Long Chain N-acyl Homoserine Lactone Production by Enterobacter sp. Isolated from Human Tongue Surfaces
Abstract
: We report the isolation of N-acyl homoserine lactone-producing Enterobacter sp. isolate T1-1 from the posterior dorsal surfaces of the tongue of a healthy individual. Spent supernatants extract from Enterobacter sp. isolate T1-1 activated the biosensor Agrobacterium tumefaciens NTL4(pZLR4), suggesting production of long chain AHLs by these isolates. High resolution mass spectrometry analysis of these extracts confirmed that Enterobacter sp. isolate T1-1 produced a long chain N-acyl homoserine lactone, namely N-dodecanoyl-homoserine lactone (C12-HSL). To the best of our knowledge, this is the first isolation of Enterobacter sp., strain T1-1 from the posterior dorsal surface of the human tongue and N-acyl homoserine lactones production by this bacterium.1. Introduction
The human oral cavity is a rich source of microorganisms [1] where a dynamic interaction exists between the host environment and the oral bacteria consortium. Although the oral cavity consists of a complex microbial environment, Streptococcus is the major genus and is well studied [2]. Other oral bacteria identified include Actinomyces spp., Capnocytophaga spp., Eikenella spp., Haemophilus spp., Prevotella spp., Propionibacterium spp., and Veillonella spp. and Fusobacterium spp. [2]. These oral bacteria interact with the environment by attaching to surfaces and establishing mixed-species communities and this routinely requires cell-to-cell communication, which often results in the formation of biofilms. It has been suggested that oral bacterial species do not use N-acyl homoserine lactone (AHL)-based cell-to-cell signalling mechanisms [2–4], but instead, the autoinducer-2 (AI-2) signalling mechanism is commonly used by most oral bacteria, which include Prevotella intermedia, Porphyromonas gingivalis, Streptococcus gordonii and Streptococcus mutans[5–8].
AHLs are arguably the most studied quorum sensing (QS) signalling molecules in proteobacteria, and are produced by a LuxI synthase that will bind to LuxR protein [9]. When the concentration of these AHLs reaches the threshold level, the AHL-luxR complex will regulate a set of genes which occur in a population density-dependent manner, leading to population driven changes in several functions including virulence determinants, antibiotic production, bioluminescence, and biofilm formation [10–13]. QS bacteria have been isolated from various sources and habitats, including the human body [14–18].
Both the Gram-positive and Gram-negative bacteria have been implicated in several systemic infections [1]. Recently, we have isolated two AHL-producing K. pneumoniae strains from the posterior dorsal surface of the tongue of a healthy individual [17]. Here we report the isolation of Enterobacter sp. isolate T1-1capable of producing C12-HSL isolated from the tongue surfaces of a healthy individual, collectively this result provide evidence that oral bacteria are not limited to AI-2 activity.
2. Experimental Section
2.1. Bacterial Strains
Agrobacterium tumefaciens NTL4(pZLR4) [19] was used as biosensor and Escherichia coli DH5α served as a host for DNA manipulations. A. tumefaciens NTL4(pZLR4) was cultured in AB medium (containing 6% (w/v) K2HPO4, 2% (w/v) KH2PO4, 2% (w/v) NH4Cl, 0.6% (w/v) MgSO4·7H20, 0.3% (w/v) KCl, 0.02% (w/v) CaCl2, and 0.005% (w/v) FeSO4·7H2O) or agar (solidified with 1.5% (w/v) bacto-agar), supplemented with 30 μg/mL gentamicin and 0.5% w/v glucose [19]. For AHL detection with A. tumefaciens NTL4(pZLR4), AB agar without gentamicin was supplemented with 20 μg/mL X-gal. A. tumefaciens NTL4(pZLR4) will cause a blue pigmentation on AB agar supplemented with X-gal in the presence of long chain AHLs. All other bacteria were cultured in Luria-Bertani (LB) medium (1% (w/v) tryptone, 0.5% (w/v) yeast extract, and 1% (w/v) NaCl), broth or agar (solidified with 1.5% (w/v) bacto-agar). All LB media were buffered with 50 mM 3-[N-morpholino]propanesulfonic acid (MOPS) to pH 5.5 to prevent opening of lactone ring of AHLs under alkali condition [20]. Where necessary, growth media were supplemented with 100 μg/mL ampicillin. E. coli cells and oral bacteria were grown at 37 °C whereas the biosensor strain was grown at 28 °C.
