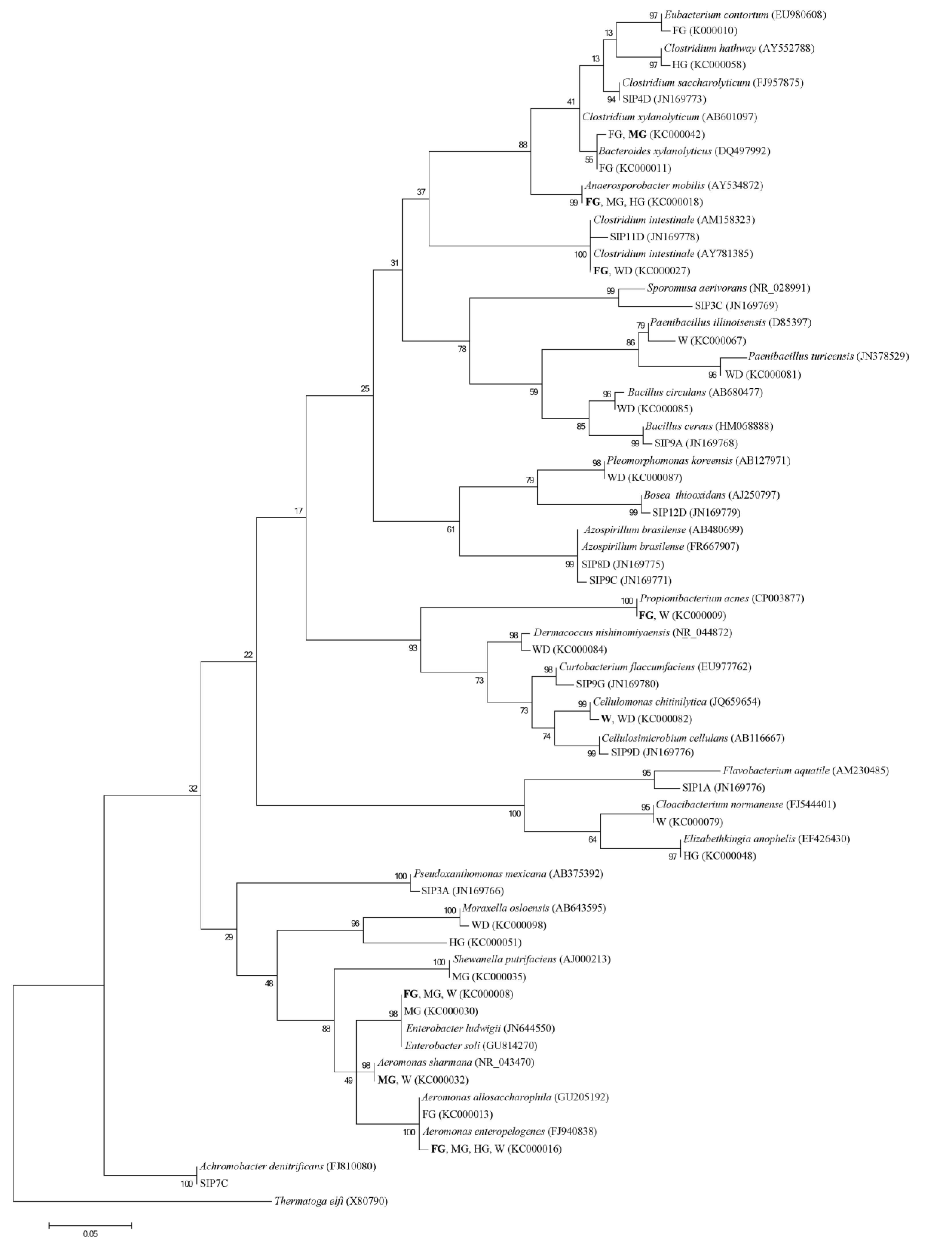
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Power, M.E. Effects of fish in river food webs. Science 1990, 250, 811–814. [Google Scholar]
- Schindler, D.E.; Carpenter, S.R.; Cole, J.J.; Kitchell, J.F.; Pace, M.L. Influence of food web structure on carbon exchange between lakes and the atmosphere. Science 1997, 277, 248–251. [Google Scholar] [CrossRef]
- Vanni, M.J. Nutrient cycling by animals in freshwater ecosystems. Annu. Rev. Ecol. Syst. 2002, 33, 341–370. [Google Scholar] [CrossRef]
- Taylor, B.W.; Flecker, A.S.; Hall, R.O. Loss of a harvested fish species disrupts carbon flow in a diverse tropical river. Science 2006, 313, 833–836. [Google Scholar] [CrossRef]
- McIntyre, P.B.; Jones, L.E.; Flecker, A.S.; Vanni, M.J. Fish extinctions alter nutrient recycling in tropical freshwaters. Proc. Natl. Acad. Sci. USA 2007, 104, 4461–4466. [Google Scholar] [CrossRef]
- Rawls, J.F.; Mahowald, M.A.; Ley, R.E.; Gordon, J.I. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell 2006, 127, 423–433. [Google Scholar] [CrossRef]
- Sullam, K.E.; Essinger, S.D.; Lozupone, C.A.; O’Connor, M.P.; Rosen, G.L.; Knight, R.O.; Kilham, S.S.; Russell, J.A. Environmental and ecological factors that shape the gut bacterial communities of fish: A meta-analysis. Mol. Ecol. 2012, 21, 3363–3378. [Google Scholar] [CrossRef]
- Rawls, J.F.; Samuel, B.S.; Gordon, J.I. Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc. Natl. Acad. Sci. USA 2004, 101, 4596–4601. [Google Scholar] [CrossRef]
- Bates, J.M.; Mittge, E.; Kuhlman, J.; Baden, K.N.; Cheesman, S.E.; Guillemin, K. Distinct signals fromthe microbiota promote different aspects of zebrafish gut differentiation. Dev. Biol. 2006, 297, 374–386. [Google Scholar] [CrossRef]
- Sugita, H.; Miyajima, C.; Deguchi, Y. Vitamin B12 producing ability of the intestinal microfora of fresh-water fish. Aquaculture 1991, 92, 267–276. [Google Scholar] [CrossRef]
- Sekirov, I.; Finlay, B.B. The role of the intestinal microbiota in enteric infection. J. Physiol. 2009, 587, 4159–4167. [Google Scholar] [CrossRef]
- Choat, J.H.; Clements, K.D. Vertebrate herbivores in marine and terrestrial environments: A nutritional ecology perspective. Annu. Rev. Ecol. Syst. 1998, 29, 375–403. [Google Scholar] [CrossRef]
- Choat, J.H.; Clements, K.D.; Robbins, W.D. The trophic status of herbivorous fishes on coral reefs 1: Dietary analyses. Mar. Biol. 2002, 140, 613–623. [Google Scholar] [CrossRef]
- Choat, J.H.; Robbins, W.D.; Clements, K.D. The trophic status of herbivorous fishes on coral reefs II: Food processing modes and trophodynamics. Mar. Biol. 2004, 145, 445–454. [Google Scholar]
- Armbruster, J.W. Phylogenetic relationships of the suckermouth armoured catfishes (Loricariidae) with emphasis on the Hypostominae and the Ancistrinae. Zool. J. Linn. Soc. 2004, 141, 1–80. [Google Scholar] [CrossRef]
