An Electrochemical DNA Microbiosensor Based on Succinimide-Modified Acrylic Microspheres
Abstract
: An electrochemical microbiosensor for DNA has been fabricated based on new acrylic microspheres modified with reactive N-acryloxysuccinimide (NAS) functional groups. Hydrophobic poly(n-butylacrylate-N-acryloxysuccinimide) microspheres were synthesized in an emulsion form with a simple one-step photopolymerization technique. Aminated DNA probe was attached to the succinimde functional group of the acrylic microspheres via covalent bonding. The hybridization of the immobilized DNA probe with the complementary DNA was studied by differential pulse voltametry using anthraquninone-2-sulfonic acid monohydrate sodium salt (AQMS) as the electroactive hybridization label. The influences of many factors such as duration of DNA probe immobilization and hybridization, pH, type of ions, buffer concentrations, ionic strength, operational temperature and non-complementary DNA on the biosensor performance were evaluated. Under optimized conditions, the DNA microbiosensor demonstrated a linear response range to target DNA over a wide concentration range of 1.0 × 10−16 and 1.0 × 10−8 M with a lower limit of detection (LOD) of 9.46 × 10−17 M (R2 = 0.97). This DNA microbiosensor showed good reproducibility with 2.84% RSD (relative standard deviation) (n = 3). Application of the NAS-modified acrylic microspheres in the construction of DNA microbiosensor had improved the overall analytical performance of the resultant DNA microbiosensor when compared with other reported DNA biosensors using other nano-materials for membranes and microspheres as DNA immobilization matrices.1. Introduction
The immobilization method and the matrix used for DNA probe immobilization are important in designing a DNA biosensor especially for achieving high sensitivity, selectivity and stablility [1–4]. Immobilizing of DNA probes onto an immobilization matrix or electrode requires strong binding between the probe and the immobilization matrix without affecting the chemical properties of the DNA probe [5]. Various methods for immobilization of DNA probes have been reported such as physical and electrochemical adsorptions [6–8], electrochemical entrapment [9] and covalent binding [10–12]. Sorption and entrapment methods are simple immobilization methods; however they produce weak molecular bonds between the biological molecules and the immobilization matrix. This results in the immobilized molecules leaching out easily and reduces the shelf life and stability of the biosensor. In general, covalent bonding and the biotin-avidin methods are found to be more suitable for DNA probe immobilization where one end of the DNA probe is attached, leaving the other end free for hybridization with target DNA to form a double-stranded DNA (dsDNA) [13]. In addition, the covalent bonding formed is stronger and will not interfere with the chemical behaviour of the DNA probe and thus it is an efficient method to yield high performance DNA biosensors in terms of sensitivity, selectivity and stability [14,15]. DNA probe immobilization via covalent bonding methods often involves the use of linker functional groups for DNA probe immobilization. Some examples are the functional groups of succinimide [16–19], aldehyde [20], and maleimido-based reactive groups [21,22]. Thus, using an immobilization matrix modified with such functional groups would create a useful immobilization surface for covalent immobilization of DNA probes.
The type of matrix for DNA probe immobilization also plays an important role affecting the performance of an electrochemical DNA biosensor. Nanoparticles used for immobilization matrices, e.g., nanoparticles and microspheres, can contribute to better performance of the resulting biosensor when compared with membrane matrices. This is due to the larger surface area of the three-dimensional structure of nanoparticles [23,24] when compared with the two-dimensional structure of a membrane. With larger surface area, DNA probe binding capacity can be improved and this further improves the DNA biosensor performance. Several polymer membranes [3,4,25–29] and microspheres, such as metal based gold and Fe2O3 [30–34] microspheres have been employed for DNA biosensor construction, but microsphere-based DNA biosensors exhibited better performance compared with polymer membrane matrices.
In this research, acrylic polymer microspheres modified with succinimide functional groups via N-acryloxysuccinimide (NAS) moieties was used as the matrix for DNA probe immobilization. As previously reported [35,36], the succinimide functional group can react with amine functional groups to form a covalent bond. The incorporation of a NAS functionality into acrylic microspheres for DNA microbiosensor application is a new idea that provides advantages of a simple preparation method where the spheres can be synthesized and functionalised via a one-step procedure using photopolymerisation in a short duration (several minutes). In addition, the microspheres have the advantage of small size and provide a large surface area for DNA probe immobilization, thus reducing the barrier to diffusion for reactants and products. This enables the improvement in the biosensor performance in terms of shorter response times and wider linear response range, which will be demonstrated in the work reported here. The mechanism of construction of DNA microbiosensing system using NAS functionalized acrylic microspheres is depicted in Figure 1.
