Genetic Diversity in Jatropha curcas L. Assessed with SSR and SNP Markers
Abstract
:1. Introduction
2. Experimental Section
| ID | World Region | Country | PE | Plants in Pool | SSR | SNP | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mda (%) | Msa (%) | Mis (%) | Mda (%) | Msa (%) | Mis (%) | |||||
| 1 | AFRICA | Gambia | P | 15 | 5.6 | 94.4 | 0.0 | 0.0 | 100.0 | 0.0 |
| 2 | AFRICA | Madagascar | P | 20 | 5.6 | 94.4 | 0.0 | 0.0 | 100.0 | 0.0 |
| 3 | ASIA | Bangladesh | P | 19 | 5.6 | 94.4 | 0.0 | 0.0 | 100.0 | 0.0 |
| 4 | ASIA | Laos | P | 8 | 5.6 | 94.4 | 0.0 | 0.0 | 100.0 | 0.0 |
| 5 | SAM | Paraguay | P | 18 | 5.6 | 94.4 | 0.0 | 0.0 | 100.0 | 0.0 |
| 6 | SAM | Argentina | P | 18 | 9.3 | 88.9 | 1.8 | 0.0 | 100.0 | 0.0 |
| 7 | CNAM | Mexico | A | 12 | 14.8 | 85.2 | 0.0 | 10.0 | 90.0 | 0.0 |
| 8 | CNAM | Mexico | P | 10 | 5.6 | 94.4 | 0.0 | 0.0 | 100.0 | 0.0 |
| 9 | CNAM | Mexico | P | 19 | 35.2 | 63.0 | 1.8 | 30.0 | 60.0 | 10.0 |
| 10 | CNAM | Mexico | A | 19 | 9.3 | 90.7 | 0.0 | 0.0 | 100.0 | 0.0 |
| 11 | CNAM | Mexico | A | 18 | 9.3 | 90.7 | 0.0 | 0.0 | 100.0 | 0.0 |
| 12 | CNAM | Mexico | A | 17 | 9.3 | 90.7 | 0.0 | 0.0 | 100.0 | 0.0 |
| 13 | CNAM | Mexico | A | 18 | 9.3 | 90.7 | 0.0 | 0.0 | 100.0 | 0.0 |
| 14 | CNAM | Mexico | A | 14 | 9.3 | 90.7 | 0.0 | 0.0 | 100.0 | 0.0 |
| 15 | CNAM | Mexico | A | 9 | 7.4 | 92.6 | 0.0 | 0.0 | 100.0 | 0.0 |
| 16 | CNAM | Mexico | A | 19 | 7.4 | 92.6 | 0.0 | 0.0 | 100.0 | 0.0 |
| 17 | AFRICA | Gambia | P | 6 | 5.6 | 94.4 | 0.0 | |||
| 18 | ASIA | Laos | P | 6 | 5.6 | 94.4 | 0.0 | |||
| 19 | SAM | Brasil | P | 6 | 5.6 | 92.6 | 1.8 | |||
| 20 | CNAM | Mexico | P | 6 | 5.6 | 92.6 | 1.8 | |||
| 21 | CNAM | Mexico | A | 6 | 9.3 | 85.2 | 5.5 | |||
| 22 | CNAM | Mexico | A | 6 | 7.4 | 92.6 | 0.0 | |||
| 23 | CNAM | Mexico | A | 6 | 7.4 | 88.9 | 3.7 | |||
| 24 | CNAM | Colombia | P | 6 | 5.6 | 83.3 | 11.1 | |||
| 25 | CNAM | Colombia | P | 6 | 5.6 | 87.0 | 7.4 | |||
| 26 | ASIA | Laos | P | 6 | 5.6 | 92.6 | 1.8 | |||
| 27 | CNAM | Colombia | P | 1 | 14.8 | 83.3 | 1.9 | |||
| 28 | CNAM | Colombia | P | 1 | 9.3 | 90.7 | 0.0 | |||
| 29 | CNAM | Guatemala | A | 1 | 14.8 | 83.3 | 1.9 | |||