- Lujan, N.K.; Winemiller, K.O.; Armbruster, J.W. Trophic diversity in the evolution and community assembly of loricariid catfishes. BMC Evol. Biol. 2012, 26, 124–136. [Google Scholar]
- Saul, W.G. An ecological study of fishes at a site in upper amazonian ecuador. Proc. Acad. Nat. Sci. USA 1975, 127, 93–114. [Google Scholar]
- Delariva, R.L.; Agostinho, A.A. Relationship between morphology and diets of six neotropical loricariids. J. Fish Biol. 2001, 58, 832–847. [Google Scholar] [CrossRef]
- Salvador, L.F., Jr.; Salvador, G.N.; Santos, G.B. Morphology of the digestive tract and feeding habits of Loricaria lentiginosa Isbrücker, 1979 in a Brazilian reservoir. Acta Zool. 2009, 90, 101–109. [Google Scholar] [CrossRef]
- Schaefer, S.A.; Stewart, D.J. Systematics of Panaque dentex species group (Siluriformes, Loricariidae) wood-eating armored catfishes from tropical South America. Ichthyol. Explor. Freshw. 1993, 4, 309–342. [Google Scholar]
- Armbruster, J.W. The Species of the Hypostomus Cochliodon Group (Siluriformes: Loricariidae). Zootaxa 2003, 249, 1–60. [Google Scholar]
- Geerinckx, T.; de Poorter, J.; Adriaens, D. Morphology and development of teeth and epidermal brushes in loricariid catfishes. J. Morphol. 2007, 268, 805–814. [Google Scholar] [CrossRef]
- Lujan, N.K.; Armbruster, J.W. Morphological and functional diversity of the mandible in suckermouth armored catfishes (Siluriformes: Loricariidae). J. Morphol. 2012, 273, 24–39. [Google Scholar] [CrossRef]
- Nonogaki, H.; Nelson, J.A.; Patterson, W.P. Dietary histories of herbivorous loricariid catfishes: evidence from δ13C values of otoliths. Environ. Biol. Fishes 2007, 78, 13–21. [Google Scholar] [CrossRef]
- German, D. Inside the guts of wood-eating catfishes: Can they digest wood? J. Comp. Physiol. B 2009, 179, 1011–1023. [Google Scholar] [CrossRef]
- Clements, K.D.; Gleeson, V.P.; Slaytor, M. Short-chain fatty acid metabolism in temperate marine herbivorous fish. J. Comp. Physiol. B 1994, 164, 372–377. [Google Scholar] [CrossRef]
- German, D.; Bittong, R. Digestive enzyme activities and gastrointestinal fermentation in wood-eating catfishes. J. Comp. Physiol. B 2009, 179, 1025–1042. [Google Scholar] [CrossRef]
- Lujan, N.K.; German, D.P.; Winemiller, K.O. Do wood-grazing fishes partition their niche?: morphological and isotopic evidence for trophic segregation in Neotropical Loricariidae. Funct. Ecol. 2011, 28, 1327–1338. [Google Scholar]
- Lynd, L.R.; Weimer, P.J.; van Zyl, W.H.; Pretorius, I.S. Microbial cellulose utilization: Fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 2002, 66, 506–577. [Google Scholar] [CrossRef]