2.2. Isolation of Bacteria from Tongue Surface Debris
We have previously reported isolation of bacteria from oral orthodontics buccal tubes and AHL-producing bacterium from human oral cavity [17,21]. Here, tongue surface debris sample was collected in 2008 from an individual with healthy oral condition at the Faculty of Dentistry, University of Malaya. This study was approved by the Ethics Committee of the Faculty. In a previous report, we have isolated oral bacteria from tongue surface debris [17]. Pure colonies were obtained by several passages on the LB agar and screened for AHL production using biosensor A. tumefaciens NTL4(pZLR4). Of the bacteria screened, isolate T1-1 which activated A. tumefaciens NTL4(pZLR4) was selected for further analysis.
2.3. Strain Identification
All DNA extraction, purification, manipulations and PCR of 16S rDNA genes were carried out as previously described [14,22]. For PCR amplification of 16S rDNA genes from the genomic DNA, the universal primer pairs 27F and 1525R were used as described before [22]. Universal primers T7, SP6, and internal primers previously designed to anneal to internal target regions of the 16S rDNA were used [23]. LASERGENE software package (DNASTAR, US) was used to edit and analyse nucleotide sequences alignments. MEGA version 4.0 [24] was used for phylogenetic analysis and trees were generated using aligned 16S rDNA gene sequences with the Neighbour-Joining algorithm. Bootstrap analyses for 1,000 resamplings were used to ensure robustness and reliability of trees constructed.
2.4. Extraction and Detection of AHLs from Bacterial Culture Supernatants
In order to screen for oral bacteria that produces AHLs, oral bacteria were cross-streaked with the biosensor A. tumefaciens NTL4(pZLR4) [25] whereby a blue pigmentation on A. tumefaciens NTL4(pZLR4) lawn suggests the presence of long chain AHLs. Bacteria cells showed positive screening results were inoculated into 100 mL of LB broth and grown overnight. Overnight grown cells were adjusted to an OD600 of 1.0 and the spent supernatant was extracted twice with equal volume of acidified ethyl acetate (0.1% v/v acetate acid). The organic layer was collected in a separation funnel, dried over excessive amounts of anhydrous magnesium sulphate, filtered, and evaporated to dryness. Residues were dissolved in 100 μL of acetonitrile and stored at −20 °C. AHLs extract was further analysed by spotting 1 μL of the AHL extract onto the A. tumefaciens NTL4(pZLR4) lawn before sending for liquid chromatography mass spectrometry.
2.5. Mass Spectrometry (MS) Analysis of AHL
High resolution mass spectrometry was performed as previously described [17,26] using the Agilent RRLC 1200 system equiped with an Agilent ZORBAX Rapid Resolution HT column (100 mm × 2.1 mm, 1.8 μm particle size). Mobile phases A and B were 0.1% v/v formic acid in water and 0.1% v/v formic acid in acetonitrile, respectively. The gradient profile is as follow (time (min), mobile phase A: mobile phase B): 0 min, 60%:40%; 5 min, 20%:80%; 7–10 min, 5%:95%; 11–13 min, 60%:40%). The high resolution ESI-MS and ESI-MS/MS analysis was performed using an Agilent 6500 Q-TOF, operated in the ESI-positive mode, with probe capillary voltage fixed at 3,000 V; desolvation temperature of 350 °C; sheath gas set at 11 mL/hour; and nebulizer pressure of 50 psi. Nitrogen gas was used as the collision gas for the MS/MS analysis, with collision energy set at 20 eV. MS data was analysed by using Agilent MassHunter.
