Symbiotic Microbes from Marine Invertebrates: Driving a New Era of Natural Product Drug Discovery
Abstract
:1. Introduction
2. β-glucosidase Inhibitors: Compounds with Anti-Diabetic Potential
3. Yondelis®, Smenamides, and Smenothiazoles: Compounds with Anti-Tumorigenic Potential
4. Manzamine, Kocurin, Norharman, Lipopeptides, and Lactones: Compounds with Anti-Pathogenic Potential
5. Constraints and Future Prospects
6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Oulhen, N.; Schulz, B.J.; Carrier, T.J. English translation of Heinrich Anton de Bary’s 1878 speech, “Die Erscheinung der Symbiose” (“De la symbiose”). Symbiosis 2016, 69, 131–139. [Google Scholar] [CrossRef]
- Staley, J.T.; Castenholz, R.W.; Colwell, R.R.; Holt, J.G.; Kane, M.D.; Pace, N.R.; Salyers, A.A.; Tiedje, J.M. The Microbial World: Foundation of the Biosphere; Colloquia Report; American Academy of Microbiology: Washington, DC, USA, 1997. [Google Scholar]
- Egerton, F.N. History of ecological sciences, part 52: Symbiosis studies. Bull. Ecol. Soc. Am. 2015, 96, 80–139. [Google Scholar] [CrossRef]
- Paracer, S.; Ahmadjian, V. Symbiosis: An introduction to biological associations; Oxford University Press: Oxford, UK, 2000; ISBN 0195118073. [Google Scholar]
- Zhang, X.; Wei, W.; Tan, R. Symbionts, a promising source of bioactive natural products. Sci. China Chem. 2015, 58, 1097–1109. [Google Scholar] [CrossRef]
- Henkel, T.; Brunne, R.M.; Müller, H.; Reichel, F. Statistical investigation into the structural complementarity of natural products and synthetic compounds. Angew. Chem. Int. Ed. 1999, 38, 643–647. [Google Scholar] [CrossRef]
- Montaser, R.; Luesch, H. Marine natural products: A new wave of drugs? Future Med. Chem. 2011, 3, 1475–1489. [Google Scholar] [CrossRef] [PubMed]
- Romano, G.; Costantini, M.; Sansone, C.; Lauritano, C.; Ruocco, N.; Ianora, A. Marine microorganisms as a promising and sustainable source of bioactive molecules. Mar. Environ. Res. 2017, 128, 58–69. [Google Scholar] [CrossRef] [PubMed]
- Harvey, A. Strategies for discovering drugs from previously unexplored natural products. Drug Discov. Today 2000, 5, 294–300. [Google Scholar] [CrossRef]
- Global Industry Analysts Inc. The Global Herbal Supplements and Remedies Market: Trends, Drivers and Projections, February 2015. Available online: www.strategyr.com/MarketResearch/Herbal_Supplements_and_Remedies_Market_Trends.asp (accessed on 17 August 2017).
- Smith, T.; Lynch, M.E.; Johnson, J.; Kawa, K.; Bauman, H.; Blumenthal, M. Herbal dietary supplement sales in US increase 6.8% in 2014. HerbalGram 2015, 107, 52–59. [Google Scholar]
- Gabay, M.; Smith, J.A.; Chavez, M.L.; Goldwire, M.; Walker, S.; Coon, S.A.; Gosser, R.; Hume, A.L.; Musselman, M.; Phillips, J.; et al. White paper on natural products. Pharmacotherapy 2017, 37, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Kong, D.X.; Jiang, Y.Y.; Zhang, H.Y. Marine natural products as sources of novel scaffolds: Achievement and concern. Drug Discov. Today 2010, 15, 884–886. [Google Scholar] [CrossRef] [PubMed]
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munro, M.H.G.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2017, 34, 235–294. [Google Scholar] [CrossRef] [PubMed]
- Bauer, A.; Brönstrup, M. Industrial natural product chemistry for drug discovery and development. Nat. Prod. Rep. 2014, 31, 35–60. [Google Scholar] [CrossRef] [PubMed]
- MarinLit: A Databse of the Marine Natural Products Literature. Available online: http://pubs.rsc.org/marinlit/ (accessed on 26 May 2017).
- Gerwick, W.H.; Moore, B.S. Lessons from the past and charting the future of marine natural products drug discovery and chemical biology. Chem. Biol. 2012, 19, 85–98. [Google Scholar] [CrossRef] [PubMed]
- National Ocean Service; National Oceanic and Atmospheric Administration; U.S. Department of Commerce. Ocean Facts: How Much of the Ocean Have We Explored? Available online: http://oceanservice.noaa.gov/facts/exploration.html (accessed on 26 May 2017).