| 30 | CNAM | Guatemala | A | 1 | 20.4 | 77.8 | 1.8 | |||
| 31 | CNAM | Colombia | P | 1 | 9.3 | 90.7 | 0.0 | |||
| 32 | CNAM | Colombia | P | 1 | 9.3 | 90.7 | 0.0 | |||
| 33 | AFRICA | Gambia | P | 4 | 5.6 | 94.4 | 0.0 | |||
| 34 | AFRICA | Chad | P | 4 | 5.6 | 94.4 | 0.0 | |||
| 35 | AFRICA | Cameroon | P | 4 | 5.6 | 94.4 | 0.0 | |||
| 36 | AFRICA | Madagascar | P | 4 | 5.6 | 94.4 | 0.0 | |||
| 37 | AFRICA | Madagascar | P | 4 | 5.6 | 94.4 | 0.0 | |||
| 38 | SAM | Argentina | P | 4 | 5.6 | 94.4 | 0.0 | |||
| 39 | SAM | Brasil | P | 4 | 5.6 | 94.4 | 0.0 | |||
| 40 | SAM | Brasil | P | 4 | 5.6 | 94.4 | 0.0 | |||
| 41 | CNAM | Mexico | A | 4 | 9.3 | 88.9 | 1.8 | |||
| 42 | CNAM | Mexico | A | 4 | 5.6 | 63.0 | 31.4 | |||
| 43 | CNAM | Mexico | A | 4 | 5.6 | 74.1 | 20.3 | |||
| 44 | CNAM | Guatemala | P | 4 | 7.4 | 88.9 | 3.7 | |||
| 45 | SAM | Argentina | P | 4 | 5.6 | 94.4 | 0.0 | |||
| 46 | CNAM | Mexico | A | 4 | 11.1 | 87.0 | 1.9 | |||
| 47 | CNAM | Mexico | A | 4 | 16.7 | 81.5 | 1.8 | |||
| 48 | AFRICA | CapeVerde | P | 4 | 5.6 | 92.6 | 1.8 | |||
| 49 | AFRICA | Madagascar | P | 1 | 5.6 | 92.6 | 1.8 | |||
| 50 | AFRICA | Madagascar | P | 1 | 5.6 | 94.4 | 0.0 | |||
| 51 | AFRICA | Madagascar | P | 1 | 5.6 | 94.4 | 0.0 | |||
| 52 | AFRICA | Madagascar | P | 1 | 5.6 | 94.4 | 0.0 | |||
| 53 | AFRICA | Madagascar | P | 1 | 5.6 | 94.4 | 0.0 | |||
| 54 | AFRICA | Madagascar | P | 1 | 5.6 | 94.4 | 0.0 | |||
| 55 | AFRICA | Madagascar | P | 1 | 5.6 | 94.4 | 0.0 | |||
| 56 | AFRICA | Madagascar | P | 1 | 5.6 | 92.6 | 1.8 | |||
| 57 | AFRICA | Madagascar | P | 1 | 5.6 | 90.7 | 3.7 | |||
| 58 | AFRICA | Madagascar | P | 1 | 5.6 | 66.7 | 27.7 | |||
| 59 | AFRICA | Madagascar | P | 1 | 5.6 | 94.4 | 0.0 | |||
| 60 | AFRICA | Madagascar | P | 1 | 5.6 | 94.4 | 0.0 | |||
| 61 | AFRICA | Madagascar | P | 1 | 5.6 | 94.4 | 0.0 | |||
| 62 | AFRICA | Madagascar | P | 1 | 5.6 | 94.4 | 0.0 | |||
| 63 | AFRICA | Madagascar | P | 1 | 5.6 | 94.4 | 0.0 | |||
| 64 | AFRICA | Madagascar | P | 1 | 5.6 | 94.4 | 0.0 | |||
| 65 | AFRICA | Madagascar | P | 1 | 5.6 | 94.4 | 0.0 | |||
| 66 | AFRICA | Madagascar | P | 1 | 5.6 | 94.4 | 0.0 | |||