- Mouchet, M.A.; Bouvier, C.; Bouvier, T.; Troussellier, M.; Escalas, A.; Mouillot, D. Genetic difference but functional similarity among fish gut bacterial communities through molecular and biochemical fingerprints. FEMS Microbiol. Ecol. 2012, 79, 568–580. [Google Scholar] [CrossRef]
- Leschine, S.B. Cellulose degradation in anaerobic environments. Ann. Rev. Microbiol. 1995, 8, 237–299. [Google Scholar]
- Wilson, D.B. Microbial diversity of cellulose hydrolysis. Curr. Opin. Microbiol. 2011, 14, 259–263. [Google Scholar]
- Breznak, J.A.; Brune, A. Role of microorganisms in the digestion of lignocellulose by termites. Annu. Rev. Entomol. 1994, 39, 453–487. [Google Scholar] [CrossRef]
- Cleveland, L.R. Symbiosis between termites and their intestinal protozoa. Proc. Natl. Acad. Sci. USA 1923, 9, 424–428. [Google Scholar] [CrossRef]
- Distel, D.L.; Beaudoin, D.J.; Morrill, W. Coexistence of multiple proteobacterial endosymbionts in the gills of the wood-boring bivalve Lyrodus pedicellatus (Bivalvia: Teredinidae). Appl. Environ. Microbiol. 2002, 68, 6292–6299. [Google Scholar] [CrossRef]
- Pellens, R.; Grandcolas, P.; da Silva Neto, I.D. A new and independently evolved case of xylophagy and the presence of intestinal flagellates in the cockroach Parasphaeria boleiriana (Dictyoptera, Blaberidae, Zetoborinae) from the remnants of the Brazilian Atlantic forest. Can. J. Zool. 2002, 80, 350–360. [Google Scholar] [CrossRef]
- Waterbury, J.B.; Turner, R.D.; Calloway, C.B. A cellulolytic nitrogen-fixing bacterium cultured from the gland of Deshayes in shipworms (Bivalvia: Teredinidae). Science 1983, 221, 1401–1403. [Google Scholar]
- Bayer, E.A.; Chanzy, H.; Lamed, R.; Shoham, Y. Cellulose, cellulases and cellulosomes. Curr. Opin. Struct. Biol. 1998, 8, 548–557. [Google Scholar] [CrossRef]
- Talbot, J.M.; Treseder, K.K. Interactions among lignin, cellulose, and nitrogen drive litter chemistry–decay relationship. Ecology 2011, 93, 345–354. [Google Scholar] [CrossRef]
- Zaldivar, J.; Nielsen, J.; Olsson, L. Ethanol fuel production from lignocellulose: A challenge for metabolic engineering and process integration. Appl. Microbiol. Biotechnol. 2001, 56, 17–34. [Google Scholar] [CrossRef]
- Ragauskas, A.J.; Williams, C.K.; Davison, B.H.; Britovsek, G.; Cairney, J.; Eckert, C.A.; Frederick, W.J.; Hallett, J.P.; Leak, D.J.; Liotta, C.L.; et al. The path forward for biofuels and biomaterials. Science 2006, 311, 484–489. [Google Scholar] [CrossRef]
- McDonald, R.; Schreier, H.J.; Watts, J.E.M. Phylogenetic Analysis of Microbial Communities in different regions of the gastrointestinal tract in Panaque nigrolineatus, a wood-eating fish. PLoS One 2012, 7, e48018. [Google Scholar] [CrossRef]
- Sowers, K.R.; Noll, K.M. Techniques for Anaerobic Growth. In Archaea: A Laboratory Manual; Robb, F.T., Sowers, K.R., DasSharma, S., Place, A.R., Schreier, H.J., Fleischmann, E.M., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1995; pp. 15–48. [Google Scholar]