2. Experimental Section
2.1. Instrumentation
All electrochemical measurements were performed with DPV (AutoLab potentiostat) in a measurement cell containing 4.5 mL of 0.05 M K-phosphate buffer at pH 7.0. The electrochemical system consists of a gold electrode (GE), a carbon pencil counter electrode and Ag/AgCl reference electrode. A Scanning Electron Microscope (SEM, LEO 1450VP) was used to determine the size and distribution of the acrylic microspheres.
2.2. Chemicals
The reagents 2-2-dimethoxy-2-phenylacetophenone (DMPP), 1,6-hexanadiol diacrylate (HDDA) and sodium dodecyl sulphate (SDS) were supplied by Fluka, Aldrich and Systerm, respectively. N-acryloxysuccinimide (NAS) and anthraqinone-2-sulfonic acid monohydrate sodium salt (AQMS) were obtained from Acros. The 20-base pair single stranded DNA was purchased from Sigma-Aldrich. The DNA sequences for this work were similar to those used in the previous study by Wong et al. [37]. These oligonucleotide DNA sequences are as follows:
Probe DNA: 5′ GGGGCAGAGCCTCACAACCT (AmC3)
Target DNA: 5′ AGGTTGTGAGGCTCTGCCCC
Non-complementary DNA: 5′ GGATGGACGAAGCGCTCAGG
Oligonucleotide stock solution (100 μM) was prepared in TE buffer solution containing 10 mM Tris-HCl and 1 mM ethylenediaminetetra-acetic acid (EDTA) at pH 7.7 and stored under −20 °C when not in use. Dissolution of oligonucleotide stock solution was carried out using 0.05 M K-phosphate buffer pH 7.0. Stock solution of 1 mM AQMS was prepared in 0.05 M K-phosphate buffer (pH 7.0).
2.3. Synthesis of NAS-Modified Acrylic Microspheres
A mixture of nBA monomers (7 mL), SDS (0.01 g), HDDA (450 μL), DMPP (0.1 g), NAS (6 mg) and H2O (15 mL) was sonicated for 10 min. The resulting emulsion solution was then photocured for 600 s with ultraviolet radiation of a wavelength of approximately 250–350 nm under a continuous nitrogen gas flow. Poly(nBA-NAS) microspheres formed were then collected by centrifugation at 4,000 rpm for 30 min and later washed three times in 0.05 M K-phosphate buffer (pH 7.0), followed by air drying.
2.4. Fabrication of DNA Biosensor by Immobilization of DNA Probe on NAS-Modified Acrylic Microspheres
About 200.0 mg of poly(nBA-NAS) microspheres were added to 5.0 μM DNA probe solution and kept at 4 °C for 24 h in order to immobilize the probes onto the microspheres. Subsequently, the microspheres were washed with 0.05 M K-phosphate buffer (pH 7.0) to remove the unbound DNA probes from the microspheres. To test the success of the DNA probe immobilization, hybridization studies were carried out by using a complementary DNA target. The acrylic microspheres immobilized with DNA probes [DNA-poly(nBA-NAS)] were first immersed in a solution containing 5.0 μM complementary DNA target and 1.0 mM AQMS. The mixture was then incubated at 4 °C for 24 h. After that, the microspheres were collected and washed with 0.05 M K-phosphate buffer at pH 7.0 to remove the free DNA target and unbound AQMS. The resultant microspheres attached with the hybridized DNA and intercalated AQMS were sonicated in 4.5 mL of 0.05 M K-phosphate buffer (pH 7.0) to dislodge the hybridized DNA and released the AQMS label from the hybridized DNA. The DPV of the AQMS release from sonication was scanned at the potential range of −0.75 V to −0.25 V using a gold electrode as working electrode, a carbon pencil counter electrode and Ag/AgCl reference electrode.
2.5. Effects of Reaction Medium on DNA Microbiosensor Response
The response of DNA microbiosensor was examined based on the effect of various parameters on the hybridization of the immobilised DNA probe. This was performed in the present of different cations (Na+, K+, Mg2+, Ca2+, Al3+), varying pH from pH 6.0–8.5 and in different Na-phosphate buffer concentrations between 0.002 M to 1.0 M. Ionic strength effect on hybridization of DNA probe and biosensor response was also examined by varying the NaCl concentration over the range of 0.02–2.0 M at pH 7.5.
2.6. Effect of Duration of Probe Immobilization and Hybridization Temperature on DNA Microbiosensor Response
The durations of probe immobilisation and temperature of hybridization with target DNA could influence the microbiosensor response. For these studies, immobilization of DNA probe was performed in 0.05 M K-phosphate buffer pH 7.0, whilst for hybridization of DNA target, it was carried out in 0.25 M Na-phosphate buffer (pH 7.5) in the presence of 0.5 M Na+ ion at 4 °C. Both the duration of probe immobilisation and hybridization were performed over a time period of 1–24 h. For temperature effect on DNA hybridization, the temperature was varied from 4–45 °C over a period of 20–160 min. In these studies, the effect on the linear response range and lower detection limit of the biosensor under each condition was examined.