| 67 | AFRICA | Madagascar | P | 1 | 7.4 | 92.6 | 0.0 | |||
| 68 | AFRICA | Madagascar | P | 1 | 5.6 | 94.4 | 0.0 | |||
| 69 | AFRICA | Madagascar | P | 1 | 5.6 | 94.4 | 0.0 | |||
| 70 | AFRICA | Madagascar | P | 1 | 5.6 | 94.4 | 0.0 | |||
| Mean | 7.6 | 90.4 | 2.0 | 0.0 | 100.0 | 0.0 | ||||
| Min | 5.6 | 63.0 | 0.0 | 0.0 | 60.0 | 0.0 | ||||
| Max | 35,2 | 94.4 | 31.5 | 30.0 | 100.0 | 10.0 | ||||
Statistical Analysis
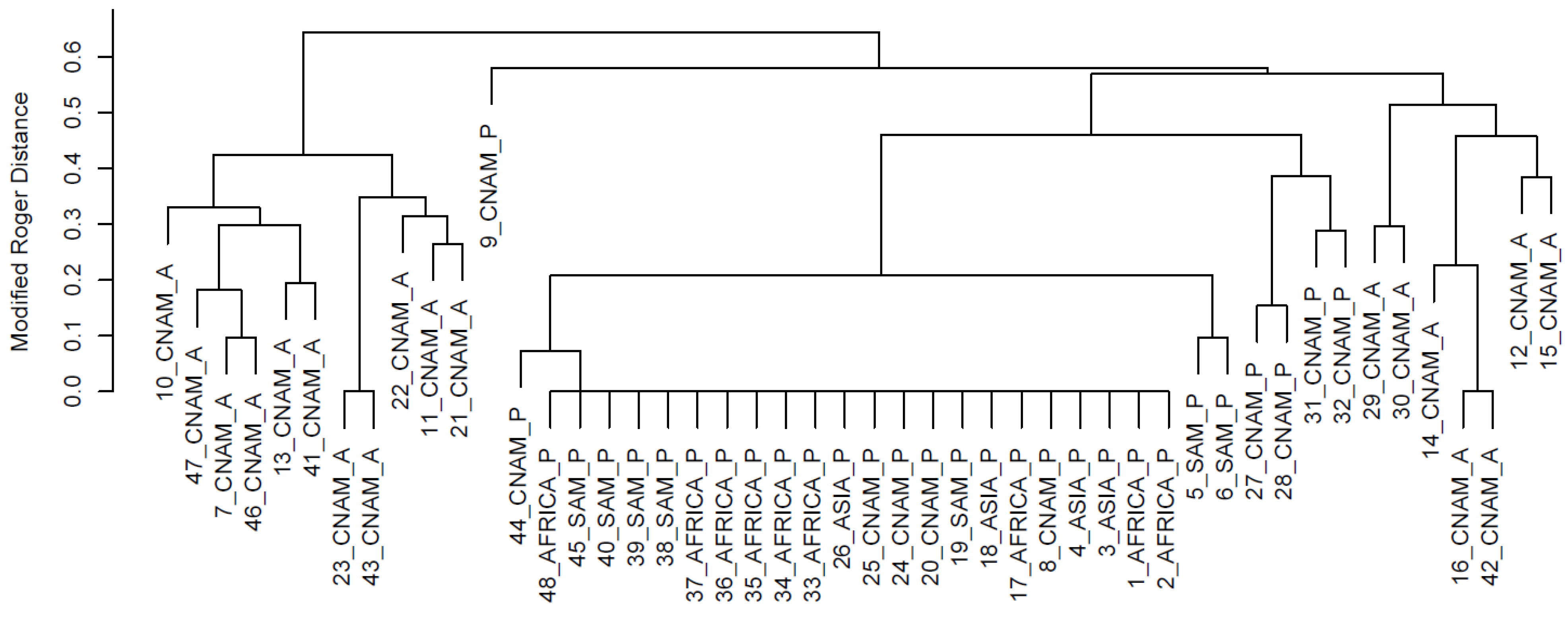
3. Results and Discussion
| Marker | Allele Proportion | Ama | PIC | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | (%) | ||
| M1 | 0.77 | 0.24 | 1.5 | 0.29 | |||||
| M2 | 0.96 | 0.04 | 0.01 | 1.4 | 0.08 | ||||
| M3 | 1.00 | 0.0 | 0.00 | ||||||
| M4 | 0.99 | 0.03 | 1.4 | 0.04 | |||||
| M5 | 0.99 | 0.02 | 0.02 | 1.5 | 0.04 | ||||
| M6 | 0.73 | 0.24 | 0.03 | 0.01 | 1.4 | 0.35 | |||
| M7 | 1.00 | 0.02 | 1.5 | 0.01 | |||||
| M8 | 0.74 | 0.10 | 0.10 | 0.04 | 0.03 | 0.02 | 2.9 | 0.42 | |
| M9 | 0.99 | 0.05 | 0.03 | 6.0 | 0.08 | ||||
| M10 | 0.81 | 0.23 | 4.3 | 0.27 | |||||
| M11 | 1.00 | 0.0 | 0.00 | ||||||
| M12 | 0.94 | 0.05 | 0.03 | 1.5 | 0.12 | ||||
| M13 | 0.94 | 0.06 | 0.02 | 1.5 | 0.11 | ||||
| M14 | 1.00 | 0.0 | 0.00 | ||||||
| M15 | 0.90 | 0.14 | 0.01 | 5.7 | 0.20 | ||||
| M16 | 1.00 | 0.90 | 0.10 | 100.0 | 0.44 | ||||
| M17 | 0.69 | 0.33 | 1.4 | 0.34 | |||||
| M18 | 0.87 | 0.20 | 7.1 | 0.24 | |||||
| M19 | 1.00 | 0.02 | 0.02 | 3.0 | 0.03 | ||||
| M20 | 1,00 | 0.0 | 0.00 | ||||||
| M21 | 0.73 | 0.27 | 0.04 | 4.3 | 0.35 | ||||
| M22 | 1.00 | 0.0 | 0.00 | ||||||
| M23 | 0.99 | 0.01 | 0.0 | 0.03 | |||||
| M24 | 1.00 | 0.04 | 0.01 | 5.7 | 0.05 | ||||
| M25 | 0.73 | 0.27 | 0.0 | 0.32 | |||||
| M26 | 0.71 | 0.10 | 0.09 | 0.07 | 0.03 | 0.01 | 0.01 | 2.9 | 0.47 |
| M27 | 0.76 | 0.24 | 0.01 | 0.01 | 2.9 | 0.34 | |||
| M28 | 1.00 | 0.0 | 0.00 | ||||||
| M29 | 1.00 | 0.0 | 0.00 | ||||||
| M30 | 1.00 | 0.01 | 1.4 | 0.01 | |||||
| M31 | 1.00 | 0.0 | 0.00 | ||||||
| M32 | 1.00 | 0.0 | 0.00 | ||||||
| M33 | 0.87 | 0.09 | 0.04 | 0.03 | 2.9 | 0.24 | |||
| M34 | 0.99 | 0.99 | 0.03 | 0.01 | 98.6 | 0.41 | |||
| M35 | 1.00 | 0.03 | 2.9 | 0.03 | |||||
