Targeting agr- and agr-Like Quorum Sensing Systems for Development of Common Therapeutics to Treat Multiple Gram-Positive Bacterial Infections
Abstract
: Invasive infection by the Gram-positive pathogen Staphylococcus aureus is controlled by a four gene operon, agr that encodes a quorum sensing system for the regulation of virulence. While agr has been well studied in S. aureus, the contribution of agr homologues and analogues in other Gram-positive pathogens is just beginning to be understood. Intriguingly, other significant human pathogens, including Clostridium perfringens, Listeria monocytogenes, and Enterococcus faecalis contain agr or analogues linked to virulence. Moreover, other significant human Gram-positive pathogens use peptide based quorum sensing systems to establish or maintain infection. The potential for commonality in aspects of these signaling systems across different species raises the prospect of identifying therapeutics that could target multiple pathogens. Here, we review the status of research into these agr homologues, analogues, and other peptide based quorum sensing systems in Gram-positive pathogens as well as the potential for identifying common pathways and signaling mechanisms for therapeutic discovery.1. Introduction to Staphylococcus aureus and agr
1.1. Introduction
The contribution of communication systems within human bacterial pathogens to gene regulation has significantly altered our comprehension of how pathogens adapt to specific niches to promote disease. Cell-to-cell communication mediated by quorum sensing (QS) within a single species coordinates cooperative behavior to enhance survival under stress, alter metabolism, and savage host tissues and immune defenses. The accessory gene regulator (agr) operon-encoded QS system in Staphylococcus aureus is one of the most well studied communication schemes of human bacterial pathogens and numerous reports have demonstrated that QS is critical to the pathogenic abilities of this Gram-positive (G+) bacterium. The sensing of, and response to, the agr-encoded auto-inducing peptide pheromone (AIP) rapidly changes the expression of hundreds of genes to promote invasive infection and virulence in host tissues [1,2]. In fact, transcriptional analyses of isolates from skin and bone abscesses clearly reveal an important role for agr in acute human infections [3]. In contrast, agr dysfunctional isolates are associated with chronic infections and represent a minority of clinical isolates [4]. While these isolates are capable of colonization [5] and nasal carriage is associated with the development of these infections and is postulated to be their source [6], agr dysfunctional isolates do not persist in natural populations, indicating that agr mutants do not contribute to transmission where S. aureus infections are endemic [7]. Additionally, when agr-deletion (Δagr) strains are tested in various infection and pathogenesis models in vivo, the bacteria may colonize but disease is attenuated [8–12], and clearance of individual S. aureus cells by host defenses is enhanced [9,13].
We and others are actively investigating host defense mechanisms that interfere with S. aureus agr-mediated communication with the goal of identifying therapeutic targets that limit disease and control infection without engendering resistance due to selective growth pressure [14–17]. Importantly, recent studies employing both traditional methods and bioinformatics techniques have revealed that G+ bacterial pathogens across the phylum of Firmicutes encode and express either homologues and analogues of the agr operon or similar QS systems that use small peptide “quormones” to regulate pathogenesis [18–24] (Table 1, see [25] for a description of the Quorumpeps database, available at http://quorumpeps.ugent.be, which provides multiple tools for investigating peptide quormones). Together these observations hint at the potential for development of anti-virulence compounds that are efficacious in numerous G+ pathogens. Whereas a single compound has been reported to inhibit common communication systems equally across multiple Gram-negative (G-) pathogens with therapeutic benefit [26], an anti-QS compound efficacious in vivo for multiple G+ pathogens has not yet been described. Anti-virulence strategies employing either drugs or vaccines could be significant adjuncts to the use of antibiotics in the treatment of infectious diseases [14,15,27,28]. Therapeutics that target virulence could aid antibiotic stewardship by limiting exposure of pathogens to antibiotics that drive resistance. To accomplish this goal, they could replace the use of prophylactic antibiotics sparing both exposure of the pathogen to antibiotic selection and disruption of the host microbiota. In addition, they could be used in lieu of antibiotics in clinical situations like uncomplicated skin and soft tissue infections in normal adults where host systems are sufficient to clear infection following surgical incision and drainage [29].
Moreover, they could aid existing antibiotics by facilitating host-dependent clearance of the pathogen rendered avirulent by the drug or antibody. Intriguingly, the development of a compound that works for multiple pathogens would increase the clinical utility of this approach. While the development of resistance to QS inhibitors has been postulated, recent studies in the G- pathogen Pseudomonas aeruginosa suggest that QS-insensitive mutants, which would be resistant to QS inhibiting anti-virulence therapies, form self-limiting populations in diseases where QS is required for pathology and dissemination [69,70] Thus, QS inhibition in G+ pathogens could be therapeutically beneficial without contributing to the spread of QS mutants. Here, we review what is currently known about the similarities in structure and function of agr, agr-like systems, and other peptide based quorum sensing systems across several human pathogens with the intent to highlight possible molecular targets for chemotherapeutic intervention against G+ bacterial quorum sensing.
1.2. Structure and Function of the agr Operon and AIP
Substantial work has gone into understanding how the various Agr components within S. aureus interact (see Novick and Geisinger 2008 [71], and Thoendel et al. 2011 [2] for in-depth reviews). Briefly, the four genes in the agr operon are read as a single polycistronic message in the transcriptional order of agrBDCA (see Figure 1). AgrD is a short polypeptide which includes the protein sequence for AIP, but AgrD undergoes significant processing before the signal peptide is released. In the model proposed by Thoendel et al. [2], AgrD associates with the inner leaflet of the plasma membrane where it serves as the ligand for AgrB [72]. The cytoplasmic face of AgrB has several functions, including a sequence-specific protease that likely recognizes conserved residues that flank the central AIP sequence in both directions. AgrB cleaves the C′ terminus of AgrD and then catalyzes the formation of the thiolactone ring that defines the AIP structure (Table 1, Figure 2(a)) [73]. AgrB has been suggested to aid in translocating the partially processed AgrD to the outer leaflet of the plasma membrane, but this remains unclear [2,74]. The type I signal peptidase SpsB completes the N′ terminal cleavage of AgrD, releasing fully formed AIP from the cell surface [75].