- Bibi, F.; Faheem, M.; Azhar, E.I.; Yasir, M.; Alvi, S.A.; Kamal, M.A.; Ullah, I.; Nasser, M.I. Bacteria from marine sponges: A source of new drugs. Curr. Drug Metab. 2016, 17, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Andersen, R.J. Sponging off nature for new drug leads. Biochem. Pharmacol. 2017, 139, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Leal, M.C.; Calado, R.; Sheridan, C.; Alimonti, A.; Osinga, R. Coral aquaculture to support drug discovery. Trends Biotechnol. 2013, 31, 555–561. [Google Scholar] [CrossRef] [PubMed]
- Horton, T.; Kroh, A.; Bailly, N.; Boury-Esnault, N.; Brandão, S.N.; Costello, M.J.; Gofas, S.; Hernandez, F.; Mees, J.; Paulay, G.; et al. World Register of Marine Species. Available online: http://marinespecies.org/ (accessed on 25 May 2017).
- Thompson, J.R.; Rivera, H.E.; Closek, C.J.; Medina, M. Microbes in the coral holobiont: Partners through evolution, development, and ecological interactions. Front. Cell. Infect. Microbiol. 2015, 4, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Peixoto, R.S.; Rosado, P.M.; de Assis Leite, D.C.; Rosado, A.S.; Bourne, D.G. Beneficial microorganisms for corals (BMC): Proposed mechanisms for coral health and resilience. Front. Microbiol. 2017, 8, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Bourne, D.G.; Morrow, K.M.; Webster, N.S. Insights into the coral microbiome: Underpinning the health and resilience of reef ecosystems. Annu. Rev. Microbiol. 2016, 70, 317–340. [Google Scholar] [CrossRef] [PubMed]
- Webster, N.S.; Thomas, T. Defining the sponge hologenome. mBio. 2016, 7, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Fuerst, J.A. Diversity and biotechnological potential of microorganisms associated with marine sponges. Appl. Microbiol. Biotechnol. 2014, 98, 7331–7347. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.W.; Radax, R.; Steger, D.; Wagner, M. Sponge-associated microorganisms: Evolution, ecology, and biotechnological potential. Microbiol. Mol. Biol. Rev. 2007, 71, 295–347. [Google Scholar] [CrossRef] [PubMed]
- Margulis, L. Symbiogenesis and symbionticism. In Symbiosis as a Source of Evolutionary Innovation: Speciation and Morphogenesis; Margulis, L., Fester, R., Eds.; MIT Press: Cambridge, MA, USA, 2001; pp. 1–14. ISBN 9780262519908. [Google Scholar]
- Reveillaud, J.; Maignien, L.; Eren, M.A.; Huber, J.A.; Apprill, A.; Sogin, M.L.; Vanreusel, A. Host-specificity among abundant and rare taxa in the sponge microbiome. ISME J. 2014, 8, 1198–1209. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, S.; Tsai, P.; Bell, J.; Fromont, J.; Ilan, M.; Lindquist, N.; Perez, T.; Rodrigo, A.; Schupp, P.J.; Vacelet, J.; et al. Assessing the complex sponge microbiota: Core, variable and species-specific bacterial communities in marine sponges. ISME J. 2012, 6, 564–576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.S.; Kwon, K.K.; Kang, S.G.; Cha, S.S.; Kim, S.J.; Lee, J.H. Approaches for novel enzyme discovery from marine environments. Curr. Opin. Biotechnol. 2010, 21, 353–357. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J. Predominately uncultured microbes as sources of bioactive agents. Front. Microbiol. 2016, 7, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Macintyre, L.; Zhang, T.; Viegelmann, C.; Martinez, I.J.; Cheng, C.; Dowdells, C.; Abdelmohsen, U.R.; Gernert, C.; Hentschel, U.; Edrada-Ebel, R.A. Metabolomic tools for secondary metabolite discovery from marine microbial symbionts. Mar. Drugs 2014, 12, 3416–3448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Egan, S.; Thomas, T.; Kjelleberg, S. Unlocking the diversity and biotechnological potential of marine surface associated microbial communities. Curr. Opin. Microbiol. 2008, 11, 219–225. [Google Scholar] [CrossRef] [PubMed]
- Abdelmohsen, U.R.; Bayer, K.; Hentschel, U. Diversity, abundance and natural products of marine sponge-associated actinomycetes. Nat. Prod. Rep. 2014, 31, 381. [Google Scholar] [CrossRef] [PubMed]
- Fenical, W. Marine pharmaceuticals. Oceanography 2006, 19, 110–119. [Google Scholar] [CrossRef]
- Bull, A.T.; Stach, J.E.M. Marine actinobacteria: New opportunities for natural product search and discovery. Trends Microbiol. 2007, 15, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Wang, G. Diversity and biotechnological potential of the sponge-associated microbial consortia. J. Ind. Microbiol. Biotechnol. 2006, 33, 545–551. [Google Scholar] [CrossRef] [PubMed]
- Barde, S.R.; Sakhare, R.S.; Kanthale, S.B.; Chandak, P.G.; Jamkhande, P.G. Marine bioactive agents: A short review on new marine antidiabetic compounds. Asian Pacific J. Trop. Dis. 2015, 5, 209–213. [Google Scholar] [CrossRef]
- World Health Organisation. The Top 10 Causes of Death. Available online: http://www.who.int/mediacentre/factsheets/fs310/en/ (accessed on 22 August 2017).