| M36 | 0.99 | 0.01 | 0.0 | 0.03 | |||||
| M37 | 0.83 | 0.09 | 0.06 | 0.03 | 0.01 | 1.4 | 0.30 | ||
| M38 | 0.91 | 0.14 | 0.05 | 0.03 | 0.02 | 0.02 | 13.8 | 0.25 | |
| M39 | 1.00 | 0.0 | 0.00 | ||||||
| M40 | 1.00 | 0.0 | 0.00 | ||||||
| M41 | 1.00 | 0.01 | 0.01 | 2.9 | 0.03 | ||||
| M42 | 0.76 | 0.14 | 0.10 | 0.01 | 1.4 | 0.37 | |||
| M43 | 1.00 | 0.0 | 0.00 | ||||||
| M44 | 1.00 | 0.0 | 0.00 | ||||||
| M45 | 1.00 | 0.0 | 0.00 | ||||||
| M46 | 1.00 | 0.0 | 0.00 | ||||||
| M47 | 1.00 | 0.0 | 0.00 | ||||||
| M48 | 0.99 | 0.01 | 0.0 | 0.03 | |||||
| M49 | 0.77 | 0.13 | 0.10 | 0.04 | 0.03 | 7.1 | 0.38 | ||
| M50 | 0.81 | 0.17 | 0.01 | 0.0 | 0.27 | ||||
| M51 | 0.81 | 0.17 | 0.01 | 0.0 | 0.27 | ||||
| M52 | 0.83 | 0.17 | 0.0 | 0.24 | |||||
| M53 | 1.00 | 0.99 | 0.29 | 0.01 | 0.01 | 100.0 | 0.50 | ||
| M54 | 1.00 | 0.24 | 23.5 | 0.19 | |||||
| Mean | 7.7 | 0.15 | |||||||
| Min | 0.0 | 0.00 | |||||||
| Max | 100.0 | 0.50 | |||||||
| Marker | Allele Proportion | Ama | PIC | |
|---|---|---|---|---|
| 1 | 2 | |||
| M1 | 0.9 | 0.1 | 0.0 | 0.11 |
| M2 | 0.5 | 0.5 | 0.0 | 0.37 |
| M3 | 0.6 | 0.4 | 6.3 | 0.11 |
| M4 | 0.9 | 0.1 | 0.0 | 0.37 |
| M5 | 0.6 | 0.5 | 6.3 | 0.37 |
| M6 | 0.9 | 0.2 | 6.3 | 0.23 |
| M7 | 0.8 | 0.3 | 0.0 | 0.30 |
| M8 | 0.7 | 0.3 | 0.0 | 0.34 |
| M9 | 0.9 | 0.1 | 0.0 | 0.11 |
| M10 | 0.9 | 0.1 | 0.0 | 0.19 |
| M11 | 0.9 | 0.1 | 0.0 | 0.11 |
| M12 | 0.7 | 0.3 | 0.0 | 0.34 |
| M13 | 0.8 | 0.2 | 0.0 | 0.29 |
| M14 | 0.6 | 0.6 | 12.5 | 0.38 |
| M15 | 0.6 | 0.4 | 6.3 | 0.37 |
| M16 | 0.7 | 0.4 | 12.5 | 0.36 |
| M17 | 0.9 | 0.2 | 6.3 | 0.23 |
| M18 | 0.8 | 0.3 | 0.0 | 0.30 |
| M19 | 0.6 | 0.4 | 0.0 | 0.37 |
| M20 | 0.8 | 0.2 | 0.0 | 0.26 |
| M21 | 0.7 | 0.3 | 0.0 | 0.34 |
| M22 | 0.9 | 0.2 | 12.5 | 0.19 |
| M23 | 0.5 | 0.5 | 0.0 | 0.37 |
| M24 | 0.6 | 0.5 | 6.3 | 0.37 |
| M25 | 0.9 | 0.1 | 0.0 | 0.20 |
| M26 | 0.7 | 0.4 | 6.3 | 0.35 |
| M27 | 0.9 | 0.1 | 6.3 | 0.16 |
| M28 | 0.9 | 0.1 | 6.3 | 0.16 |
| M29 | 0.6 | 0.5 | 6.3 | 0.37 |
| M30 | 0.6 | 0.5 | 6.3 | 0.37 |
| M31 | 0.9 | 0.1 | 0.0 | 0.11 |
| M32 | 0.9 | 0.1 | 0.0 | 0.11 |
| M33 | 0.9 | 0.1 | 0.0 | 0.11 |
| M34 | 0.6 | 0.5 | 6.3 | 0.37 |
