Automated Signal Processing Applied to Volatile-Based Inspection of Greenhouse Crops
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Dataset
2.2. The Experimental Equipment and the Instrumental Settings
2.3. Manual Processing of Data
2.4. Automated Processing of Data
3. Results
3.1. Data pre-processing with MetAlign
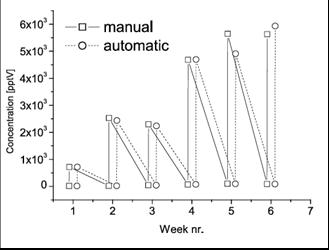
3.2. Manual Processing Versus Automated Processing: Detection and Concentration of VOCs
3.4. Manual vs. Automated Processing: Time Needed for Analysis
4. Discussion
5. Conclusions
Acknowledgments
References
- Huang, R; Jarvis, WR. Greenhouse Crop Losses (diseases). In Encyclopedia of pest management; Pimentel, D, Ed.; Marcel Dekker, Inc: New York, NY, USA, 2002; Volume 1, pp. 348–350. [Google Scholar]
- Jacobson, RJ. Integrated Pest Management (IPM) in Glasshouses. In Thrips as Crop Pests; Lewis, T, Ed.; CAB International: Wallingford, UK, 1997; pp. 639–666. [Google Scholar]
- Jansen, RMC; Miebach, M; Kleist, E; van Henten, EJ; Wildt, J. Release of lipoxygenase products and monoterpenes by tomato plants as an indicator of Botrytis cinerea-induced stress. Plant Biol 2009, 11, 859–868. [Google Scholar]
- Toome, M; Randjärv, P; Copolovici, L; Niinemets, Ü; Heinsoo, K; Luik, A; Noe, SM. Leaf rust induced volatile organic compounds signalling in willow during the infection. Planta 2010, 232, 235–243. [Google Scholar]
- Wei, J; Wang, L; Zhu, J; Zhang, S; Nandi, O; Kang, L. Plants attract parasitic wasps to defend themselves against insect pests by releasing hexenol. PLOS one 2007, 2, e852. [Google Scholar]
- Miresmailli, S; Gries, R; Gries, G; Zamar, RZ; Isman, MB. Herbivore-induced plant volatiles allow detection of Trichoplusia ni (lepidoptera: Noctuidae) infestation on greenhouse tomato plants. Pest Manag. Sci 2010, 66, 916–924. [Google Scholar]
- Schütz, S; Weissbecker, B; Koch, U; Hummel, HE. Erfassung von Pflanzenschäden durch die messung von verletzungsbedingt freigesetzten volatilen verbindugne mittels eines mobilen EAG-Geräts. Mitt. Dtsch. Ges. Allg. Angew. Entomol 1995, 10, 231–236. [Google Scholar]
- Jansen, RMC; Hofstee, JW; Wildt, J; Verstappen, FWA; Bouwmeester, HJ; Posthumus, MA; van Henten, EJ. Health monitoring of plants by their emitted volatiles: trichome damage and cell membrane damage are detectable at greenhouse scale. Ann. Appl. Biol 2009, 154, 441–452. [Google Scholar]
- Santos, FJ; Galceran, MT. The application of gas chromatography to environmental analysis. TrAC 2002, 21, 672–682. [Google Scholar]
- Matz, G; Loogk, M; Lennemann, F. On-line gas chromatography-mass spectrometry for process monitoring using solvent-free sample preparation. J. Chromatogr 1998, 819, 51–60. [Google Scholar]
- Smith, PA; Sng, MT; Eckenrode, BA; Leow, SY; Koch, D; Erickson, RP; Lepage, CRJ; Hook, GL. Towards smaller and faster gas chromatography-mass spectrometry systems for field chemical detection. J. Chromatogr 2005, 1067, 285–294. [Google Scholar]
- Eckenrode, BA. Environmental and forensic applications of field-portable GC-MS: An overview. J. Am. Soc. Mass Spectrom 2001, 12, 683–693. [Google Scholar]
- Smith, JN; Keil, A; Likens, J; Noll, RJ; Cooks, RG. Facility monitoring of toxic industrial compounds in air using an automated, fieldable, miniature mass spectrometer. Analyst 2010, 135, 994–1003. [Google Scholar]
- Malmquist, LMV; Olson, RR; Hansen, AB; Anderson, E; Christensen, JH. Assessment of oil weathering by gas chromatography-mass spectrometry, time warping and principal component analysis. J. Chromatogr 2007, 1164, 262–270. [Google Scholar]
- Katajamaa, M; Orešič, M. Data processing for mass spectrometry-based metabolomics. J. Chromatogr 2007, 1158, 318–328. [Google Scholar]
- Li, XN; Liang, YZ; Chau, FT. Smoothing methods applied to dealing with heteroscedastic noise in GC/MS. Chemometrics Intellig. Lab. Syst 2002, 63, 139–153. [Google Scholar]
- Skov, T; van den Berg, F; Tomasi, G; Bro, R. Automated alignment of chromatographic data. J. Chemometrics 2007, 20, 484–497. [Google Scholar]
- Daviss, B. Growing pains for metabolomics. The Scientist 2005, 19, 25–28. [Google Scholar]
- Jonsson, P; Gullberg, J; Nordström, A; Kusano, M; Kowalczyk, M. A strategy for identifying differences in large series of metabolomic samples analyzed by GC/MS. Anal. Chem 2004, 76, 1738–1745. [Google Scholar]
- McMaster, MC. GC/MS, A Practical User's Guide, 2nd ed; John Wiley & Sons, Inc: Hoboken, NJ, USA, 2008; p. 180. [Google Scholar]