- Lauritano, C.; Ianora, A. Marine organisms with anti-diabetes properties. Mar. Drugs 2016, 14, 220. [Google Scholar] [CrossRef] [PubMed]
- Pandey, S.; Sree, A.; Dash, S.S.; Sethi, D.P.; Chowdhury, L. Diversity of marine bacteria producing beta-glucosidase inhibitors. Microb. Cell. Fact. 2013, 12, 35. [Google Scholar] [CrossRef] [PubMed]
- Pandey, S.; Sree, A.; Dash, S.S.; Sethi, D.P. A novel method for screening beta-glucosidase inhibitors. BMC Microbiol. 2013, 13, 55. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, D.D.; Frommer, W.; Müller, L.; Truscheit, E. Glucosidase inhibitoren aus Bazillen. Naturwissenschaften 1979, 66, 584–585. [Google Scholar] [CrossRef] [PubMed]
- Asano, N. Glycosidase inhibitors: Update and perspectives on practical use. Glycobiology 2003, 13, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Niwa, T.; Inouye, S.; Tsuruoka, T.; Koaze, Y.; Niida, T. “Nojirimycin” as a potent inhibitor of glucosidase. Agric. Biol. Chem. 1970, 34, 966–968. [Google Scholar] [CrossRef]
- Rath, C.M.; Janto, B.; Earl, J.; Ahmed, A.; Hu, F.Z.; Hiller, L.; Dahlgren, M.; Kreft, R.; Yu, F.; Wolff, J.J.; et al. Meta-omic characterization of the marine invertebrate microbial consortium that produces the chemotherapeutic natural product ET-743. ACS Chem. Biol. 2011, 6, 1244–1256. [Google Scholar] [CrossRef] [PubMed]
- Esposito, G.; Teta, R.; Miceli, R.; Ceccarelli, L.S.; Della Sala, G.; Camerlingo, R.; Irollo, E.; Mangoni, A.; Pirozzi, G.; Costantino, V. Isolation and assessment of the in vitro anti-tumor activity of Smenothiazole A and B, chlorinated thiazole-containing peptide/polyketides from the Caribbean sponge, Smenospongia aurea. Mar. Drugs 2015, 13, 444–459. [Google Scholar] [CrossRef] [PubMed]
- Teta, R.; Irollo, E.; Della Sala, G.; Pirozzi, G.; Mangoni, A.; Costantino, V. Smenamides A and B, chlorinated peptide/polyketide hybrids containing a dolapyrrolidinone unit from the Caribbean sponge Smenospongia aurea. Evaluation of their role as leads in antitumor drug research. Mar. Drugs 2013, 11, 4451–4463. [Google Scholar] [CrossRef] [PubMed]
- Rao, K.V.; Santarsiero, B.D.; Mesecar, A.D.; Schinazi, R.F.; Tekwani, B.L.; Hamann, M.T. New Manzamine alkaloids with activity against infectious and tropical parasitic diseases from an indonesian sponge. J. Nat. Prod. 2003, 66, 823–828. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, J.; Tsuda, M. Structures and biogenesis of Manzamines and related alkaloids. Heterocycles 1997, 46, 765–794. [Google Scholar] [CrossRef]
- Magnier, E.; Langlois, Y. Manzamine alkaloids, syntheses and synthetic approaches. Tetrahedron 1998, 54, 6201–6258. [Google Scholar] [CrossRef]
- Yousaf, M.; El Sayed, K.A.; Rao, K.V.; Lim, C.W.; Hu, J.F.; Kelly, M.; Franzblau, S.G.; Zhang, F.; Peraud, O.; Hill, R.T.; et al. 12,34-Oxamanzamines, novel biocatalytic and natural products from manzamine producing Indo-Pacific sponges. Tetrahedron 2002, 58, 7397–7402. [Google Scholar] [CrossRef]
- Palomo, S.; González, I.; De La Cruz, M.; Martín, J.; Tormo, J.R.; Anderson, M.; Hill, R.T.; Vicente, F.; Reyes, F.; Genilloud, O. Sponge-derived Kocuria and Micrococcus spp. as sources of the new thiazolyl peptide antibiotic Kocurin. Mar. Drugs 2013, 11, 1071–1086. [Google Scholar] [CrossRef] [PubMed]
- Montalvo, N.F.; Mohamed, N.M.; Enticknap, J.J.; Hill, R.T. Novel actinobacteria from marine sponges. Antonie Van Leeuwenhoek 2005, 87, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Chen, H.; Han, X.; Lin, W.; Yan, X. Antimicrobial screening and active compound isolation from marine bacterium NJ6-3-1 associated with the sponge Hymeniacidon perleve. World J. Microbiol. Biotechnol. 2005, 21, 201–206. [Google Scholar] [CrossRef]
- Pabel, C.T.; Vater, J.; Wilde, C.; Franke, P.; Hofemeister, J.; Adler, B.; Bringmann, G.; Hacker, J.; Hentschel, U. Antimicrobial activities and matrix-assisted laser desorption/ionization mass spectrometry of Bacillus isolates from the marine sponge Aplysina aerophoba. Mar. Biotechnol. 2003, 5, 424–434. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.A.; Shao, C.L.; Gu, Y.C.; Blum, M.; Gan, L.S.; Wang, K.L.; Chen, M.; Wang, C.Y. Antifouling and fungicidal resorcylic acid lactones from the sea anemone-derived fungus Cochliobolus lunatus. J. Agric. Food Chem. 2014, 62, 3183–3191. [Google Scholar] [CrossRef] [PubMed]
- ChemSpider ID 58738. Nojirimycin. Available online: http://www.chemspider.com/Chemical-Structure.58738.html?rid=8ee6c8fc-333c-4358-896f-d569bf9c95a9&page_num=0 (accessed on 17 October 2017).