| M35 | 0.8 | 0.3 | 0.0 | 0.30 |
| M36 | 0.6 | 0.4 | 0.0 | 0.36 |
| M37 | 0.6 | 0.4 | 0.0 | 0.37 |
| M38 | 0.8 | 0.2 | 0.0 | 0.26 |
| M39 | 0.9 | 0.1 | 0.0 | 0.11 |
| M40 | 0.8 | 0.2 | 0.0 | 0.26 |
| M41 | 0.8 | 0.3 | 6.7 | 0.29 |
| M42 | 0.6 | 0.4 | 0.0 | 0.37 |
| M43 | 0.9 | 0.1 | 0.0 | 0.19 |
| M44 | 0.8 | 0.2 | 0.0 | 0.26 |
| M45 | 0.8 | 0.3 | 12.5 | 0.30 |
| M46 | 0.8 | 0.2 | 0.0 | 0.26 |
| M47 | 0.9 | 0.2 | 6.3 | 0.23 |
| M48 | 0.9 | 0.1 | 0.0 | 0.19 |
| M49 | 0.7 | 0.3 | 0.0 | 0.35 |
| M50 | 0.9 | 0.1 | 0.0 | 0.11 |
| M51 | 0.9 | 0.1 | 0.0 | 0.11 |
| M52 | 0.5 | 0.5 | 0.0 | 0.37 |
| M53 | 0.9 | 0.1 | 0.0 | 0.11 |
| M54 | 0.9 | 0.1 | 0.0 | 0.20 |
| M55 | 0.6 | 0.4 | 0.0 | 0.36 |
| M56 | 0.9 | 0.1 | 0.0 | 0.11 |
| M57 | 0.6 | 0.4 | 7.1 | 0.36 |
| M58 | 0.7 | 0.4 | 6.3 | 0.35 |
| M59 | 0.6 | 0.5 | 6.3 | 0.37 |
| M60 | 0.7 | 0.4 | 6.3 | 0.35 |
| M61 | 0.6 | 0.5 | 6.3 | 0.37 |
| M62 | 0.6 | 0.4 | 0.0 | 0.37 |
| M63 | 0.6 | 0.5 | 6.3 | 0.37 |
| M64 | 0.9 | 0.1 | 6.3 | 0.16 |
| M65 | 0.6 | 0.4 | 0.0 | 0.35 |
| M66 | 0.8 | 0.2 | 0.0 | 0.26 |
| M67 | 0.9 | 0.1 | 0.0 | 0.20 |
| M68 | 0.9 | 0.1 | 0.0 | 0.19 |
| M69 | 0.9 | 0.1 | 0.0 | 0.11 |
| M70 | 0.9 | 0.1 | 0.0 | 0.11 |
| M71 | 0.6 | 0.4 | 6.3 | 0.37 |
| M72 | 0.8 | 0.2 | 0.0 | 0.24 |
| M73 | 0.8 | 0.3 | 0.0 | 0.30 |
| M74 | 0.8 | 0.3 | 6.3 | 0.32 |
| M75 | 0.5 | 0.5 | 0.0 | 0.38 |
| M76 | 0.8 | 0.3 | 6.3 | 0.28 |
| M77 | 0.8 | 0.3 | 6.3 | 0.32 |
| M78 | 0.6 | 0.5 | 6.3 | 0.37 |
| M79 | 0.8 | 0.2 | 0.0 | 0.26 |
| M80 | 0.7 | 0.3 | 0.0 | 0.34 |
| M81 | 0.6 | 0.5 | 6.3 | 0.37 |
| M82 | 0.6 | 0.4 | 0.0 | 0.37 |
| M83 | 0.8 | 0.2 | 0.0 | 0.23 |
| M84 | 0.9 | 0.1 | 0.0 | 0.12 |
| M85 | 0.8 | 0.2 | 0.0 | 0.26 |
| M86 | 0.5 | 0.5 | 0.0 | 0.38 |
| M87 | 0.9 | 0.1 | 6.7 | 0.16 |
| M88 | 0.6 | 0.5 | 6.3 | 0.37 |
| M89 | 0.6 | 0.4 | 0.0 | 0.37 |
| M90 | 0.6 | 0.5 | 6.3 | 0.37 |
| M91 | 0.7 | 0.4 | 6.3 | 0.35 |
| M92 | 0.9 | 0.1 | 0.0 | 0.11 |
| M93 | 0.7 | 0.3 | 0.0 | 0.34 |
| M94 | 0.6 | 0.5 | 6.3 | 0.37 |
| M95 | 0.7 | 0.3 | 0.0 | 0.35 |
| M96 | 0.9 | 0.1 | 0.0 | 0.11 |
| M97 | 0.8 | 0.2 | 0.0 | 0.26 |
| M98 | 0.6 | 0.4 | 0.0 | 0.37 |
| M99 | 0.5 | 0.5 | 0.0 | 0.38 |