The receptor for AIP, AgrC, is an integral membrane protein, and a member of the class 10 receptor histidine protein kinases (HPKs) with homology to members of the LytST/R two-component regulatory system (2CRS) family. AgrC has a high affinity for AIP, with activation EC50 values in the low nanomolar range. This exquisite sensitivity may serve as a defense mechanism for S. aureus, as we have previously shown that a single cell enclosed in a small space, such as the phagosome of a macrophage, can secrete sufficient AIP within a short time to trigger the agr-mediated QS transcriptional program [76]. AgrC can dimerize without binding AIP and binding even a single AIP molecule is sufficient to activate the receptor complex [77]. The two cytoplasmic HPK tails of AgrC cross-phosphorylate to allow AgrC to in turn activate the response regulator module AgrA.
Like AgrC, the transcription factor AgrA shares significant homology with LytST/R family members, and the consensus binding sequence has been identified for AgrA, with unsurprising similarities to the sequence for LytTR binding [78]. The best known target for AgrA binding, and the region most important for virulence regulation in S. aureus, is the divergent promoter region P2/P3 which controls transcription of the agr operon and the RNAIII/hld gene, respectively. Due to a two-nucleotide substitution in the consensus binding sequence in the P3 promoter, phospho-AgrA displays a preference for P2 at low concentrations, promoting an autoactivation circuit for agr [78]. Such positive feedback loops have been identified in other examples of QS in numerous species, and these circuits foster accelerated accumulation of the QS pheromone and rapid activation of the target transcriptional program. At high concentrations, active AgrA may slightly prefer P3 over P2 [79], but both agr and RNAIII are actively transcribed.
The sequence of the agr operon is not the same in all S. aureus strains. Four common variants, designated agr I through agr IV (Table 1), have been identified in clinical isolates and these are individually associated with various clusters of S. aureus isolates according to pulsed-field gel electrophoresis (PFGE) type; for example, the most common CA-MRSA isolate worldwide, PFGE type USA300, is an agr I strain (Figure 2(a)), while the PFGE type USA400 strain MW2 is agr type III. These types have arisen due to the presence of a hypervariable region in the agr operon, which encompasses the 3′ region of agrB, all of agrD, and part of the 5′ region of agrC [43]. Recent work from Geisinger et al. established that the four agr genotypes function with different kinetics, in that each agr allele activates gene regulation at distinct times during log phase growth and by varying degrees [80].
The AIPs encoded by the four agr types have slightly different lengths and peptide sequences (Table 1). The conserved central cysteine residue required for formation of the thiolactone bond is followed by four more amino acids to the C′ terminus (Figure 2(a)). The length of the N-terminal “tail” varies and all known AIP variants in S. aureus and other species range from 7 to 9 residues in toto. Of the four AIP molecules, AIP 1 and AIP 4 differ by a single amino acid at position 5 (Table 1) and have some cross-reactivity for their cognate AgrC receptors. However, the differences in peptide sequence between AIP 1/4, 2 and 3 allows these molecules to function as cross-type antagonists for AgrC activation, with low nanomolar IC50 values, apparently granting competitive advantages to competing strains of S. aureus [30,81,82]. The therapeutic potential inherent in these natural peptide analogs will be discussed below.
1.3. Gene Regulation by agr and RNAIII
The agr/RNAIII virulence regulon is one of the largest and most complex prokaryotic transcriptional programs currently known and reports continue to emerge of new virulence factors expressed under the agr regulatory scheme. Broadly, activation of agr/RNAIII triggers the transition from production of surface-bound proteins to the secretion of soluble exotoxins and degradative enzymes. Much of this complexity stems from the chromosomal juxtaposition of the agr QS system and the RNAIII molecule, combining the regulatory network of a 2CRS with a pleiotropic regulatory RNA (Figure 1, [1,83]). Appropriate to such an energetically taxing transcriptional program, upstream regulation of the agr operon is temporally, nutritionally, and spatially complex.
The two key transcription factors that positively regulate expression of the agr operon are staphylococcal accessory regulator A (SarA) and AgrA itself [84–86]. Deletion of sarA dramatically alters both the level and timing of RNAIII production in response to AIP signaling by markedly reducing transcription of the agr operon and deletion of agrA completely abolishes both RNAIII and agr mRNA [86]. This finding demonstrates that AgrA binds the P2 sequence at constitutively low levels even in the absence of an active AIP-driven signal through AgrC and that AgrA may be the most important element in the initiation of transcription at P2. MgrA, a transcription factor involved in control of multidrug resistance and cell wall turnover, along with other sar family members SarU and SarZ, all contribute positively to agr expression [87–89]. MgrA also promotes expression of SarX, a repressor of agr, giving S. aureus an “off-switch” for the agr signaling cascade [88]. The metabolic sensor and transcriptional repressor CodY prevents agr transcription under conditions of enriched nutrition; once key nutrients are depleted below a threshold, CodY can no longer bind to the P2/P3 region, derepressing agr [90,91]. σB, an alternative transcription factor produced as part of the broad stress response in S. aureus, strongly promotes biofilm production and is an additional inhibitor of agr, although its mechanism of action remains unclear [2,92,93]. In addition, redox signaling controls AgrA-DNA interactions via oxidation of a critical cysteine residue within AgrA that terminates its transcriptional activity [94].