- ChemSpider ID 97236. Trabectedin. Available online: http://www.chemspider.com/Chemical-Structure.97236.html?rid=41381930-9347-4a57-a7e8-32fbbe0a8256 (accessed on 17 October 2017).
- Schöffski, P.; Dumez, H.; Wolter, P.; Stefan, C.; Wozniak, A.; Jimeno, J.; Van Oosterom, A.T. Clinical impact of trabectedin (ecteinascidin-743) in advanced/metastatic soft tissue sarcoma. Expert Opin. Pharmacother. 2008, 9, 1609–1618. [Google Scholar] [CrossRef] [PubMed]
- Janssen Products Ltd. Yondelis®. Available online: https://www.yondelis.com/ (accessed on 21 August 2017).
- ChemSpider ID 35516725. Smenothiazole A. Available online: http://www.chemspider.com/Chemical-Structure.35516725.html?rid=5c041417-c159-4802-bfad-31e03bd22214 (accessed on 17 October 2017).
- ChemSpider ID 35516726. Smenothiazole B. Available online: http://www.chemspider.com/Chemical-Structure.35516726.html?rid=b3eaff42-e7ed-4f12-9568-64fa222c485c (accessed on 17 October 2017).
- Sakai, R.; Higa, T.; Jefford, C.W.; Bernardinelli, G. Manzamine A, a novel antitumor alkaloid from a sponge. J. Am. Chem. Soc. 1986, 108, 6404–6405. [Google Scholar] [CrossRef]
- ChemSpider ID 5006904. Manzamine A. Available online: http://www.chemspider.com/Chemical-Structure.5006904.html?rid=bc7b7a2e-93b4-4408-a08f-0d78ec044124 (accessed on 17 October 2017).
- ChemSpider ID 29215512. Kocurin. Available online: http://www.chemspider.com/Chemical-Structure.29215512.html?rid=29adbdad-a08f-4d47-a6de-0a9c86532abd&page_num=0 (accessed on 17 October 2017).
- ChemSpider ID 58486. Norharman. Available online: http://www.chemspider.com/Chemical-Structure.58486.html?rid=2d9b1de5-41ec-43d4-9220-f2fad643b678 (accessed on 17 October 2017).
- ChemSpider ID 24672437. Zeaenol. Available online: http://www.chemspider.com/Chemical-Structure.24672437.html?rid=822b22bb-71c1-4292-afe1-85c89ddc4e3b&page_num=0 (accessed on 17 October 2017).
- Munro, M.H.G.; Blunt, J.W.; Dumdei, E.J.; Hickford, S.J.H.; Lill, R.E.; Li, S.; Battershill, C.N.; Duckworth, A.R. The discovery and development of marine compounds with pharmaceutical potential. Prog. Ind. Microbiol. 1999, 35, 15–25. [Google Scholar] [CrossRef]
- Kijjoa, A.; Sawangwong, P. Drugs and cosmetics from the sea. Mar. Drugs 2004, 2, 73–82. [Google Scholar] [CrossRef]
- Sagar, S.; Kaur, M.; Minneman, K.P. Antiviral lead compounds from marine sponges. Mar. Drugs 2010, 8, 2619–2638. [Google Scholar] [CrossRef] [PubMed]
- Bergmann, W.; Feeney, R.J. The isolation of a new thymine pentoside from sponges. J. Am. Chem. Soc. 1950, 72, 2809–2810. [Google Scholar] [CrossRef]
- Essai Inc. Halaven®. Available online: http://www.halaven.com/metastatic-breast-cancer (accessed on 21 August 2017).