| M100 | 0.7 | 0.3 | 0.0 | 0.34 |
| M101 | 0.8 | 0.2 | 0.0 | 0.26 |
| M102 | 0.9 | 0.1 | 0.0 | 0.11 |
| M103 | 0.8 | 0.2 | 0.0 | 0.28 |
| M104 | 0.5 | 0.5 | 0.0 | 0.38 |
| M105 | 0.6 | 0.4 | 0.0 | 0.36 |
| M106 | 0.8 | 0.2 | 0.0 | 0.26 |
| M107 | 0.7 | 0.3 | 0.0 | 0.34 |
| M108 | 0.9 | 0.1 | 0.0 | 0.11 |
| M109 | 0.8 | 0.3 | 6.3 | 0.32 |
| M110 | 0.9 | 0.1 | 0.0 | 0.19 |
| M111 | 0.9 | 0.1 | 6.3 | 0.16 |
| M112 | 0.5 | 0.5 | 0.0 | 0.38 |
| M113 | 0.8 | 0.3 | 0.0 | 0.30 |
| M114 | 0.8 | 0.2 | 0.0 | 0.26 |
| M115 | 0.8 | 0.3 | 6.3 | 0.28 |
| M116 | 0.8 | 0.2 | 0.0 | 0.26 |
| M117 | 0.9 | 0.2 | 12.5 | 0.19 |
| M118 | 0.6 | 0.4 | 0.0 | 0.37 |
| M119 | 0.9 | 0.1 | 0.0 | 0.11 |
| M120 | 0.7 | 0.3 | 0.0 | 0.34 |
| Mean | 2.4 | 0.27 | ||
| Min | 0.0 | 0.11 | ||
| Max | 12.5 | 0.38 | ||


| Source of Variation | Df a | Df b | SSR a | SNP a | SSR b |
|---|---|---|---|---|---|
| World region | 3 | 3 | 50.0 | 47.5 | 32.6 |
| Countries within world region | 3 | 9 | 0.0 | 0.0 | 19.3 |
| Accessions within countries and world region | 9 | 57 | 50.0 | 52.5 | 48.0 |
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ceasar, S.A.; Ignacimuthu, S. Applications of biotechnology and biochemical engineering for the development of jatropha and biodiesel: A review. Renew. Sustain. Energy Rev. 2011, 15, 5176–5185. [Google Scholar] [CrossRef]
- Divakara, B.N.; Upadhyaya, H.D.; Wani, S.P.; Laxmipathi Gowda, C.L. Biology and genetic improvement of Jatropha curcas L.: A review. Appl. Energy 2010, 87, 732–742. [Google Scholar] [CrossRef]
- Francis, G.; Edinger, R.; Becker, K. A concept for simultaneous wasteland reclamation, fuel production, and socio-economic development in degraded areas in India. Need, potential and perspectives of Jatropha plantations. Nat. Resour. Forum 2005, 29, 12–24. [Google Scholar] [CrossRef]
- Fairless, D. The little shrub that could—Maybe. Nature 2007, 449, 652–655. [Google Scholar] [CrossRef]
- Sanderson, K. Wonder weed plans fail to flourish. Nature 2009, 461, 328–329. [Google Scholar] [CrossRef]