- Lommen, A. MetAlign: An interface-driven, versatile metabolomics tool for hyphenated full-scan MS data pre-processing. Anal. Chem 2009, 81, 3079–3086. [Google Scholar]
- Jansen, RMC; Hofstee, JW; Verstappen, FWA; Bouwmeester, HJ; van Henten, EJ. A method to detect baseline emission and plant damage induced volatile emission in a greenhouse. Acta Horticulturae 2008, 108, 1415–1422. [Google Scholar]
- Lommen, A; van der Weg, G; van Engelen, MC; Bor, G; Hoogenboom, LAP; Nielen, MWF. An untargeted metabolomics approach to contaminant analysis: Pinpointing potential unknown compounds. Anal. Chim. Acta 2007, 584, 43–49. [Google Scholar]
- Tikunov, Y; Lommen, A; de Vos, CHR; Verhoeven, HA; Bino, RJ; Hall, RD. A novel approach for nontargeted data analysis for metabolomics. large-scale profiling of tomato fruit volatiles. Plant Physiol 2005, 139, 1125–1137. [Google Scholar]
- Vos, RC; de Moco, S; Lommen, A; Keurentjes, JJ; Bino, RJ; Hall, RD. Untargeted large-scale plant metabolomics using liquid chromatography coupled to mass spectrometry. Nat. Protoc 2007, 2, 778–791. [Google Scholar]
- Eilers, PHC. Parametric time warping. Anal. Chem 2004, 76, 404–411. [Google Scholar]
- Christensen, JH; Mortensen, J; Hansen, AB; Andersen, O. Chromatographic preprocessing of GC-MS data for analysis of complex chemical mixtures. J. Chromatogr 2005, 1062, 113–123. [Google Scholar]
- Nielsen, NPV; Carstensen, JM; Smedsgaard, J. Aligning of single and multiple wavelength chromatographic profiles for chemomentric data analysis using correlation optimised warping. J. Chromatogr 1998, 805, 17–35. [Google Scholar]
- Tomasi, G; van den Berg, F; Andersson, C. Correlation optimized warping and dynamic time warping as preprocessing methods for chromatographic data. J. Chemometrics 2004, 18, 231–241. [Google Scholar]
- Lin, SM; Haney, RP; Campa, MJ; Fitzgerald, MC; Patz, EF. Characterising phase variations in MALDI-TOF data and correcting them by peak alignment. Cancer Inform 2005, 1, 32–40. [Google Scholar]
- Varns, JL; Glynn, MT. Detection of disease in stored potatoes by volatile monitoring. Am. Potato J 1979, 56, 185–197. [Google Scholar]
- Waterer, DR; Pritchard, MK. Production of volatile metabolites in potatoes infected by Erwinia carotovora var. carotovora and E. carotovora var. atroseptica. Can. J. Plant Pathol 1984, 7, 47–51. [Google Scholar]
- Vikram, A; Lui, LH; Hossain, A; Kushalappa, AC. Metabolic fingerprinting to discriminate diseases of stored carrots. Ann. Appl. Biol 2006, 10, 1–10. [Google Scholar]
- Hettinga, KA; van Valenberg, HJF; van Hooijdonk, ACM. Quality control of raw cows' milk by headspace analysis. Int. Dairy J 2008, 18, 506–513. [Google Scholar]
- Moalemiyan, M; Vikram, A; Kushalappa, AC; Yaylayan, V. Metabolic profiling to discriminate stem-end rot and anthracnose diseases of Tommy Atkins mangoes. Plant Pathol 2006, 55, 792–802. [Google Scholar]
- Vikram, A; Prithivaraj, B; Hamzehzarghani, H; Kushalappa, AC. Volatile metabolite profiling to discriminate diseases of McInthosh apple inoculated with fungal pathogens. J. Sci. Food Agric 2004, 84, 1333–1340. [Google Scholar]





| Setting | Value |
|---|---|
| Retention begin (scan nr.) | 0 |
| Retention end (scan nr.) | 15,000 |
| Maximum amplitude | 200,000,000 |
| Peak slope factor | 0.5 |
| Peak threshold factor | 1 |
| Average peak width at half height | 20 |
| Scaling | Marker peak |
| Nominal mass | 128 at scan nr. 9520 |
| Initial peak search criteria : maximum shift begin of 1st region | 15 |
| Initial peak search criteria : maximum shift end of 1st region | 50 |
| Maximum shift per 100 scans | 35 |
| Pre-align processing | Iterative |
| Minimum S/N ratio | 10 |
© 2010 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Jansen, R.; Hofstee, J.W.; Bouwmeester, H.; Henten, E.v. Automated Signal Processing Applied to Volatile-Based Inspection of Greenhouse Crops. Sensors 2010, 10, 7122-7133. https://doi.org/10.3390/s100807122
Jansen R, Hofstee JW, Bouwmeester H, Henten Ev. Automated Signal Processing Applied to Volatile-Based Inspection of Greenhouse Crops. Sensors. 2010; 10(8):7122-7133. https://doi.org/10.3390/s100807122
Chicago/Turabian StyleJansen, Roel, Jan Willem Hofstee, Harro Bouwmeester, and Eldert van Henten. 2010. "Automated Signal Processing Applied to Volatile-Based Inspection of Greenhouse Crops" Sensors 10, no. 8: 7122-7133. https://doi.org/10.3390/s100807122