For all of the regulatory checks and balances upstream of agr, AgrA as a transcription factor directly regulates only a few promoter sequences in addition to P2/P3 (Figure 1), notably the α- and β-phenol soluble modulins (PSMs) [1]. PSMs are alpha-helical, amphipathic cytolytic peptides that range in size from ∼20 to 50 aa. They are genome encoded and most strains produce PSMs. S. aureus produces 4 PSMα peptides encoded in the psma operon that are homologous to the RNAIII-encoded δ-toxin and two PSMβ peptides encoded in the psmb operon [9]. This RNAIII-independent but agr-dependent regulation of these multifunctional cytolytic peptides suggests that QS regulation of PSMs evolved prior to the addition of a wider control of virulence to the agr regulon [1]. This may well have occurred by expansion of the RNAIII-encoding region around the initial gene for δ-toxin. While these data have been confirmed primarily in the strain MW2, they suggest that the RNAIII-independent but agr-dependent suppression of carbohydrate metabolism confirmed by microarray may reflect a wider contribution of agr, possibly through AgrA, to metabolism [1].
RNAIII is one of the largest known prokaryotic regulatory RNA (rRNA) molecules and facilitates the greater part of agr-dependent virulence via interactions mediated by specific regions of RNAIII secondary structure (for a recent review see Felden et al. [83]). The only protein translated from RNAIII is the small peptide δ-toxin, as the hld gene is encoded within the first half of the sequence of RNAIII, but the protein is not produced until significantly after RNAIII has been transcribed [95]. For some genes, like α-hemolysin, RNAIII binding derepresses translation initiation by opening the ribosomal binding site in the target mRNA. Various structures within RNAIII also work via antisense RNA mechanisms to prevent translation initiation in target mRNAs which often accelerates mRNA turnover by RNAse digestion of extended dsRNA structures. Many of the target genes shut down by RNAIII through post-transcriptional mechanisms are key members of other regulatory pathways, like repressor of toxins (rot), members of the Sar and MarR families of transcriptional regulators, and multiple 2CRS modules that can variously promote and repress expression of hundreds of toxins, proteases, adhesion factors and metabolic pathways [1,2,83,96]. RNAIII also directly affects the transcripts of many genes whose expression is regulated by Rot and other transcription factors, so that total cellular mRNA turnover increases dramatically after AIP signaling. This combination of direct and indirect regulation by RNAIII rapidly transforms S. aureus cells from a sessile, stationary state into an aggressive planktonic form capable of invasive infection.
Microarray analyses of the agr-regulated transcriptome in various S. aureus strains have identified core virulence factors that depend either on RNAIII or AgrA for regulation [1,31,32]. These include genes for PSMs, capsule production, α-hemolysin, cytolytic toxins, proteases and lipases. However, there is considerable variability in the total number of genes regulated as well as their contribution to genetic programs that regulate growth and survival. This fact suggests that inclusion of genes into the agr-regulated transcriptome could represent an adaptive response to alter not only virulence factors but also metabolic pathways required for survival within distinct niches.
1.4. agr in S. aureus Disease
Agr involvement in disease is complex, and numerous groups have worked to clarify the role of agr in S. aureus disease, but varying infection and disease models, and variable patient populations, cloud the issue. Nevertheless, it is becoming clear that agr plays an important role in promoting acute and aggressive infection in both animals and humans. For example, murine dermonecrosis is an acute model of invasive skin and skin structure infection characterized by development of a subcutaneous abscess and a superficial ulcer after subdermal injection of S. aureus. Deletion of agr blocks ulcer formation and reduces abscess size [10], a result also observed when PSMαs are deleted [9] or α-hemolysin is blocked by vaccination [97]; both gene products are tightly regulated by agr in many S. aureus strains. Of all the agr-regulated toxins, the PSMs are among the most important in enhancing the survival and dissemination of S. aureus in invasive infection. PSMαs are cytolytic against neutrophils, although this enhances expression of inflammatory molecules in mouse models of dermonecrosis [9] Agr is also required for development of invasive, necrotizing pneumonia in mice [10,98,99], although the inflammatory response in the lung does not require agr function [98]. The pathology of S. aureus osteomyelitis in rabbits is reduced in the absence of agr [11], and the early colonization of heart valves is markedly reduced in rabbits intravenously injected with a Δagr strain [100].
These findings derive from acute infection models, with readouts in hours or days after infection, and they contrast with the literature about chronic S. aureus infections in humans. Patients suffering from persistent S. aureus bacteremia are more likely to be carrying agr-deficient strains, a pool of point mutations which is also associated with a higher incidence of chronic infective endocarditis [101]. Similar studies confirmed that S. aureus pools from chronically infected patients accumulate mutations rendering agr inoperative, and these isolates are associated with increased hospitalization rates [5,7,102]. Another group reported that loss of expression of Panton-Valentine leukocidin and increased expression of protein A, a phenotype correlating to a loss of agr function, was associated with well-established, active S. aureus infection [3]. A recent report by Gagnaire et al. found a strong association between the duration of chronic infections, the loss of agr function, and an increase in glycopeptide resistance [4]. Overall, agr deficiency is more commonly found in hospital-acquired methicillin resistant S. aureus (HA-MRSA) than in community acquired MRSA (CA-MRSA) isolates [5,102] which may be due in part to the finding that expression of methicillin-resistance gene mecA, commonly found in HA-MRSA isolates, inhibits agr function [103].