3. Results and Discussion
3.1. Confirmation of DNA Probe Immobilization on NAS-Modified Acrylic Microspheres
A typical scanning electron micrograph image of the as prepared acrylic microspheres (Figure 2) demonstrated that the size of the microspheres was of a diameter approximately in the μm range, with a rather homogenous size distribution. These microspheres that have been modified with NAS functional group were used for DNA probe immobilization via the aminated end of the DNA probe.
Figure 3 shows the DPV peak at −0.5 V for AQMS that has been intercalated into DNA probe immobilized on poly(nBA-NAS) microspheres after hybridization with complementary DNA. However, the current at peak −0.5V was small and non-observable for events that did not involved hybridization such as in the presence of non-complementary DNA, in the absence of DNA and blank microspheres (no immobilised DNA probes; Experiments 2–4). The higher current response observed for Experiment 1 compared with Experiment 2, i.e., when non-complementary DNA was introduced, indicated that the immobilized DNA probe was selective towards complementary DNA and it was indicated by AQMS intercalation into dsDNA formed as has been previously reported [3,38–40]. Control Experiments 3 and 4 were also performed to determine whether any non-specific adsorption of AQMS on ds-DNA probe or blank acrylic microsphere. The relatively small currents observed in Experiments 3 and 4 compared with Experiment 1 implied that the non-specific adsorption of AQMS was negligible. The slight adsorption of AQMS onto DNA probe (ssDNA) was consistent with observation previously reported [41]. The highest current observed for the complementary DNA (Experiment 1) thus indicated hybrization where intercalation of AQMS had occurred in dsDNA formed on the microsphere surface.
3.2. Effect of DNA Probe Loading on DNA Microbiosensor Response
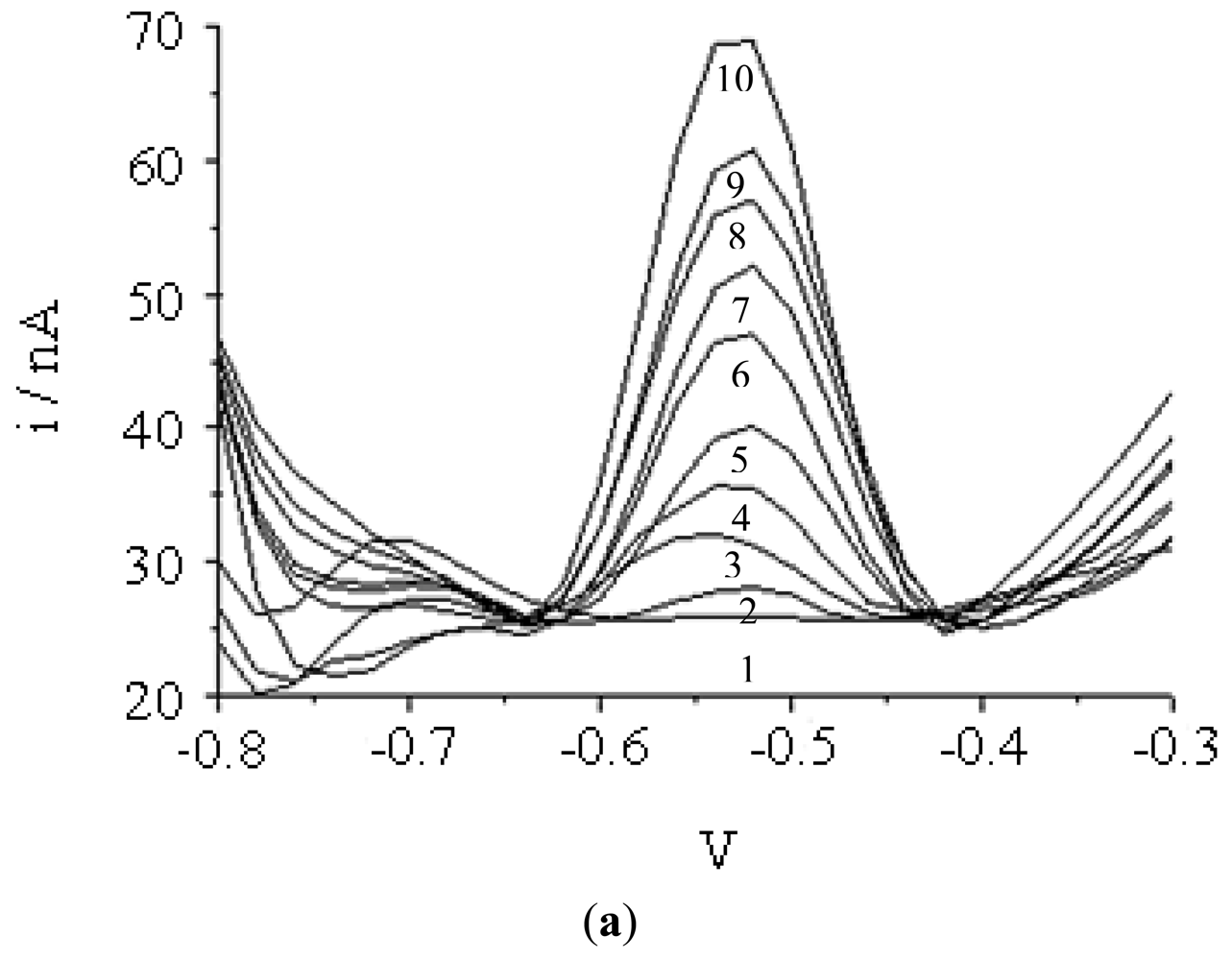
Figure 4 represents the DNA microbiosensor response with various concentrations of DNA probe immobilized onto the acrylic microspheres after hybridization with complementary DNA and followed by intercalation of AQMS. DNA microbiosensor response increased proportionally with increasing concentration of DNA probe immobilised. This suggests that the capacity of immobilized DNA probe to hybridize with complementary DNA has increased with an increase in DNA probe attached onto the acrylic microspheres. The dependence of DNA hybridization on DNA probe concentration has also been reported previously [15,42,43].