- Hirata, Y.; Ljemura, D. Halichondrins-antitumor polyether Macrolides from a marine sponge. Pure Appl. Chem. 1986, 58, 701–710. [Google Scholar] [CrossRef]
- Menis, J.; Twelves, C. Eribulin (Halaven): A new, effective treatment for women with heavily pretreated metastatic breast cancer. Breast Cancer Targets Ther. 2011, 3, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Seattle Genetics Inc. ADCETRIS®. Available online: http://www.seattlegenetics.com/products/adcetris-us (accessed on 21 August 2017).
- Pettit, G.R.; Kamano, Y.; Herald, C.L.; Tuinman, A.A.; Boettner, F.E.; Kizu, H.; Schmidt, J.M.; Baczynskyj, L.; Tomer, K.B.; Bontems, R.J. The isolation and structure of a remarkable marine animal antineoplastic constituent: Dolastatin 10. J. Am. Chem. Soc. 1987, 109, 6883–6885. [Google Scholar] [CrossRef]
- Katz, J.; Janik, J.E.; Younes, A. Brentuximab Vedotin (SGN-35). Clin. Cancer Res. 2011, 17, 6428–6436. [Google Scholar] [CrossRef] [PubMed]
- Rinehart, K.L. Antitumor compounds from tunicates. Med. Res. Rev. 2000, 20, 1–27. [Google Scholar] [CrossRef]
- Corey, E.J.; Gin, D.Y.; Kania, R.S. Enantioselective total synthesis of Ecteinascidin 743. J. Am. Chem. Soc. 1996, 118, 9202–9203. [Google Scholar] [CrossRef]
- Moss, C.; Green, D.H.; Perez, B.; Velasco, A.; Henriquez, R.; McKenzie, J.D. Intracellular bacteria associated with the ascidian Ecteinascidia turbinata: Phylogenetic and in situ hybridisation analysis. Mar. Biol. 2003, 143, 99–110. [Google Scholar] [CrossRef]
- Pérez-Matos, A.E.; Rosado, W.; Govind, N.S. Bacterial diversity associated with the Caribbean tunicate Ecteinascidia turbinata. Antonie van Leeuwenhoek 2007, 92, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Penesyan, A.; Kjelleberg, S.; Egan, S. Development of novel drugs from marine surface associated microorganisms. Mar. Drugs 2010, 8, 438–459. [Google Scholar] [CrossRef] [PubMed]
- Bhatnagar, I.; Kim, S.K. Immense essence of excellence: Marine microbial bioactive compounds. Mar. Drugs 2010, 8, 2673–2701. [Google Scholar] [CrossRef] [PubMed]
- Kumar Jha, R.; Zi-Rong, X. Biomedical compounds from marine organisms. Mar. Drugs 2004, 2, 123–146. [Google Scholar] [CrossRef]
- Edrada, R.A.; Proksch, P.; Wray, V.; Witte, L.; Müller, W.E.G.; van Soest, R.W.M. Four new bioactive Manzamine-type alkaloids from the Philippine marine sponge Xestospongia ashmorica. J. Nat. Prod. 1996, 59, 1056–1060. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, H.; Deng, S.; Kobayashi, J.; Ohizumi, Y.; Tomotake, Y.; Matsuzaki, T.; Hirata, Y. Keramamine-A and -B, novel antimicrobial alkaloids from the Okinawan marine sponge Pellina sp. Tetrahedron Lett. 1987, 28, 621–624. [Google Scholar] [CrossRef]
- Ang, K.K.H.; Holmes, M.J.; Higa, T.; Hamann, M.T.; Kara, U.A.K. In vivo antimalarial activity of the β-carboline alkaloid Manzamine A. Antimicrob. Agents Chemother. 2000, 44, 1645–1649. [Google Scholar] [CrossRef] [PubMed]
- El Sayed, K.A.; Kelly, M.; Kara, U.A.K.; Ang, K.K.H.; Katsuyama, I.; Dunbar, D.C.; Khan, A.A.; Hamann, M.T. New manzamine alkaloids with potent activity against infectious diseases. J. Am. Chem. Soc. 2001, 123, 1804–1808. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Global TB Report 2016 Facts. Available online: http://www.who.int/tb/publications/factsheet_global.pdf?ua=1 (accessed on 22 August 2017).