Aggressive CA-MRSA infections have been hypothesized to require agr expression to maximize host-to-host transmission and early colonization [7,8], but that HA-MRSA infections may not rely on agr for transmission because surgical and other medical procedures can bypass human infection barriers such as the skin and mucus membranes. Alternatively, constant activation of the agr regulon is suggested to be energetically expensive. Therefore, transition to a metabolically favorable agr-negative status is beneficial for long-term survival once innate host defenses have been eluded [8,104,105].
One noted phenotype associated with the loss of agr is the development of thicker biofilms [93,106–108], a behavior that likely stems from agr regulation of PSMβ and/or protease production. PSMβs are crucial to biofilm maturation and dissemination in S. aureus and foci of agr-mediated PSM promoter activation can be observed within static biofilm structures [109]. Single S. aureus cells can produce sufficient AIP to activate quorum sensing within confined spaces [76], and this ability may explain how agr-mediated QS controls S. aureus dissemination from within mature biofilms. Thicker, agr-deficient biofilms with limited dissemination may serve as incubators for the development of antibiotic resistant strains [110,111]. In cases of S. aureus biofilm formation on medical implants and indwelling lines, loss of agr could prove somewhat beneficial as this would inhibit biofilm maturation and retard bacterial dissemination [106,109], thus reducing the spread of infection.
1.5. Host and Molecular Antagonism of S. aureus agr
Mounting evidence suggests that the story of S. aureus and humans is one of co-evolution and co-adaptation [112,113]. While S. aureus may thrive in the anterior nares as a commensal, colonization elsewhere in the body is met with vigorous immune response, as demonstrated by the development of defenses that target many of the effectors of S. aureus pathogenesis. Our adaptive immune system works to thwart chronic or recurrent infections, often by producing antibodies against capsular polysaccharides or exotoxins. Ongoing efforts to develop suitable vaccine epitopes derived from many virulence factors have met with mixed success [97,114–116]; their failures to protect against a broad assortment of S. aureus strains are likely due to the marked genetic variability and pathogenic profiles between isolates. As a small peptide, AIP is not particularly immunogenic and thus the host adaptive immune system does not regulate QS. To address whether adaptive immunity could provide protection by targeting QS, Park et al. demonstrated that a synthetic analog of AIP-4, when linked to a strongly immunogenic hapten, could be used to develop an agr-blocking antibody in mice. This antibody prevented tissue damage in a mouse model of dermonecrosis when it was passively administered at the time of infection with the AIP 4 strain RN4850 [117]. This finding confirmed that specific targeting of agr-mediated QS is a valid therapeutic approach to limit pathogenesis, although it requires development of a multi-type vaccine to be broadly effective against agr-mediated diseases.
We have other innate immune defenses that work to quench or disable the agr-driven signaling cascade. While low pH is an effective physical barrier and is also used by phagocytes to digest many bacteria, S. aureus is notoriously acid tolerant, with an acid shock response that allows it to survive prolonged exposure to low pH [118]. However, it was observed 20 years ago that low environmental pH inhibits agr-mediated pathogenesis [119] due to the acid shock transcriptional program down-regulating agr expression [120]. Phagocytes, neutrophils and macrophages also produce reactive oxygen species like hypochlorite and superoxide which, in addition to damaging bacterial membranes and extracellular proteins, also oxidize and inactivate AIP-1 [121], the agr type most commonly found in S. aureus infections. In addition, there are two common components in blood which can quench AIP signaling: hemoglobin and apolipoprotein B (ApoB) [122,123]. ApoB found in low- and very-low-density lipoproteins (LDL/VLDL) binds AIP directly and sequesters it thus preventing ligand activation of AgrC [123]. The α- and β-chains of hemoglobin also antagonize agr signaling, most likely by inhibiting AIP secretion following hemoglobin-mediated membrane disruption [122,124,125]. These innate defense mechanisms are employed when inflammatory cytokines induce leaky vascular endothelium and serum components enter the site of infection.
Besides host mechanisms that counter agr-mediated QS, there are numerous examples of molecules produced by other strains of S. aureus and by other species of bacteria that interfere with agr signaling. As described above, different AIP types can antagonize non-cognate AgrC receptors to prevent activation of the agr regulon, even after only a single application of inhibitory AIP [2,30,126]. Many other Staphylococcus species are also agr+ (Section 2.1) and some research has suggested that S. epidermidis AIP molecules antagonize several of the S. aureus AIP genotypes in vitro [127,128], but in vivo experiments have not yet confirmed this finding [129]. Some bacterial species disrupt agr-mediated QS in S. aureus using molecules distinctly unlike AIP. Several reports demonstrate that members of the Lactobacillus genus produce anti-QS molecules against both G- and G+ organisms, and Li et al. recently reported on a dicyclic peptide from L. reuteri that quenches agr-mediated production of toxic shock syndrome toxin across multiple agr genotypes [130], although the mechanism of interference is currently unknown. Pseudomonas aeruginosa produces at least two molecules that interfere with agr-mediated QS in S. aureus; one is a long-chain acylhomoserine lactone (AHL) that P. aeruginosa employs for its own QS processes [131], and another is an oxidized quinoline [132]. AHL binds to a saturable but unknown receptor to inhibit AIP signaling, and the quinoline may function through destabilizing membrane proteins to interrupt cell functions. In light of the dawning appreciation that humans are host to polymicrobial communities across all of our diverse epithelia, these observations support the concept of probiotic treatments and therapies derived from inter-species bacterial warfare.