3.3. Dependence of Biosensor Response on pH and Ionic Strength
Figure 5 illustrates the pH effect on DNA hybridization response of the biosensor. The DPV current increased abruptly at pH 7.5, after which a sharp decline in current response was observed under more alkaline conditions. The increased response of the DNA microbiosensor at pH 7.5 indicates that more DNA probes were hybridized with complementary DNA at this pH. Previous studies [44] have reported that the rate of DNA hybridization reaction can be influenced by the pH of the solution. At a more acidic environment, the protonation reaction of the phosphodieter of the DNA can reduce the solubility of the DNA molecule, which eventually decreases the DNA hybridization [3]. Under a more basic medium, DNA hybridization also decreased and hence the response of DNA biosensor was also lower [45]. Therefore, the optimize pH of DNA hybridization was selected at pH 7.5 using Na-phosphate buffer solution for subsequent biosensor studies.
Positively charged ions such as Li+, Na+, K+, and Mg2+ ions can interact with the negatively charge phosphodiester chain of the DNA. This ionic reaction will neutralize the charge of the DNA molecule and thus decreases the electrostatic repulsions between DNA molecules. The absent of electrostatic repulsion eases the DNA hybridization reaction [46–48]. Figure 6 depicts the effect of some cations on the DNA hybridization reaction of the biosensor. The DNA hybridization reaction rate increased in the present of positively charged ion in the order of Na+ > K+ > Al3+≈Ca2+≈Mg2+. The presence of Ca2+ and Al3+ ions had reduced the DNA hybridization compared with that of Na+ and K+ ions because the ionic interactions of Ca2+ and Al3+ ions with phosphate ions from the buffer lead to formation of insoluble phosphate compounds. Thus, this reduces the ionic content of the medium and increases the electrostatic repulsion between DNA molecules. Under such conditions, hybridization is more difficult to achieve and the biosensor response declined. The higher DNA hybridization current as indicated by the biosensor response in the presence of Na+ ion was due to the smaller size and stronger affinity of Na+ ion towards DNA phosphodieter chain to reduce the electrostatic repulsion between DNA molecules as compared with K+ and Mg2+ ions. As the use of Na+ ion demonstrated better biosensor signal than K+, Mg2+, Ca2+ and Al3+ ions, therefore Na+ ion was used in further DNA biosensor studies based on modified acrylic microspheres.
In Figure 7(a,b), both Na-phosphate buffer (pH fixed at 7.5) concentration and ionic strength have effect on the biosensor response. In both cases, there is an optimum value where the biosensor gave the highest current response or the highest degree of hybridization. Thus, 0.25 M Na-phosphate buffer (pH 7.5) and 2.0 M ionic strength (from NaCl) were found to be optimum for the biosensor response. This may be explained by at certain amount of ionic content of the solution, the electrostatic repulsion between DNA molecules decreases and thus improving the DNA hybridization reaction. In the presence of too low or too high ionic content, the presence of electrostatic repulsion becomes dominant and the hybridization of DNA molecules become difficult [46].
3.4. Influence of the Duration of DNA Immobilization on Microbiosensor Response
For the duration of immobilisation, the microbiosensor response showed a current increase from 1.0–4.0 h of immobilisation time, after which there was no obvious change in the current measured (Figure 8). Longer immobilization times resulted in the higher amount of DNA probes immobilized onto the microspheres. After 4.0 h of exposure to the DNA probes, the active sites of microspheres are presumably fully attached with DNA probes.