- King, A.J.; He, W.; Cuevas, J.A.; Freundenberger, M.; Ramiaramanana, D.; Graham, I.A. Potential of Jatropha curcas as source of renewable oil and animal feed. J. Exp. Bot. 2009, 60, 2897–2905. [Google Scholar] [CrossRef]
- Sujatha, M. Genetic diversity, molecular markers and marker assisted breeding in Jatropha. In Jatropha, Challenges for a New Energy Crop, 1st ed.; Carels, N., Sujatha, M., Bahadur, B., Eds.; Springer: New York, NY, USA, 2012; Volume 2, pp. 395–422. [Google Scholar]
- Sun, Q.; Li, L.; Li, Y.; Wu, G.; Ge, X. SSR and AFLP markers reveal low genetic diversity in the biofuel plant Jatropha curcas in China. Crop Sci. 2008, 48, 1865–1971. [Google Scholar] [CrossRef]
- Rosado, T.B.; Laviola, B.G.; Faria, D.A.; Pappas, M.R.; Bhering, L.L.; Quirino, B.; Grattapaglia, D. Molecular markers reveal limited genetic diversity in a large germplasm collection of the biofuel crop Jatropha curcas L. in Brazil. Crop Sci. 2010, 50, 2372–2382. [Google Scholar] [CrossRef]
- Ambrosi, D.G.; Gala, G.; Purelli, M.; Barbi, T.; Fabbri, A.; Lucretti, S.; Sharbel, T.F.; Barccacia, G. DNA markers and FCSS analyses shed light on the genetic diversity and reproductive strategy of Jatropha cruces L. Diversity 2010, 2, 810–836. [Google Scholar] [CrossRef]
- Luo, C.W.; Li, K.; Chen, Y.; Sun, Y.Y. Floral display and breeding system of Jatropha curcas L. For. Stud. China 2007, 9, 114–119. [Google Scholar] [CrossRef]
- Luo, C.W.; Huang, Z.Y.; Chen, X.M.; Li, K.; Chen, Y.; Sun, Y.Y. Contribution of diurnal and nocturnal insects to the pollination of Jatropha curcas (Euphorbiaceae) in southwestern China. J. Econ. Entomol. 2011, 104, 149–154. [Google Scholar] [CrossRef]
- Bressan, E.A.; Sebbenn, A.M.; Ferreira, R.R.; Lee, T.S.G.; Figueira, A. Jatropha curcas L. (Euphorbiaceae) exhibits a mixed mating system, high correlated mating and apomixes. Tree Genet. Genomes 2013, 9, 1089–1097. [Google Scholar] [CrossRef]
- Devappa, R.K.; Makkar, H.P.S.; Becker, K. Jatropha toxicity—A Review. J. Toxicol. Environ. Health 2010, 13, 476–507. [Google Scholar] [CrossRef]