2. agr and Analogous Peptide Quormone Systems
2.1. agr Homologues
Elegant in its organization and compact in its function, agr is the prototype for small peptide quormone QS systems in G+ bacteria. As described previously, the agr operon encodes for the complete set of functions necessary for a density-dependent master regulator of bacterial programming. In S. aureus, agr activation effects a marked shift in the transcriptome and in the metabolic capability of responding cells, and this is reflected in the reported effects of agr signaling in those species with agr homologues, as described below. In comparison, non-agr peptide quormone systems do not trigger changes in species behavior as dramatic as those seen in S. aureus, even though these other systems play an important role in promoting virulence. Below we describe the role of agr-mediated QS processes in controlling virulence behaviors in several species.
2.1.1. Staphylococcal agr
The agr operon is found throughout the genus of Staphylococcus in pathogenic, commensal, and environmental species with genetic variation in the operons closely following the 16S rRNA phylogenetic tree [24]. Although the function of agr has not been explored in all of these species, there are several notable staphylococcal pathogens besides S. aureus where expression of virulence factors is regulated by agr. Of these Staphylococcus species, S. epidermidis is currently recognized as the most clinically important nosocomial pathogen, due in part to its ubiquitous presence on human skin and mucus membranes [133]. Unlike S. aureus, S. epidermidis is rarely associated with invasive infection. Instead, S. epidermidis forms tenacious biofilms growing on in-dwelling lines and surgical implants, so that most infections are sub-acute or chronic. However, agr is important for S. epidermidis virulence [34,35], and there are at least three different AIP types for S. epidermidis (AIP-Se, Table 1). Agr signaling in S. epidermidis induces production of RNAIII/δ-toxin, exoproteases, a lipase, and PSMs [19,34,36], but also promotes the expression of numerous adhesins including polysaccharide intercellular adhesin PIA [33]. An agr-deleted strain was unable to produce PSMs and elicited less neutrophil migration and inflammatory cytokine production compared to wild-type S. epidermidis [36]. Many of the genes affected by AIP-Se signaling belong to metabolic pathways, suggesting that agr-mediated QS prepares S. epidermidis for the stationary, sessile lifestyle within a biofilm [19,38]. Intriguingly, in addition to increasing biofilm deposition agr regulation also enhances biofilm remodeling, maturation and dissemination through expression of PSMβs [134]. In contrast, Dai et al. recently reported that S. epidermidis isolates recovered from in-dwelling catheters produced thicker biofilms and exhibited more autolytic behavior in concert with down-regulation or deletion of the agr operon and upregulation of the autolysin atlE [135] suggesting that agr regulation of biofilm formation is carefully counterbalanced by other factors. From these reports it is clear that agr regulates pathogenesis in S. epidermidis, making the agr QS system an attractive target for pharmacotherapeutic control of S. epidermidis infection.
S. lugdunensis is a commensal skin organism but is also a highly virulent, opportunistic pathogen responsible for endocarditis, septicemia, osteomyelitis, and skin and soft tissue infections. Known virulence factors include δ-toxin, lipase and a polysaccharide capsule, and some reports suggest that different strains are positive for a membrane-bound coagulase or α-toxin [136,137]. S. saprophyticus is an important pathogen in urinary tract infections and it expresses numerous adhesins that bind to extracellular matrix proteins although it produces no known toxins. Both species produce AIP (Table 1) and their agr/RNAIII are organized similarly to those of S. aureus and S. epidermidis, but the hld gene for δ-toxin is not embedded within RNAIII in S. lugdunensis or S. saprophyticus [40,43,138]. At this time there are no animal models of pathogenesis for either species, so the relevance of agr-mediated QS to in vivo virulence regulation is unknown.
The S. intermedius group includes three species that, like S. aureus, straddle the commensal-pathogen divide in their respective hosts. The best studied of these, S. pseudintermedius, is a gastrointestinal commensal in dogs, but it is the major cause of canine pyoderma and can very rarely infect humans [139]. Along with recent taxonomic reclassification separating S. pseudintermedius from S. intermedius [42], a complete sequence of the S. pseudintermedius genome has recently been published, demonstrating that it carries genes for multiple toxins, many of which are highly homologous to similar toxins in S. aureus, including leukotoxins and α-, β-, and δhemolysins [140]. S. pseudintermedius AIP was first identified in 2003, and biochemical characterization determined that instead of forming a thiolactone like that found in AIP-Sa, the nonamer AIP-Si relied on a central serine residue to form a lactone ring (Table 1, Figure 2(b)) [141]. This molecule is functional for QS in S. pseudintermedius, and substitution of a cysteine for the key serine molecule reduced the expression of RNAIII [41]. Three other genetic variants of AIP have since been identified in S. pseudintermedius (Table 1, [41,42]), but whether agr-mediated QS controls pathogenesis in this species remains to be determined.
Genomic and proteomic analyses of other Staphylococcus species have demonstrated that numerous clinical and environmental isolates include an agr operon and likely produce their own version of AIP [18,30]. However, lacking medical or veterinary impact, the role of QS in modulating pathogenesis in these other species has not been examined.
2.1.2. Clostridial agr
Species in the order Clostridiales generate some of the most potent bacterial toxins known, ranging from the severe neurotoxins of C. botulinum and C. tetani, to a range of aggressive proteases produced by C. perfringens. While the role of agr in virulence programming in some clostridial species has yet to be determined, it is evident that agr controls toxin production in C. botulinum and C. perfringens. Pathogenic Clostridium species are generally resistant to multiple antibiotics, thanks to the slow growth of most species, obligate anaerobic life style, and their ability to form endospores, a non-metabolizing and durable cell form highly resistant to heat, desiccation, radiation, oxidation and numerous other chemical means of microbial control. Therefore the role of QS in controlling toxin production and metabolic alteration in these species is receiving a great deal of attention and several recent reports have demonstrated that agr-mediated signaling regulates both toxigenesis and sporulation (Table 1).