3.5. Hybridization Duration and Temperature Effects on Microbiosensor Response
Temperatures appear to affect the time taken for the maximum DPV current response of the microbiosensor. When temperatures were raised from 4 °C and 45 °C, the time taken for maximum DNA hybridization reduces to just about 40 min at 45 °C (Figure 9).
However at 4 °C, there was no obvious increase in the current response, even up to 140 min hybridization time. The increase in DNA hybridization rate at higher temperatures was attributed to a greater mass transfer rate and rate of reaction of DNA molecules, as well as an increase in the solubility of the DNA molecules under higher temperature conditions [46]. The microbiosensor response also dependent on the duration allowed for hybridization to occur. The increase in DPV current with hybridization time indicated that more DNA hybridization reactions were occurring [32]. The optimum microbiosensor response (Figure 9) is dependent on the temperature and reached at a shorter time span for higher hybridization temperatures. The length of DNA hybridization time obtained herein is similar to that reported elsewhere [41]. Even at room temperature of 25 °C, the current response of the microbiosensor is large enough for further biosensor studies.
3.6. Performance of the DNA Microbiosensor Based on NAS-Modified Acrylic Microspheres
Figure 10 demonstrates the effect of different complementary DNA concentrations on the biosensor response. The current response increases with increasing DNA concentrations indicates more DNA hybridization. This current response was linear towards the DNA concentration from 1.0 × 10−16 and 1.0 × 10−8 M with a LOD of 9.46 × 10−17 M. Using two DNA concentrations of 1.0 × 10−13 M and 1.0 × 10−10 M, the DNA microbiosensor yielded a good reproducibility with the RSD value of 2.86% (n = 3). The performance of the DNA microbiosensor based on NAS-modified acrylic microspheres yielded a wider linear response range and lower detection limit when compared with a DNA microbiosensor employing the same sequence of DNA probes self-assembled on a mercaptol modified gold electrode, which yielded a linear response range from 5.0 × 10−10 to 1.0 × 10−6 M with detection limit at 10−12 M level [37].
3.7. Comparison with Other Reported DNA Microbiosensors
A comparison of the DNA microbiosensor based on NAS-modified acrylic microspheres with other electrochemical DNA biosensors reported in the literature was carried out (Table 1). For most DNA biosensors based on electrochemical transduction, membranes doped with nanomaterials were often used. For example, the use of chitosan films doped with carbon nanotubes or CeO2 [3,28].
In the literature, DNA microbiosensors based on microspheres normally require the microspheres to be immobilized in some form of matrices, e.g., polyaniline nanofibers doped with Fe2O3 microspheres or Au microspheres supported by polyaniline nanofiber. This clearly demonstrates the advantage of the acrylic microspheres where the acrylic polymeric microspheres can function as a DNA microbiosensor alone after attachment of DNA probe. Overall, the DNA microbiosensor from this work using acrylic microspheres has shown large improvement in terms of linear response range and detection limit where the detection limit is at least 1,000 times lower that those DNA biosensors designed from nanomaterials or other microsphere materials.
4. Conclusions
Acrylic microspheres for use as a DNA immobilization matrix has been synthesized using the methods of emulsion and photopolymerization simultaneously. Covalent immobilization of DNA probes onto the microspheres was possible via succinimide functional groups incorporated during the synthesis step. The designed DNA microbiosensor based on acrylic microspheres showed good performance with a wider linear response range and the capability of detecting DNA targets at fM concentration. In addition, there is no specific adsorption of AQMS on the acrylic microspheres. The performance of the new DNA biosensor using succinimide functional group-modified acrylic microspheres was better compared with the other electrochemical transduction-based DNA biosensors.
Acknowledgments
We would like to thank the National Biotechnology Directorate of the Ministry of Science, Techonology and Innovation Malaysia for a research grant and Universiti Kebangsaan Malaysia for financial support via research operational grants OUP-2012-132 and UKM-DLP-2011-014.