3. Results and Discussion
3.1. Enrichment and Characterization of Bacteria
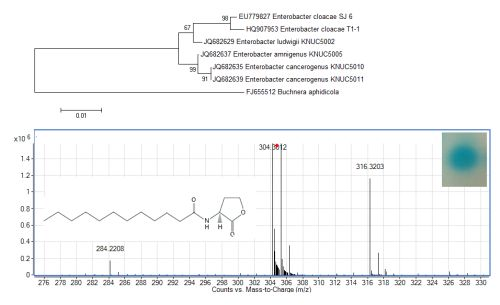
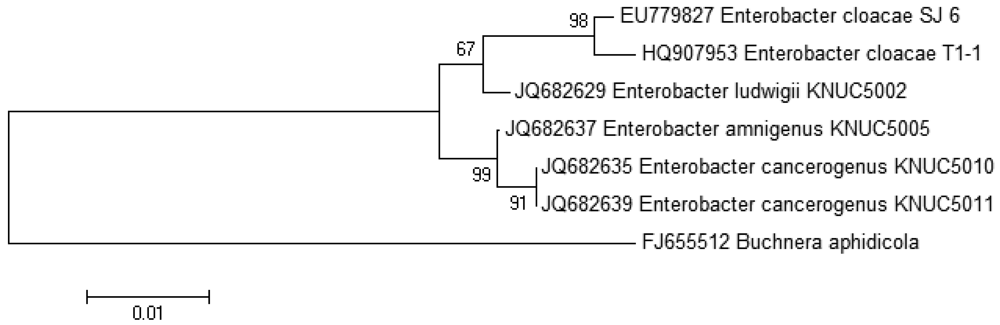
The growth medium became turbid at the first cycle (after 48 hour) and in the following cycles, suggesting that bacterial growth occurred during the enrichment procedure. Enrichment was in vitamin-free KG medium [16] containing C7-HSL. We use the comparatively rare odd number N-acyl side chain namely C7-HSL as previously reported [17]. For each enrichment cycle, cell culture was serially diluted and spread onto LB agar to obtain pure colonies. Bacterial colonies with distinctive morphologies were obtained after several successive streaks on LB agar and isolate T1-1 was selected for further analysis. Enterobacter sp. isolate T1-1 was molecularly identified by their 16S rDNA. Phylogenetic analysis of isolate T1-1 was performed by analyzing the partial sequence its 16S rDNA gene sequences (647 nucleotides), which has been deposited into GenBank, with accession number of HQ907953. Web-based search indicated T1-1 was Enterobacter sp. and further phylogenetic analysis confirmed that T1-1 was Enterobacter sp. closely related to the E. cloacae strain SJ6 (Figure 1).
3.2. Detection of AHLs by Enterobacter sp. Isolate T1-1
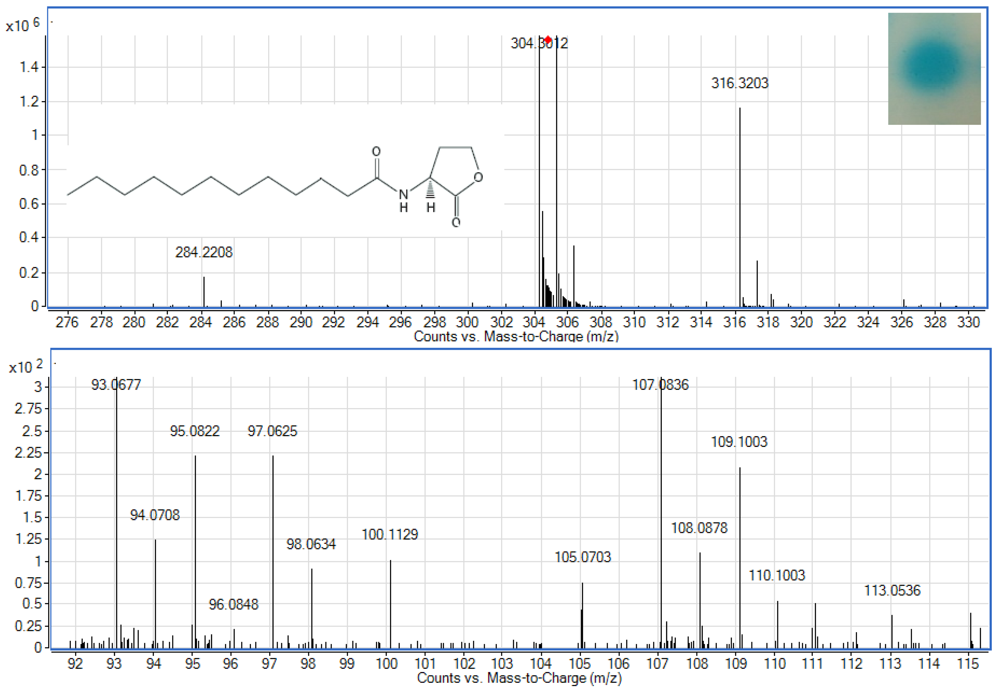
To examine whether Enterobacter sp. isolate T1-1 produced AHLs, spent culture supernatant was extracted with acidified ethyl acetate (0.1% (v/v) acetate acid) and bioassay was used to detect AHLs in the concentrated extracts. A. tumefaciens NTL4(pZLR4) was used to detect AHLs in extracts obtained from the spent supernatant where the culture medium were acidic KG medium amended with MOPS to prevent lactonolysis by alkali. The biosensor was activated as evident by the blue pigmentation formed. In cross-streaked assay, oral bacterial isolate T1-1 activated the biosensor A. tumefaciens NTL4(pZLR4) suggested the production of long chain AHLs by isolate T1-1 (Figure 2, inset).