Botulism intoxication in healthy adults is not normally due to infection with C. botulinum, although colonization of the gut is the accepted mechanism for infant botulism. In adults, rather, consuming preserved food initially contaminated with botulism spores leads to ingestion of the botulism neurotoxin, of which there are several types. Cooksley et al. demonstrated that Group 1 C. botulinum expresses two AgrBD homologues (Table 1) and the two signaling peptides play different roles in controlling pathogenesis [44]. mRNA expression of both agrBD1 and agrBD2 peaks late in exponential growth and markedly decreases in stationary phase growth, typical of genes involved in QS. But while agrD1 expression regulates sporulation, agrD2 controls toxin production during growth in vitro. The cognate sensors or 2CRS for these two agrD homologues have not yet been described.
C. difficile infection is associated with severe diarrhea due to clostridial overgrowth and tissue adhesion after the patient's normal gastrointestinal flora has been eliminated due to antibiotic therapy. Sequencing the complete C. difficile genome has revealed the presence of both a complete agrBDCA operon as well as a second copy of agrBD ([45,142], Table 1), but currently their roles in pathogenesis in vivo are unknown.
Infection by C. perfringens can also lead to gastrointestinal distress, but this species is also the causative agent for gas gangrene. Deep wounds can give rise to anaerobic conditions, and thus provide an ideal environment for infection by environmentally ubiquitous C. perfringens endospores. Germinating C. perfringens produces numerous proteases and toxins to digest host tissues, which in turn supply necessary amino acids and nutrients for which C. perfringens lacks the necessary biosynthetic pathways. This exotoxin-mediated release of nutrients in situ rapidly accelerates bacterial growth, further complicating the treatment of C. perfringens infection. Several reports have demonstrated that agrBD (Table 1) is important for toxin production and sporulation in many of the pathogenic groups of C. perfringens, including both human and veterinary isolates [21,46,47,143,144]. Epsilon-toxin and beta 2 toxin production by ΔagrB Type B strains was reduced compared to wild-type bacteria in in vitro assays using enterocyte-like CaCo2 cells [47,144], and agrB controls in vivo toxicity and colitis in a rabbit intestinal loop model with a Type C strain [143]. Remarkably, in at least one Group A strain agrBD regulates expression of genes on a plasmid [21], a novel result previously unheralded in the agr literature. The cognate receptor for C. perfringens AIP has not been conclusively identified, but in Type A strains the 2CRS virRS, which regulates expression of several toxins and virulence factors through the regulatory RNA molecule VR-RNA [145,146], may be involved in agr-mediated QS and pathogenesis [46]. However, the link between agr and vir is unclear in other strain types, as VirRS does not appear to be involved in agrBD-mediated toxin production in Type B or D strains of C. perfringens [47]. While more research into the role of QS in C. difficile pathogenesis is required, it is clear that developing treatments to inhibit agr-controlled pathogenesis in C. perfringens and C. botulinum could prove beneficial to reducing disease severity.
2.1.3. Enterococcus
E. faecalis resides in the human gastrointestinal tract as a member of the endogenous flora, but it can persist in the face of numerous environmental hazards, and vancomycin-resistant strains are nosocomial pathogens of increasing importance. E. faecalis infections of the urinary and seminal tracts include urethritis, prostatitis and nephritis, and it can also invade the kidneys, giving rise to bacteremia and endocarditis. Virulence factors, several of which are regulated by QS processes, include numerous adhesins and biofilm components but the best characterized are proteases, gelatinase (GelE) and a serine protease (SprE), that enable E. faecalis to invade host tissues. In particular, gelatinase, a metalloprotease, aids in digesting host extracellular matrix proteins and also promotes biofilm dissemination [147,148]. The fsr operon regulates QS-mediated pathogenesis through control of GelE and SprE expression, down-regulation of biofilm components, and alterations in metabolic function [149–151] (Table 1). FsrD, the propeptide for the GBAP quormone, (Table 1) is cleaved, cyclized and exported by FsrB [50,152]. Fsr organization differs from agr as fsrD is contained entirely within the sequence of fsrB in a different frame, and is read from a distinct transcriptional start site [48]. Furthermore, GBAP forms a lactone ring four residues larger than the prototypical AIP thiolactone (Figure 2(c)). However, FsrA and C are highly homologous to AgrA and C, respectively, and there are two genes adjacent to fsr that are FsrA targets [50,152], echoing the operon and regulon structure of agr. Deletion or disruption of fsr appears to trigger a stress-response phenotype [153], enhances host survival in a mouse model of infectious peritonitis [50] and reduces tissue damage in a rabbit model of experimental endopthalmitis, suggesting that disruption of fsr regulation could have therapeutic benefits [148].
2.1.4. Listeria
Listeria monocytogenes is the etiologic agent of listeriosis, a food-borne infection that causes a wide range of severe illnesses in the elderly, neonates and immunocompromised patients, and the agr operon assists this pathogen in persisting through harsh environments. L. monocytogenes forms biofilms on a broad variety of industrial abiotic surfaces including stainless steel and plastic, and these biofilms provide reservoirs for colonization of raw or partially-processed foodstuffs. Virulence factors include the toxins listeriolysin O (LLO), hemolysin, phospholipase, and several cell-surface proteins involved in promoting phagocyte internalization and actin polymerization. L. monocytogenes evades host immune defenses by breaching the vesicle membrane of a phagocytosing cell, where ActA-catalyzed formation of actin “tails” propels the bacteria through the cytosol and into a neighboring cell.