References
- Chiorcea, P.A.M.; Diculescu, V.C.; Oretskaya, T.S.; Oliveira, B.A.M. AFM and electroanalytical studies of synthetic oligonucleotide hybridization. Biosens. Bioelectron. 2004, 20, 933–944. [Google Scholar]
- Ilaria, M.; Maria, M.; Sara, T.; Ronghui, W.; Maria, M.S.; Mascini, M. Direct immobilization of DNA probes for the development of affinity biosensors. Bioelectrochemistry 2005, 66, 129–138. [Google Scholar]
- Feng, K.J.; Yang, Y.H.; Wang, Z.J.; Jiang, J.H.; Shen, G.L.; Yu, R.Q. A nano-porous CeO2/Chitosan composite film as immobilization matrix for colorectal cancer DNA sequence-selective electrochemical biosensor. Talanta 2006, 70, 561–565. [Google Scholar]
- Arora, K.; Prabhakar, N.; Chand, S.; Malhotra, B.D. Immobilization of single stranded DNA probe onto polypyrrole-polyvinyl sulfonate for application to DNA hybridization biosensor. Sens. Actuat. B Chem. 2007, 126, 655–663. [Google Scholar]
- Xu, S.; Tu, G.; Peng, B.; Han, X. Self-assembling gold nanoparticles on thiol-functionalized poly(styrene-co-acrylic acid) nanosphere for fabrication of a mediatorless biosensor. Anal. Chim. Acta 2006, 570, 151–157. [Google Scholar]
- Arora, K.; Chand, S.; Malhotra, B.D. Recent developments in bio-molecular electronics techniques for food pathogen. Anal. Chim. Acta 2006, 568, 259–274. [Google Scholar]
- Arora, K.; Chaubey, A.; Singhal, S.; Sigh, R.P.; Samanta, R.B.; Pandey, M.K.; Chand, S.; Malhotra, B.D. Application of electrochemically prepared polypyrrole-polyvinyl sulphonate films to DNA biosensor. Biosens. Bioelectron. 2006, 21, 1777–1783. [Google Scholar]
- Lucarelli, F.; Palchetti, I.; Marrazza, G.; Mascini, M. Electrochemical DNA biosensor as screening tool for the detection of toxicants in water and wastewater samples. Talanta 2002, 56, 949–957. [Google Scholar]
- Mian, J.; Joseph, W. Recognition and detection of oligonucleotides in the presence of chromosomal DNA based on entrapment within conducting-polymer networks. J. Electroanal. Chem. 2001, 500, 584–589. [Google Scholar]
- Zhou, X.C.; Huang, L.Q.; Li, S.F.Y. Microgravimetric DNA sensor based on quartz microbalance: Comparison of oligonucleotide immobilization method and application in genetic diagnosis. Biosens. Bioelectron. 2001, 16, 85–95. [Google Scholar]
- Mouffouk, F.; Higgins, S.J. A biotin-functionalised poly(3,4-ethylene dioxy thiophene) coated microelectrode which responds electrochemically to avidin binding. Electrochem. Commun. 2006, 8, 15–20. [Google Scholar]
- Youssoufi, H.K.; Markrouf, B. Electrochemical biosensor of DNA hybridization by ferrocenyl functionalized polypyrrole. Anal. Chim. Acta 2002, 469, 85–92. [Google Scholar]
- Feng, L.; Wei, C.; Shusheng, Z. Development of DNA electrochemical biosensor based on covalent immobilization of DNA probe by direct coupling of sol-gel and self-assembly technologies. Biosens. Bioelectron. 2008, 24, 781–786. [Google Scholar]
- Ronghui, W.; Sara, T.; Maria, M.; Maria, M.S.; Mascini, M. Immobilization of DNA probes for the development of SPR-based sensing. Biosens. Bioelectron 2004, 20, 967–974. [Google Scholar]
- Oscar, A.L.; Susana, C.; Maria, P.; Jose, M.P. DNA sensor base on an Escherichia coli lac Z gene probe immobilization at self-assembled monolayers-modified gold electrodes. Talanta 2007, 73, 838–844. [Google Scholar]
- Lisa, M.S.; Guadalupe, A.R.; Esther, V.B. Morphological studies of ligodeoxyribonucleotides probes covalently immobilized at polystyrene modified surfaces. J. Biotechnol. 2005, 118, 233–245. [Google Scholar]
- Kerman, K.; Ozkan, D.; Kara, P.; Meric, B.; Gooding, J.J.; Ozsoz, M. Voltammetric determination of DNA hybridization using methylene blue and self-assembled alkanethiol monolayer on gold electrodes. Anal. Chim. Acta 2002, 462, 39–47. [Google Scholar]
- Ozkan, D.; Erdem, A.; Kara, P.; Kerman, K.; Gooding, J.J.; Nielsen, P.E.; Ozsoz, M. Electrochemical detection of hybridization using peptide nucleic acids and methylene blue on self-assembled alkanethiol monolayer modified gold electrodes. Electrochem. Commun. 2002, 4, 796–802. [Google Scholar]