AHLs extracted from the spent supernatant of Enterobacter sp. isolate T1-1 was spotted onto the biosensor A. tumefaciens NTL4(pZLR4) lawn and a clear blue spot was observed (Figure 2). To further verify the presence of AHLs in the extracts, we used high resolution mass spectrometry that provides unequivocal determination of these AHL molecules in the spent supernatants. The mass spectra data indicated long chain AHLs have been detected. High resolution mass spectrometry analysis confirmed the presence of C12-HSL (m/z 284.2208) (Figure 2, upper panel) in the spent supernatant of T1-1 (Enterobacter sp. isolate T1-1). The ESI-MS/MS spectrum of C12-HSL shows fragments (m/z 95.0822, 109.1003) (Figure 2, lower panel) typical of a lactone-moiety [26]. Taken the biosensor and mass spectra data together, it is unequivocal confirming the presence of C12-HSL in the spent supernatant of Enterobacter sp. (T1-1).
It has been reported that Enterobacter aerogenes, K. pneumoniae and K. oxytoca all show AHL production, but these isolates only produce AHLs when grown microaerophilically in LB medium [27]. Interestingly, Enterobacter sp. isolate T1-1 isolated in the present work showed AHL production when grown aerobically in low-pH defined LB medium. Similar to our previous finding that reports the production of AHL by K. pneumoniae from oral cavity [17], here our result shows the quorum sensing activity of Enterobacter sp. isolate T1-1. Acid tolerance test indicated that Enterobacter sp. isolate T1-1 was viable when grown for 24 h in LB buffered at pH 5 remained viable at pH 3 (data not shown). Since QS regulates many important phenotypes including virulence production, hence study on QS in oral bacteria may lead to better understanding of how these oral bacteria colonise oral cavity. Natural products isolated from plants that show anti-QS activity may provide solution to prevent oral infection caused by QS pathogens [28].
In this study, Enterobacter sp. isolate T1-1 was isolated from the posterior dorsal surface of the tongue. In a study of the oral prevalence of aerobic and facultatively anaerobic Gram-negative rods and yeasts, it is reported that the oral prevalence of these microorganisms are 41.7% and Enterobacteriaceae species represents 73% of all these isolates. The most commonly found species are E. cloacae and K. pneumoniae[29]. Our finding on the isolation of strain T1-1 is in agreement with this report [29] but this is the first report of AHL-producing Enterobacter sp. isolated from the human tongue surface. In contrast, several genera of oral bacteria have been reported to produce AI-2, synthesized by LuxS [2,3,5,7]. Our work has expand the QS research in oral bacteria from AI-2 to AHL, currently we are performing whole genome sequencing of this bacterium to study the AHL synthase genes in Enterobacter sp. isolate T1-1.
4. Conclusions/Outlook
This work is the first documentation of AHL-producing Enterobacter sp. isolated from the human tongue surface. This enables us to have a deeper understanding on the oral bacterial communication and might be providing useful information for the AHL-based quorum sensing mechanism in human oral cavity.
Acknowledgments
This work was supported by grants from the University of Malaya (High Impact Research Grant (A000001-50001) to Kok-Gan Chan, and Kathivaran P (RG033/09HTM) which are gratefully acknowledged. We thank Yeun-Mun Choo for mass spectrometry analysis.