- King, A.J.; Montes, L.R.; Clarke, J.G.; Affleck, J.; Li, Y.; Witsenboer, H.; van der Vossen, E.; van der Linde, P.; Tripathi, Y.; Tavares, E.; et al. Linkage mapping in the oilseed crop jatropha curcas L. reveals a locus controlling the biosynthes is of phorbol esters which cause seed toxicity. Plant Biotechnol. J. 2013, 11, 986–996. [Google Scholar] [CrossRef]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar]
- Reif, J.C.; Melchinger, A.E.; Frisch, M. Genetical and Mathematical Properties of Similarity and Dissimilarity Coefficients Applied in Plant Breeding and Seed Bank Management. Crop Sci. 2005, 45, 1–7. [Google Scholar] [CrossRef]
- R Core Team. 2013: R: A language and environment for statistical computing. R Foundation for Statistical Computing: Vienna, Austria. Available online: http://www.R-project.org/ (accessed on 3 June 2014).
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; Henry, M.; Stevens, H.; et al. Vegan: Community Ecology Package. R package version 2.0–7. Available online: http://CRAN.R-project.org/package=vegan (accessed on 3 June 2014).
- Paradis, E. PEGAS: An R package for population genetics with an integrated-modular approach. Bioinformatics 2010, 26, 419–420. [Google Scholar] [CrossRef]
- Salvador-Figueroa, M.; Magana-Ramos, J.; Vazquez-Ovando, J.A.; Adriano-Anaya, M.L.; Ovando-Medina, I. Genetic diversity and structure of Jatropha curcas L. in its centre of origin. Plant Genet. Resour. 2014. [Google Scholar] [CrossRef]
- Wang, C.M.; Liu, P.; Yi, C.; Gu, K.; Sun, F.; Lei, L.; Lo, L.C.; Liu, X.; Feng, F.; Lin, G.; et al. A first generation microsatellite- and SNP-based linkage map of Jatropha. PLoS One 2011. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Montes, J.M.; Technow, F.; Martin, M.; Becker, K. Genetic Diversity in Jatropha curcas L. Assessed with SSR and SNP Markers. Diversity 2014, 6, 551-566. https://doi.org/10.3390/d6030551
Montes JM, Technow F, Martin M, Becker K. Genetic Diversity in Jatropha curcas L. Assessed with SSR and SNP Markers. Diversity. 2014; 6(3):551-566. https://doi.org/10.3390/d6030551
Chicago/Turabian StyleMontes, Juan M., Frank Technow, Matthias Martin, and Klaus Becker. 2014. "Genetic Diversity in Jatropha curcas L. Assessed with SSR and SNP Markers" Diversity 6, no. 3: 551-566. https://doi.org/10.3390/d6030551