- Rojas, J.L.; Martín, J.; Tormo, J.R.; Vicente, F.; Brunati, M.; Ciciliato, I.; Losi, D.; Van Trappen, S.; Mergaert, J.; Swings, J.; et al. Bacterial diversity from benthic mats of Antarctic lakes as a source of new bioactive metabolites. Mar. Genomics 2009, 2, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Sharma, G.M.; Burkholder, P.R. Studies on antimicrobial substances of sponges. Isolation, purification, and properties of a new bromine-containing antibacterial substance. J. Antibiot. (Tokyo) 1967, 20, 200–203. [Google Scholar] [PubMed]
- Zhang, H.; Zhao, Z.; Wang, H. Cytotoxic natural products from marine sponge-derived microorganisms. Mar. Drugs 2017, 15, 68. [Google Scholar] [CrossRef] [PubMed]
- Abad, M.J.; Bedoya, L.M.; Bermejo, P. Marine compounds and their antimicrobial activities. In Science against Microbial Pathogens: Communicating Current Research and Technological Advances; Mendez-Vilas, A., Ed.; Formatex Research Center: Badajoz, Spain, 2011; pp. 1293–1306. ISBN 978-84-939843-2-8. [Google Scholar]
- Indraningrat, A.A.G.; Smidt, H.; Sipkema, D. Bioprospecting sponge-associated microbes for antimicrobial compounds. Mar. Drugs 2016, 14, 87. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.; Canal, S. Review of natural products from marine organisms in the Red Sea. Int. J. Pharm. Sci. Res. 2017, 8, 940–974. [Google Scholar] [CrossRef]
- Gomes, N.G.M.; Dasari, R.; Chandra, S.; Kiss, R.; Kornienko, A. Marine invertebrate metabolites with anticancer activities: Solutions to the “supply problem”. Mar. Drugs 2016, 14. [Google Scholar] [CrossRef] [PubMed]
- Daletos, G.; Ancheeva, E.; Chaidir, C.; Kalscheuer, R.; Proksch, P. Antimycobacterial metabolites from marine invertebrates. Arch. Pharm. (Weinheim) 2016, 349, 763–773. [Google Scholar] [CrossRef] [PubMed]
- Cheung, R.C.F.; Ng, T.B.; Wong, J.H.; Chen, Y.; Chan, W.Y. Marine natural products with anti-inflammatory activity. Appl. Microbiol. Biotechnol. 2016, 100, 1645–1666. [Google Scholar] [CrossRef] [PubMed]
- Pimentel-Elardo, S.M.; Kozytska, S.; Bugni, T.S.; Ireland, C.M.; Moll, H.; Hentschel, U. Anti-parasitic compounds from Streptomyces sp. strains isolated from Mediterranean sponges. Mar. Drugs 2010, 8, 373–380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia, M.; Monzote, L. Marine products with anti-protozoal activity: A review. Curr. Clin. Pharmacol. 2014, 9, 258–270. [Google Scholar] [CrossRef] [PubMed]
- Watts, K.R.; Tenney, K.; Crews, P. The structural diversity and promise of antiparasitic marine invertebrate-derived small molecules. Curr. Opin. Biotechnol. 2011, 21, 808–818. [Google Scholar] [CrossRef] [PubMed]
- Tullius Scotti, M.; Scotti, L.; Ishiki, H.; Fávaro Ribeiro, F.; Marques Duarte da Cruz, R.; Pedrosa de Oliveira, M.; Jaime Bezerra Mendonça, F. Natural products as a source for antileishmanial and antitrypanosomal agents. Comb. Chem. High. Throughput Screen. 2016, 19, 537–553. [Google Scholar] [CrossRef]
- Brinkmann, C.; Marker, A.; Kurtböke, D. An overview on marine sponge-symbiotic bacteria as unexhausted sources for natural product discovery. Diversity 2017, 9, 40. [Google Scholar] [CrossRef]
- Epstein, S. The phenomenon of microbial uncultivability. Curr. Opin. Microbiol. 2013, 16, 636–642. [Google Scholar] [CrossRef] [PubMed]
- Leal, M.C.; Calado, R. Marine natural products: Biodiscovery, biodiversity, and bioproduction. In Bioactive Natural Products: Chemistry and Biology; Brahmachari, G., Ed.; Wiley: Hoboken, NJ, USA, 2015; pp. 473–490. ISBN 9783527684403. [Google Scholar]
- Leal, M.; Sheridan, C.; Osinga, R.; Dionísio, G.; Rocha, R.; Silva, B.; Rosa, R.; Calado, R. Marine microorganism-invertebrate assemblages: Perspectives to solve the “supply problem” in the initial steps of drug discovery. Mar. Drugs 2014, 12, 3929–3952. [Google Scholar] [CrossRef] [PubMed]
- Sweet, M.J.; Bulling, M.T. On the importance of the microbiome and pathobiome in coral health and disease. Front. Mar. Sci. 2017, 4, 9. [Google Scholar] [CrossRef]
- Sweet, M.J.; Smith, D.; Bythell, J.C.; Craggs, J. Changes in microbial diversity associated with two coral species recovering from a stressed state in a public aquarium system. J. Zoo Aquarium Res. 2013, 1, 52–60. [Google Scholar]
- MaCuMBA Group. MaCuMBA Project Legacy Brochure. Available online: http://www.macumbaproject.eu/images/MACUMBA/Media/Public_Deliverables/Legacy_Brochure_FINAL_HR.pdf (accessed on 23 August 2017).