The L. monocytogenes genome encodes for a complete agr operon (Table 1), which plays a critical role in promoting biofilm production, expression of adhesion factors, and internalins, but does not affect growth (Table 1 [52,53]). Listeria lacks RNAIII, so the agr-mediated QS transcriptional program is controlled by direct AgrA binding of promoter sequences, and there is evidence that several dozen genes are both positively and negatively regulated [52]. A role for agr-mediated QS in virulence is less clear and may depend upon the growth phase of the inoculum administered. Mouse tail vein injections of a ΔagrA mutant grown to mid-exponential phase had attenuated virulence compared to the parental strain [51], whereas infection with a similar mutant grown to stationary phase demonstrated that loss of AgrA had no effect upon systemic virulence [154]. In contrast, in vitro cell invasion assays demonstrated that agr plays no role in maintenance of the intracellular life cycle that characterizes L. monocytogenes disease [51,154]. Many L. monocytogenes isolates are agr+ [23] suggesting that this locus plays an important role somewhere in the Listeria life cycle even if it is not essential for human infection, and that application of anti-agr therapeutics in industrial settings associated with food processing could limit the spread of listeriosis.
2.1.5. Lactobacillus
No Lactobacillus species are yet identified as pathogens of either humans or animals, but they are of great interest in both food production and the burgeoning field of probiotic therapy. This approach aims to prevent pathogenic infections by prophylactically colonizing tissues with transient or potentially commensal bacteria with antimicrobial properties. Numerous studies have investigated the ability of sundry Lactobacillus species and similar lactic acid bacteria to generate anti-bacterial peptides such as bacteriocins, and other compounds like reuterin and nisin [155–157]. Among these, at least one species, L. plantarum, encodes multiple agr homologues [158]. The lam operon, structured as lamBDCA, is an agr homologue that produces a thiolactone peptide (Table 1) and is involved in bacterial adhesion to abiotic surfaces [54]. QS-mediated control of bacteriocin production in Lactobacillus species is regulated by other analogous systems, which will be discussed below.
2.2. Other Peptide Quormone Systems
While a few G+ pathogens are reported to employ AHL-based QS systems to regulate growth and toxin expression, the rest of the pathogens in the Firmicutes with QS-mediated control of virulence rely on small peptides as the quormone molecules. Outside of the agr family of homologues described above, there are primarily two other families of signaling systems found to regulate virulence. The RNPP family, best studied in Bacillus species and in Enterococcus faecalis, controls the production of numerous toxins and extracellular virulence factors [159]. The com family of parallel QS systems found in Streptococcus species triggers significant changes in bacterial lifestyle and behavior that can significantly enhance or promote virulence. Although components in individual examples of each of these systems may lack homology to members of the agr family, there are striking functional and behavioral similarities across these three different systems that suggest targeting QS-mediated control of virulence in these species could prove fruitful in inhibiting disease in vivo.
2.2.1. Bacillus Systems
Bacillus anthracis produces a tri-partite cytotoxin and an anti-phagocytic capsule, and several strains of B. cereus are toxigenic and can cause brief but severe gastrointestinal distress. Research into the role of QS in virulence regulation in these pathogens is in its initial stages, but the action of peptide quormones in these species is not as clear-cut as with the agr operons. LuxS, a QS system best characterized in G- bacteria that uses AHLs as signaling molecules, plays a role in bacterial growth, and this system affects the expression of virulence factors in B. anthracis but its loss does not affect virulence [160–162]. In addition to the luxS system, numerous members of the Bacillus genus express members of the RNPP (Rap, NprR, PlcR, and PrgX) family of quorum sensing operons [56,60,163]. RNPP and agr share several crucial features, as both QS systems encode for a propeptide that is cleaved and secreted, and in the case of many RNPP members, the quormone is a heptapeptide [61,164]. Additionally, the cognate receptor has strong specificity for the co-evolved peptide [22,55]. But the two systems differ in that the RNPP peptides are linear, the processing enzyme(s) for the propeptide are not encoded in the same operon [165], and the signaling peptides must be re-imported into the cell via the Opp transporter before binding the cytoplasmic receptor [55,61].
In B. cereus, RNPP members regulate growth, sporulation, and toxin production (Table 1, [22,55,56]). The RNPP families regulate virulence in B. cereus, as it has been shown that a PlcR mutant strain caused less retinal epithelial cell death compared to wild-type bacteria in an in vitro model of endopthalmitis [57]. As shown in Table 1, there are five known B. cereus PapR signal peptide “pherotypes” analogous to the four agr types in S. aureus, and one report demonstrated that four of these pherotypes can cross-inhibit each other [164]. The fifth peptide, PapRa, was recently reported by Huillet et al. and plays a role in the oxidative stress response and sulfur metabolism during stationary phase [58] but its role in virulence, and its ability to interfere with other Pap quormones are yet to be determined. In B. thuringinensis, which is not pathogenic to humans but produces an insecticidal toxin, deletion of papR, which encodes the signal propeptide, reduced toxin production and the killing of susceptible insects [55]. Of note, NprR expression in B. thuringinensis is required for vegetative bacterial growth, biofilm formation and sporulation in insect cadavers [59], a novel finding demonstrating a role for quorum sensing behavior in post-pathogenic behavior. In light of this extant family of virulence-regulating QS systems, it remains to be determined whether an agr-like of QS system operon in Bacillus species operates in parallel to the RNPP family in regulating bacterial pathogenesis. Together, these studies suggest that at least 3 pathways in 2 QS systems contribute to pathogenic gene regulation in Bacillus species with significant potential for exploiting common strategies for modulation of biologic function.