- Lane, D.J. 16S/23S rRNA Sequencing. In Nucleic Acid Techniques in Bacterial Systematics; Stackebrandt, E., Ed.; John Wiley and Sons Ltd.: Cambridge, UK, 1991; pp. 115–175. [Google Scholar]
- Ferris, M.J.; Muyzer, G.; Ward, D.M. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl. Environ. Microbiol. 1996, 62, 340–346. [Google Scholar]
- Warshaw, J.E.; Leschine, S.B.; Canale-Parola, E. Anaerobic cellulolytic bacteria from wetwood of living trees. Appl. Environ. Microbiol. 1985, 50, 807–811. [Google Scholar]
- Neufeld, J.D.; Vohra, J.; Dumont, M.G.; Lueders, T.; Manefield, M.; Friedrich, M.W.; Murrell, J.C. DNA stable-isotope probing. Nat. Protocols 2007, 2, 860–866. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar]
- Swidsinski, A.; Weber, J.; Loening-Baucke, V.; Hale, L.P.; Lochs, H. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J. Clin. Microbiol. 2005, 43, 3380–3389. [Google Scholar] [CrossRef]
- Rogers, G.M.; Baecker, A.A.W. Clostridium xylanolyticum sp. nov., an anaerobic xylanolytic bacterium from decayed Pinus patula wood chips. Int. J. Syst. Bacteriol. 1991, 41, 140–143. [Google Scholar]
- Jeong, H.; Lim, Y.W.; Yi, H.; Sekiguchi, Y.; Kamagata, Y.; Chun, J. Anaerosporobacter mobilis gen. nov., sp. nov., isolated from forest soil. Int. J. Syst. Evol. Microbiol. 2007, 57, 1784–1787. [Google Scholar]
- Manter, D.; Hunter, W.; Vivanco, J. Enterobacter soli sp. nov.: A lignin-degrading γ-Proteobacteria isolated from soil. Curr. Microbiol. 2011, 62, 1044–1049. [Google Scholar]
- Yoon, M.-H.; Ten, L.N.; Im, W.-T.; Lee, S.-T. Cellulomonas chitinilytica sp. nov., a chitinolytic bacterium isolated from cattle-farm compost. Int. J. Syst. Evol. Microbiol. 2008, 58, 1878–1884. [Google Scholar]
- Kim, C.H. Characterization and substrate specificity of an endo-beta-1,4-D-glucanase I (Avicelase I) from an extracellular multienzyme complex of Bacillus circulans. Appl. Environ. Microbiol. 1995, 61, 959–965. [Google Scholar]
- Singh, B.; Bhat, T.K.; Sharma, O.P.; Kanwar, S.S.; Rahi, P.; Gulati, A. Isolation of tannase-producing Enterobacter ludwigii GRT-1 from the rumen of migratory goats. Small Rumin. Res. 2012, 102, 172–176. [Google Scholar] [CrossRef]
- Buchman, A.L.; Pickett, M.J.; Mann, L.; Ament, M.E. Central venous catheter infection caused by Moraxella osloensis in a patient receiving home parenteral nutrition. Diagn. Microbiol. Infect. Dis. 1993, 17, 163–166. [Google Scholar] [CrossRef]
- Debelian, G.J.; Olsen, I.; Tronstad, L. Profiling of Propionibacterium acnes recovered from root canal and blood during and after endodontic treatment. Endod. Dent. Traumatol. 1992, 8, 248–254. [Google Scholar] [CrossRef]
- Figueira, V.; Vaz-Moreira, I.; Silva, M.; Manaia, C.M. Diversity and antibiotic resistance of Aeromonas spp. in drinking and waste water treatment plants. Water Res. 2011, 45, 5599–5611. [Google Scholar] [CrossRef]