- Trindade, M.; van Zyl, L.J.; Navarro-Fernández, J.; Elrazak, A.A. Targeted metagenomics as a tool to tap into marine natural product diversity for the discovery and production of drug candidates. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
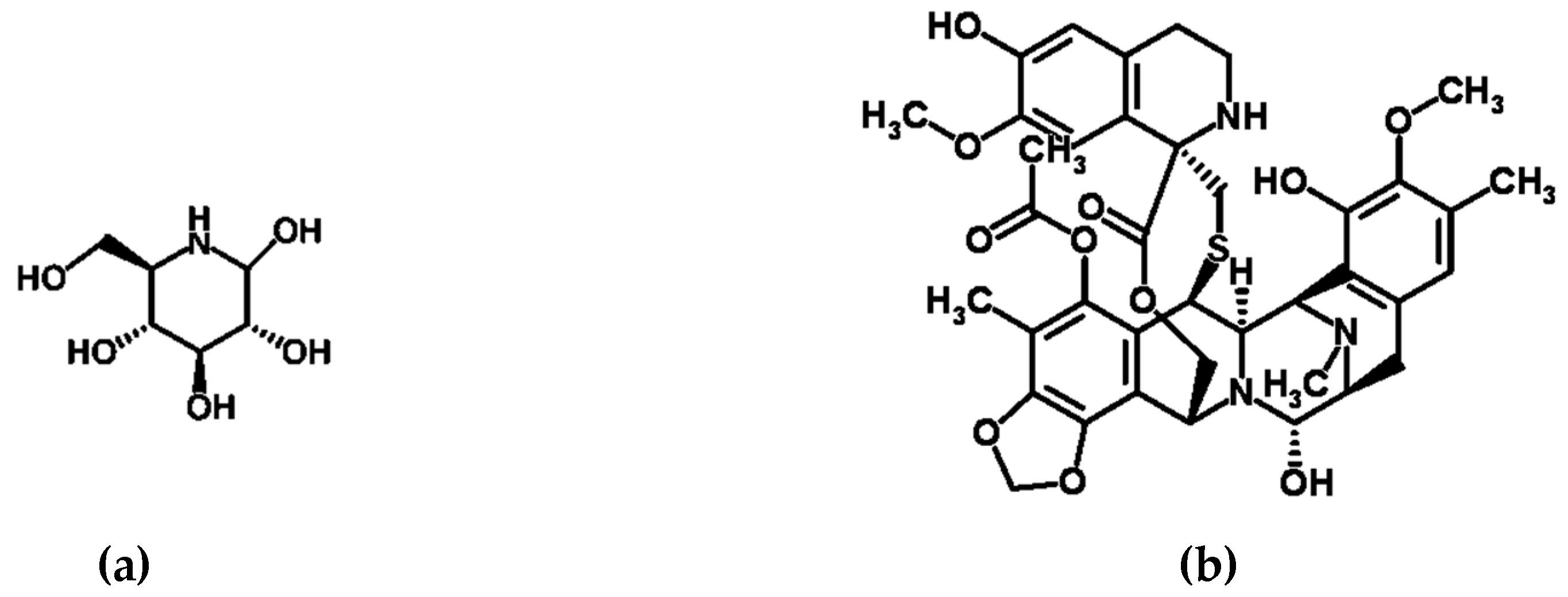
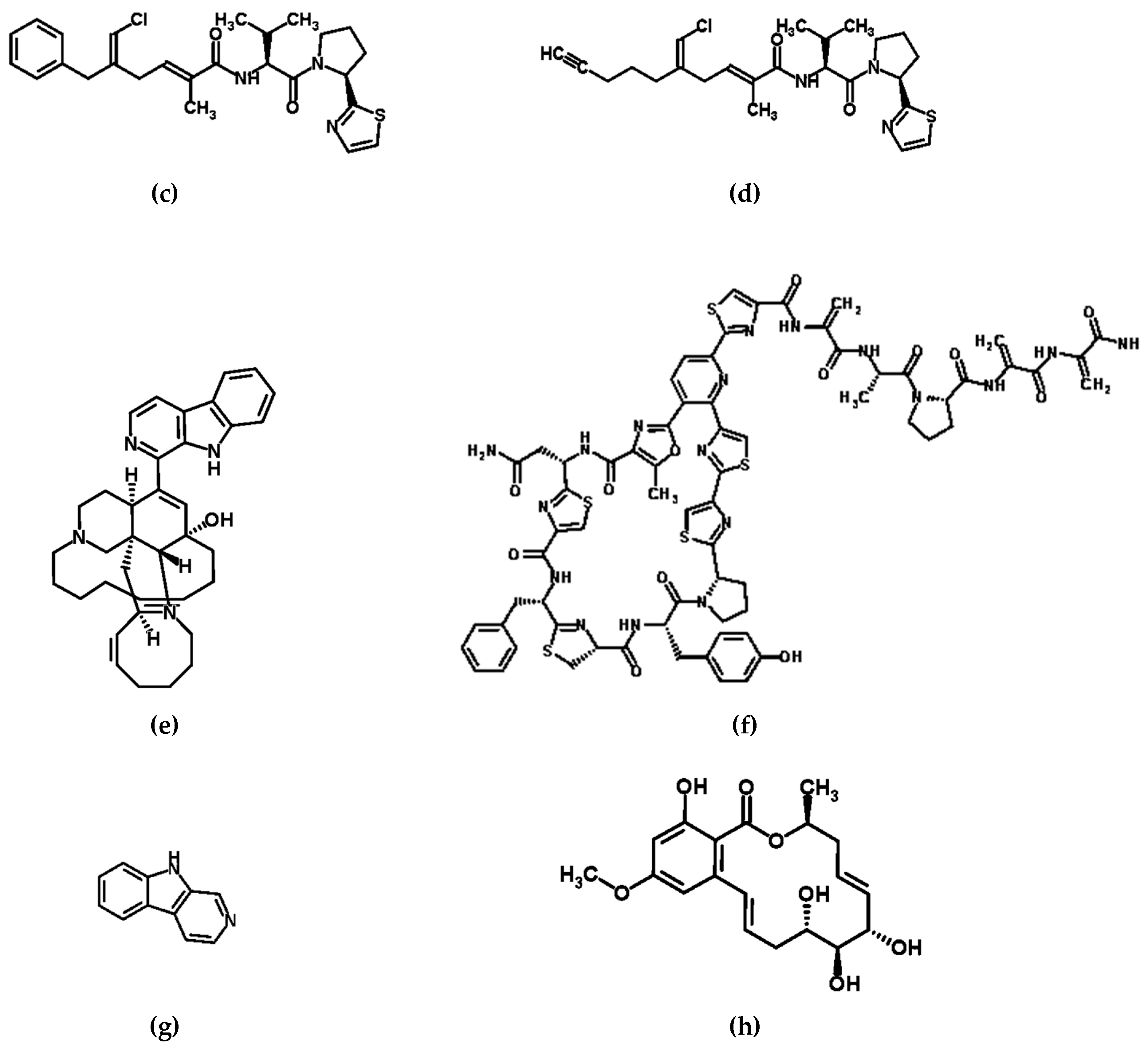
| Microbial Species | Host | Location | Compound/s | Compound Activity |
|---|---|---|---|---|
| Halomonas sulfidaeris and Bacillus tequilensis | Sponge Sarcotragus fasciculatus | Bay of Bengal | β-glucosidase inhibitors | Anti-diabetic potential [43] |
| Bacillus sp., Leucobacter chromiiresistens, Planococcus rifitoensis, Bacillus stratosphericus, Bacillus amyloliquefaciens subsp. amyloliquefaciens, and Streptomyces rangoonensis | Sponge Aka coralliphaga | Bay of Bengal | β-glucosidase inhibitors | Anti-diabetic potential [43] |
| Candidatus Endoecteinascidia frumentensis | Tunicate Ecteinascidia turbinata | Carribean Sea | Yondelis® (ecteinascidin 743/ trabectedin) | Anti-tumerogenic drug [48] |
| Synechococcus spongiarum | Sponge Smenospongia aurea | Bahamas | Smenamide A/B and smenothiazoles A/B | Anti-tumerogenic activity [49,50] |
| Unknown: Various species suspected including a-proteobacteria and Actinomycetes | Sponge Genera Petrosiidae, Ircinia, Amphimedon, Haliclona, Xestospongia, Pachypellina, and Prianos | Global | Manzamine-type alkaloids | Antitumor, antibacterial, cytotoxic, and immunestimulatory activities [51,52,53,54] |