References
- Aas, J.A.; Paster, B.J.; Stokes, L.N.; Olsen, I.; Dewhirst, F.E. Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 2005, 43, 5721–5732. [Google Scholar]
- Kolenbrander, P.E.; Andersen, R.N.; Blehert, D.S.; Egland, P.G.; Foster, J.S.; Palmer, R.J., Jr. Communication among oral bacteria. Microbiol. Mol. Biol. Rev. 2002, 66, 486–505. [Google Scholar]
- Frias, J.; Olle, E.; Alsina, M. Periodontal pathogens produce quorum sensing signal molecules. Infect. Immun. 2001, 69, 3431–3434. [Google Scholar]
- Whittaker, C.J.; Klier, C.M.; Kolenbrander, P.E. Mechanisms of adhesion by oral bacteria. Annu. Rev. Microbiol. 1996, 50, 513–552. [Google Scholar]
- Burgess, N.A.D.; Kirke, D.F.; Williams, P.; Winzer, K.; Hardie, K.R.; Meyers, N.L.; Aduse-Opoku, L.; Curtis, M.A.; Cámara, M. LuxS-dependent quorum sensing in Porphyromonas gingivalis modulates protease and haemagglutinin activities but is not essential for virulence. Microbiology 2002, 148, 763–772. [Google Scholar]
- Chung, W.O.; Park, Y.; Lamont, R.J.; McNab, R.; Barbieri, B.; Demuth, D.R. Signaling system in Porphyromonas gingivalis based on a LuxS protein. J. Bacteriol. 2001, 183, 3903–3909. [Google Scholar]
- Fong, K.P.; Chung, W.O.; Lamont, R.J.; Demuth, D.R. Intra- and interspecies regulation of gene expression by Actinobacillus actinomycetemcomitans LuxS. Infect. Immun. 2001, 69, 7625–7634. [Google Scholar]
- Merritt, J.; Qi, F.; Goodman, S.D.; Anderson, M.H.; Shi, W. Mutation of luxS affects biofilm formation in Streptococcus mutans. Infect. Immun. 2003, 71, 1972–1979. [Google Scholar]
- Fuqua, C.; Winans, S.C.; Greenberg, E.P. Census and consensus in bacterial ecosystems: The LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu. Rev. Microbiol. 1996, 50, 727–751. [Google Scholar]
- Fuqua, C.; Parsek, M.R.; Greenberg, E.P. Regulation of gene expression by cell-to-cell communication: Acyl-homoserine lactone quorum sensing. Annu. Rev. Genet. 2001, 35, 439–468. [Google Scholar]
- Williams, P.; Winzer, K.; Chan, W.; Cámara, M. Look who's talking: Communication and quorum sensing in the bacterial world. Philos. Trans. Roy. Soc. Lond. B Biol. Sci. 2007, 362, 1119–1134. [Google Scholar]
- Horng, Y.T.; Deng, S.C.; Daykin, M.; Soo, P.C.; Wei, J.R.; Luh, K.T.; Ho, S.W.; Swift, S.; Lai, H.C.; Williams, P. The LuxR family protein SpnR functions as a negative regulator of N-acylhomoserine lactone-dependent quorum sensing in Serratia marcescens. Mol. Microbiol. 2002, 45, 1655–1671. [Google Scholar]
- Dunlap, P.V. Quorum regulation of luminescence in Vibrio fischeri. J. Mol. Microbiol. Biotechnol. 1999, 1, 5–12. [Google Scholar]
- Chan, K.G.; Atkinson, S.; Mathee, K.; Sam, C.K.; Chhabra, S.R.; Cámara, M.; Koh, C.L.; Williams, P. Characterization of N-acylhomoserine lactone-degrading bacteria associated with the Zingiber officinale (ginger) rhizosphere: Co-existence of quorum quenching and quorum sensing in Acinetobacter and. Burkholderia. BMC Microbiol. 2011, 11. [Google Scholar] [CrossRef]
- Chan, K.G.; Puthucheary, S.D.; Chan, X.Y.; Yin, W.F.; Wong, C.S.; See Too, W.S.; Chua, K.H. Quorum sensing in Aeromonas species isolated from patients in Malaysia. Curr. Microbiol. 2010, 62, 167–172. [Google Scholar]
- Wong, C.S.; Yin, W.F.; Choo, Y.M.; Sam, C.K.; Koh, C.L.; Chan, K.G. Coexistence of quorum-quenching and quorum-sensing in tropical marine Pseudomonas aeruginosa strain MW3A. World J. Microbiol. Biotechnol. 2011, 28, 453–461. [Google Scholar]