- Teh, H.F.; Gong, H.; Dong, X.D.; Zeng, X.; Tan, A.L.K.; Yang, X.; Tan, S.N. Electrochemical biosensing of DNA with capture probe covalently immobilized onto glassy carbon surface. Anal. Chim. Acta 2005, 551, 23–29. [Google Scholar]
- Zammatteo, N.; Jeanmart, L.; Hamels, S.; Courtois, S.; Louette, P.; Hevesi, L.; Remacle, J. Comparison between different strategies of covalent attachment of DNA to glass surfaces to build DNA microarrays. Anal. Biochem. 2000, 280, 143–150. [Google Scholar]
- Jyoti, C.; Kumar, P.; Gupta, K.C. N-(3-Triethoxysilylpropyl)-6-(N-maleimido)-hexanamide: An eYcient heterobifunctional reagent for the construction of oligonucleotide microarrays. Anal. Biochem. 2006, 357, 240–248. [Google Scholar]
- Wang, Y.; Prokein, T.; Hinz, M.; Seliger, H.; Goedel, W.A. Immobilization and hybridization of oligonucleotides on maleimido-terminated self-assembled monolayers. Anal. Biochem. 2005, 344, 216–223. [Google Scholar]
- Yang, H.; Zhu, Y. Size dependence of SiO2 particles enhanced glucose biosensor. Talanta 2006, 68, 569–574. [Google Scholar]
- Merkoci, A. Nanoparticles-based strategies for DNA, protein and cell sensors. Biosens. Bioelectron. 2010, 26, 1164–1177. [Google Scholar]
- Jiang, C.; Yang, T.; Jiao, K.; Gao, H. A DNA electrochemical sensor with poly-l-lysine/single-walled carbon nanotubes films and its application for the highly sensitive EIS detection of PAT gene fragment and PCR amplification of NOS gene. Electrochim. Acta. 2008, 53, 2917–2924. [Google Scholar]
- Niu, S.; Zhao, M.; Hu, L.; Zhang, S. Carbon nanotube-enhanced DNA biosensor for DNA hybridization detection using rutin-Mn as electrochemical indicator. Sens. Actuators B Chem. 2008, 135, 200–205. [Google Scholar]
- Uygun, A. DNA hybridization electrochemical biosensor using a functionalized polythiophene. Talanta 2009, 79, 194–198. [Google Scholar]
- Sun, W.; Qin, P.; Gao, H.; Li, G.; Jiao, K. Electrochemical DNA biosensor based on chitosan/nano-V2O5/MWCNTs composite film modified carbon ionic liquid electrode and its application to the LAMP product of Yersinia enterocolitica gene sequence. Biosens. Bioelectron. 2010, 25, 1264–1270. [Google Scholar]
- Yang, J.; Yang, T.; Feng, Y.; Jiao, K. A DNA electrochemical sensor based on nanogold-modiWed poly-2, 6-pyridinedicarboxylic acid Wlm and detection of PAT gene fragment. Anal. Biochem. 2007, 365, 24–30. [Google Scholar]
- Horak, D.; Španova, A.; Tvrdikova, J.; Rittich, B. Streptavidin-modified magnetic poly (2-ydroxyethyl methacrylate-coglycidyl methacrylate) microspheres for selective isolation of bacterial DNA. Eur. Polym. J. 2010, 47, 1090–1096. [Google Scholar]
- Lai, G.S.; Zhang, H.L.; Han, D.Y. A novel hydrogen peroxide biosensor based on hemoglobin immobilized on magnetic chitosan microspheres modified electrode. Sens. Actuators B Chem. 2008, 129, 497–503. [Google Scholar]
- Zhang, W.; Yang, T.; Li, X.; Wang, D.; Jiao, K. Conductive architecture of Fe2O3 microspheres/self-doped polyaniline nanofibers on carbon ionic liquid electrode for impedance sensing of DNA hybridization. Biosens. Bioelectron. 2009, 25, 428–434. [Google Scholar]
- Wang, X.; Yang, T.; Li, X.; Jiao, K. Three-step electrodeposition synthesis of self-doped polyaniline nanofiber-supported flower-like Au microspheres for high-performance biosensing of DNA hybridization recognition. Biosens. Bioelectron. 2011, 26, 2953–2959. [Google Scholar]
- Fan, H.; Ju, P.; Ai, S. Controllable synthesis of CdSe nanostructures with tunable morphology and their application in DNA biosensor of Avian Influenza Virus. Sens. Actuat. B Chem. 2010, 149, 98–104. [Google Scholar]
- Chen, J.P.; Chiu, S.H. A poly (N-isopropylacrylamide-co-N-acryloxysuccinimede-co-2-hydroxyethyl methacrylate) composite hydrogel membrane for urease immobilization to enhance urea hydrolysis rate by temperature swing. Enzym. Micro. Technol. 2000, 26, 359–367. [Google Scholar]