| Bacterial genera Micrococccus and Kocuria | Sponge genera Xestospongia | Florida Keys | Kocurin | Thiazolyl peptide antibiotic [55,56] |
| Pseudoalteromonas piscicida | Sponge Hymeniacidon perleve | Eastern China Sea | Norharman (β-carboline alkaloid) | Anti-microbial [57] |
| Bacillus subtilis (Strain A184) | Sponge Aplysina aerophoba | Mediterran | Lipoproteins | Anti-microbial [58] |
| Cochliobolus lunatus (Strain TA26- 46) | Sea anemone Palythoa haddoni | South China Sea | Resorcylic acid lactones | Anti-fungal [59] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Blockley, A.; Elliott, D.R.; Roberts, A.P.; Sweet, M. Symbiotic Microbes from Marine Invertebrates: Driving a New Era of Natural Product Drug Discovery. Diversity 2017, 9, 49. https://doi.org/10.3390/d9040049
Blockley A, Elliott DR, Roberts AP, Sweet M. Symbiotic Microbes from Marine Invertebrates: Driving a New Era of Natural Product Drug Discovery. Diversity. 2017; 9(4):49. https://doi.org/10.3390/d9040049
Chicago/Turabian StyleBlockley, Alix, David R. Elliott, Adam P. Roberts, and Michael Sweet. 2017. "Symbiotic Microbes from Marine Invertebrates: Driving a New Era of Natural Product Drug Discovery" Diversity 9, no. 4: 49. https://doi.org/10.3390/d9040049