- Yin, W.F.; Purmal, K.; Chin, S.; Chan, X.Y.; Koh, C.L.; Sam, C.K.; Chan, K.G. N-acyl homoserine lactone production by Klebsiella pneumoniae isolated from human tongue surface. Sensors 2012, 12, 3472–3483. [Google Scholar]
- Yin, W.F.; Tung, H.J.; Sam, C.K.; Koh, C.L.; Chan, K.G. Quorum quenching Bacillus sonorensis isolated from soya sauce fermentation brine. Sensors 2012, 12, 4065–4073. [Google Scholar]
- Farrand, S.K.; Hwang, I.; Cook, D.M. The tra region of the nopaline-type Ti plasmid is a chimera with elements related to the transfer systems of RSF1010, RP4, and F. J. Bacteriol. 1996, 178, 4233–4247. [Google Scholar]
- Yates, E.A.; Philipp, B.; Buckley, C.; Atkinson, S.; Chhabra, S.R.; Sockett, R.E.; Goldner, M.; Dessaux, Y.; Cámara, M.; Smith, H.; Williams, P. N-acylhomoserine lactones undergo lactonolysis in a pH-, temperature-, and acyl chain length-dependent manner during growth of Yersinia pseudotuberculosis and Pseudomonas aeruginosa. Infect. Immun. 2002, 70, 5635–5646. [Google Scholar]
- Purmal, K.; Chin, S.; Pinto, J.; Yin, W.F.; Chan, K.G. Microbial contamination of orthodontic buccal tubes from manufacturers. Int. J. Mol. Sci. 2010, 11, 3349–3356. [Google Scholar]
- Chong, T.M.; Koh, C.L.; Sam, C.K.; Choo, Y.M.; Yin, W.F.; Chan, K.G. Characterization of quorum sensing and quorum quenching soil bacteria isolated from Malaysian tropical montane forest. Sensors 2012, 12, 4846–4859. [Google Scholar]
- Lane, D.L.; Pace, B.; Olsen, G.J.; Stahl, D.A.; Sogin, M.L.; Pace, N.R. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc. Natl. Acad. Sci. USA 1985, 82, 6955–6959. [Google Scholar]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar]
- Steindler, L.; Venturi, V. Detection of quorum-sensing N-acyl homoserine lactone signal molecules by bacterial biosensors. Fed. Microbiol. Soc. 2006, 266, 1–9. [Google Scholar]
- Ortori, C.A.; Atkinson, S.; Chhabra, S.R.; Cámara, M.; Williams, P.; Barret, D.A. Comprehensive profiling of N-acylhomoserine lactones produced by Yersinia pseudotuberculosis using liquid chromatography coupled to hybrid quadrupole-linear ion trap mass spectrometry. Anal. Bioanal. Chem. 2007, 387, 497–511. [Google Scholar]
- Wang, H.; Cai, T.; Weng, M.; Zhou, J.; Cao, H.; Zhong, Z.; Zhu, J. Conditional production of acyl-homoserine lactone-type quorum-sensing signals in clinical isolates of enterobacteria. J. Medical. Microbiol. 2006, 55, 1751–1753. [Google Scholar]
- Chong, Y.M.; Yin, W.F.; Ho, C.Y.; Mustafa, M.R.; Hadi, A.H.A.; Awang, K.; Narrima, P.; Koh, K.L.; Appleton, D.R.; Chan, K.G. Malabaricone C from Myristica cinnamomea exhibits anti-quorum sensing activity. J. Nat. Prod. 2011, 74, 2261–2264. [Google Scholar]
- Sedgley, C.M.; Samaranayake, L.P. The oral prevalence of aerobic and facultatively anaerobic gram-negative rods and yeasts in Hong Kong Chinese. Arch. Oral Biol. 1994, 39, 459–466. [Google Scholar]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Yin, W.-F.; Purmal, K.; Chin, S.; Chan, X.-Y.; Chan, K.-G. Long Chain N-acyl Homoserine Lactone Production by Enterobacter sp. Isolated from Human Tongue Surfaces. Sensors 2012, 12, 14307-14314. https://doi.org/10.3390/s121114307
Yin W-F, Purmal K, Chin S, Chan X-Y, Chan K-G. Long Chain N-acyl Homoserine Lactone Production by Enterobacter sp. Isolated from Human Tongue Surfaces. Sensors. 2012; 12(11):14307-14314. https://doi.org/10.3390/s121114307
Chicago/Turabian StyleYin, Wai-Fong, Kathiravan Purmal, Shenyang Chin, Xin-Yue Chan, and Kok-Gan Chan. 2012. "Long Chain N-acyl Homoserine Lactone Production by Enterobacter sp. Isolated from Human Tongue Surfaces" Sensors 12, no. 11: 14307-14314. https://doi.org/10.3390/s121114307