- Chaix, C.; Pacard, E.; Elaissari, A.; Hilaire, J.F.; Picot, C. Surface functionalization of oil-in-water nanoemulsion with a reactive copolymer: Colloidal characterization and peptide immobilization. Coll. Surf. B Biointer. 2002, 29, 39–52. [Google Scholar]
- Wong, E.L.S.; Gooding, J.J. Charge transfer through DNA: A selective electrochemical DNA biosensor. Anal. Chem. 2006, 78, 2138–2144. [Google Scholar]
- Wong, E.L.S.; Mearns, F.J.; Gooding, J.J. Further development of an electrochemical DNA hybridization biosensor based on long-range electron transfer. Sens. Actuat. B Chem. 2005, 111–122, 515–521. [Google Scholar]
- Xu, S.; Tu, G.; Peng, B.; Han, X. Self-assembling gold nanoparticles on thiol-functionlaizad poly(styrene-co-acrylic acid) nanosphere for fabrication of a mediatorless biosensor. Anal. Chim. Acta 2006, 570, 151–157. [Google Scholar]
- Wong, E.L.S.; Chow, E.; Gooding, J.J. The electrochemical detection of cadmium using surface-immobilized DNA. Electrochem. Commun. 2007, 9, 845–849. [Google Scholar]
- Wong, E.L.S.; Erohkin, P.; Gooding, J.J. A comparison of cationic and anionic intercalators or the electrochemical transduction of DNA hybridization via long range electron transfer. Electrochem. Commun. 2004, 6, 648–654. [Google Scholar]
- De la Escosura-Muniz, A.; Gonzalez-Garcia, M.B.; Costa-Garcis, A. DNA hybridization sensor based on aurothiomalate electroactive label on glassy carbon electrodes. Biosens. Bioelectron. 2007, 22, 1048–1054. [Google Scholar]
- Kerman, K.; Morita, Y.; Takamura, Y.; Ozsoz, M.; Tamiya, E. Modification of Escharichia coli singles for electrochemical detection of DNA hybridization. Anal. Chim. Acta. 2004, 510, 169–174. [Google Scholar]
- Hames, B.B.; Higgins, S.J. Nucleic Acid Hybridisation—A Practical Approach; Oxford University Press: New York, NY, USA, 1985; p. 78. [Google Scholar]
- Jin, P. Voltametric detection of DNA hybridization using a non-competitive enzyme linked assay. Biochem. Eng. J. 2007, 35, 183–190. [Google Scholar]
- Metzenberg, S. Working with DNA: The Basics; Taylor & Francis Group: Florence, KY, USA, 2007. [Google Scholar]
- Zhu, N.; Cai, H.; He, P.; Fang, Y. Tris(2,2′-bipyridyl)cobalt(III)-doped silica nanoparticles DNA probe for electrochemical detection DNA hybridization. Anal. Chem. 2005, 481, 181–189. [Google Scholar]
- Teles, F.R.R.; Fonseca, L.P. Trend in DNA biosensors. Talanta 2008, 77, 606–623. [Google Scholar]










| Immobilization matrix | Transduction method | Dynamic range (M) | Detection limit (M) | Reproducibility (RSD%) | Ref. |
|---|---|---|---|---|---|
| Acrylic microspheres | Amperometry | 1.0 × 10−16 to 1.0 × 10−8 | 9.46 × 10−17 | 2.86 | This work |
| CeO2/chitosan composite film | Amperometry | 1.59 × 10−11 to 1.16 × 10−7 | 1.0 × 10−11 | 4.04 | [3] |
| Poly-l-lysine films | Electrochemical impedance | 1.0 × 10−12 to 1.0 × 10−7 | 3.1 × 10−13 | 3.16 | [25] |
| Chitosan/nano-V2O5/MWCNTs | Amperometry | 1.0 × 10−11 to 1.0 × 10−6 | 1.76 × 10−12 | 3.0 | [28] |
| Poly-2,6-pyridine-dicarboxylic acid film | Amperometry | 1.0 × 10−10 to 1.0 × 10−5 | 12.4 × 10−11 | - | [29] |
| Fe2O3 microspheres/polyaniline nanofibers | Electrochemical impedance | 1.0 × 10−13 to 1.0 × 10−7 | 2.1 × 10−14 | 3.58 | [32] |
| Polyaniline nanofiber/ Au microspheres | Amperometry | 1.0 × 10−13 to 1.0 × 10−6 | 1.9 × 10−14 | - | [33] |
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Ulianas, A.; Heng, L.Y.; Hanifah, S.A.; Ling, T.L. An Electrochemical DNA Microbiosensor Based on Succinimide-Modified Acrylic Microspheres. Sensors 2012, 12, 5445-5460. https://doi.org/10.3390/s120505445
Ulianas A, Heng LY, Hanifah SA, Ling TL. An Electrochemical DNA Microbiosensor Based on Succinimide-Modified Acrylic Microspheres. Sensors. 2012; 12(5):5445-5460. https://doi.org/10.3390/s120505445
Chicago/Turabian StyleUlianas, Alizar, Lee Yook Heng, Sharina Abu Hanifah, and Tan Ling Ling. 2012. "An Electrochemical DNA Microbiosensor Based on Succinimide-Modified Acrylic Microspheres" Sensors 12, no. 5: 5445-5460. https://doi.org/10.3390/s120505445