- Wang, X.; Feng, Y.; Wang, H.; Qu, Y.; Yu, Y.; Ren, N.; Li, N.; Wang, E.; Lee, H.; Logan, B.E. Bioaugmentation for electricity generation from corn stover biomass using microbial fuel cells. Environ. Sci. Technol. 2009, 43, 6088–6093. [Google Scholar] [CrossRef]
- Lednická, D.; Mergaert, J.; Cnockaert, M.C.; Swings, J. Isolation and identification of cellulolytic bacteria involved in the degradation of natural cellulosic fibres. Syst. Appl. Microbiol. 2000, 23, 292–299. [Google Scholar] [CrossRef]
- Halsall, D.M.; Gibson, A.H. Comparison of two cellulomonas strains and their interaction with azospirillum brasilense in degradation of wheat straw and associated nitrogen fixation. Appl. Environ. Microbiol. 1986, 51, 855–861. [Google Scholar]
- Gößner, A.S.; Küsel, K.; Schulz, D.; Trenz, S.; Acker, G.; Lovell, C.R.; Drake, H.L. Trophic interaction of the aerotolerant anaerobe Clostridium intestinale and the acetogen Sporomusa rhizae sp. nov. isolated from roots of the black needlerush Juncus roemerianus. Microbiology 2006, 152, 1209–1219. [Google Scholar] [CrossRef]
- Chen, Y.; Vohra, J.; Murrell, J.C. Applications of DNA-Stable Isotope Probing in Bioremediation Studies. In Bioremediation, Methods in Molecular Biology; Cummings, S.P., Ed.; Humana Press: New York, NY, USA, 2010; pp. 129–138. [Google Scholar]
- Leadbetter, J.R. Cultivation of recalcitrant microbes: Cells are alive, well and revealing their secrets in the 21st century laboratory. Curr. Opin. Microbiol. 2003, 6, 274–281. [Google Scholar] [CrossRef]
- Doolittle, W.F. Phylogenetic classification and the universal tree. Science 1999, 284, 2124–2128. [Google Scholar] [CrossRef]
- Roeselers, G.; Mittge, E.K.; Stephens, W.Z.; Parichy, D.M.; Cavanaugh, C.M.; Guillemin, K.; Rawls, J.F. Evidence for a core gut microbiota in the zebrafish. ISME J. 2011, 5, 1595–1608. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Watts, J.E.M.; McDonald, R.; Daniel, R.; Schreier, H.J. Examination of a Culturable Microbial Population from the Gastrointestinal Tract of the Wood-Eating Loricariid Catfish Panaque nigrolineatus. Diversity 2013, 5, 641-656. https://doi.org/10.3390/d5030641
Watts JEM, McDonald R, Daniel R, Schreier HJ. Examination of a Culturable Microbial Population from the Gastrointestinal Tract of the Wood-Eating Loricariid Catfish Panaque nigrolineatus. Diversity. 2013; 5(3):641-656. https://doi.org/10.3390/d5030641
Chicago/Turabian StyleWatts, Joy E. M., Ryan McDonald, Rachelle Daniel, and Harold J. Schreier. 2013. "Examination of a Culturable Microbial Population from the Gastrointestinal Tract of the Wood-Eating Loricariid Catfish Panaque nigrolineatus" Diversity 5, no. 3: 641-656. https://doi.org/10.3390/d5030641