2.2.2. Streptococcal Systems
Streptococcus pyogenes, also known as Group A β-hemolytic streptococcus (GAS), is a commonly encountered G+ pathogen, and it gives rise to suppurative infections that range from mild (pharyngitis) to severe (necrotizing fasciitis). Different isolates of GAS display distinct preferences for either the skin or the throat as their site of infection, and this bias may be affected by the particular peptide quormone QS system encoded by a given isolate. GAS produce a wide range of toxins and virulence factors, including streptolysins, superantigens, a polysaccharide capsule, and proteases, and production of several of these virulence factors is affected by peptide quormone signaling.
The streptococcal com system, historically the first QS-mediated behavioral program described in the literature [166], controls bacterial competence in numerous α-hemolytic streptocci. It consists of two com operons; the first encoding a protease and dedicated peptide export channel and the second including a propeptide and a 2CRS sensor and regulatory modules [64,167]. ComC is cleaved to release the C′ terminal competence stimulating peptide (CSP), which is a 16-mer linear peptide (Table 1). Other α-hemolytic streptococci may also carry com with species-specific variations [167], but it has been suggested that several S. pyogenes isolates may have evolved without the ability to detect CSP in order to limit genetic variability [64]. In S. mutans, CSP signaling also regulates the production of biofilms and bacteriocins, enhancing both pathogenicity and competitive survival in the oral cavity [168–170]. ComRS constitutes a second QS-regulated competence system in S. pyogenes and in non-pyogenic streptococci that promotes competence in stationary phase cultures [67,171]. After export from the cytosol, the short polypeptide ComS is cleaved into XIP (Table 1) and then imported into the cell where XIP binds ComR. This protein is part of the Rgg family of transcription factors activated by binding small signal peptides. Once bound to XIP, ComR in turn promotes transcription of both ComS and SigX, an alternative sigma factor that promotes competence. The XIP quormone promotes competence in chemically defined media, but in more nutritionally complex environments, XIP signaling may promote bacterial cell death [172], possibly enhancing biofilm structure and mass through release of structural components such as extracellular DNA.
While competence is not directly related to bacterial pathogenesis in the majority of α-hemolytic streptococci, the ability to utilize exogenous genetic material clearly enhances the ability of streptococcal species to survive in a wide range of environments and under stressful conditions. This concept is supported by the finding that non-opsonizing antibodies against S. pneumoniae polysaccharide capsular antigens enhance transformation efficiency in pneumococci [173], potentially increasing bacterial pathogenesis and improving bacterial viability. However, currently there is no evidence the com operon or streptococcal competence play direct roles in GAS pathogenesis either in vitro or in vivo.
Genes in the streptococcal fas operon share homology with the 2CRS components of agr, as FasA shares sequence homology with AgrA, and FasB and C are strongly related to AgrC, although the ligand has not yet been identified for FasB or C [63]. Significantly, activated FasA promotes transcription of FasX, a small, regulatory RNA that binds the 5′ untranslated region of the streptokinase A (SKA) mRNA to increase transcript stability and message half-life [65]. SKA is a well-characterized virulence factor for skin-trophic GAS isolates that converts human plasminogen into the fibrin-cleaving protein plasmin. FasX, or possibly FasA, also appears to increase bacterial adhesion to human epithelial cells, promote internalization, and enhances host cell apoptosis and infection-mediated expression of the pro-inflammatory cytokine IL-8 [174]. Whether fas is involved in QS-mediated transcriptional programming is currently unclear, as the environmental/nutritional sensor CodY positively regulates FasA production [175]. This link suggests that early steps in control of S. pyogenes pathogenesis may be regulated more by locale within the host rather than bacterial density. This observation is in contrast to the finding that CodY represses agr function in S. aureus [90,91].
The streptococcal invasion locus (sil) uses a peptide pheromone and a 2CRS to limit the ability of Group A and Group G streptococci to invade host tissues while it enhances Streptococcus survival in the host. Sil is characterized by three transcripts covering two operons and a standalone gene. The locus sits between two of its major regulatory targets, the blpM and blpU operons that encode bacteriocin-like peptides [62,176]. The first operon, silAB, comprises the regulatory and sensor-HPK modules of the 2CRS. The second, silEDCR, includes two parts of an ATP-binding cassette transporter (SilD and E) and SilCR is a propeptide homologous to ComC that is processed into a 17-residue linear pheromone peptide (Table 1). SilCR signals through SilB to activate SilA, which increases transcription of silEDCR, blpM and blpU [62,177]. SilA also promotes expression of the virulence factors streptolysin S, iron transporter SiaA, and serine protease ScpC in a growth-phase dependent fashion [178]. The third sil mRNA is silC, which overlaps much of silCR and is transcribed in the opposite direction [62,176,177], but expression of silC is blocked in response to SilCR signaling and silEDCR transcription [177]. SilC contains a com-box [176], so it should be activated in the late exponential growth phase after com-mediated QS occurs and may promote systemic invasion of the host as part of a regulatory circuit with silCR [177]. In mouse models of necrotizing fasciitis, SilCR signaling appears to reduce lesion size and attenuate acute lethality due to systemic infection [176,179] but retards host healing of the initial lesion due to decreased clearance of GAS [178]. SilCR is able to simultaneously signal multiple isolates of both GAS and GGS [62] and there may be another, non-SilB receptor for the autoinducing peptide [176]. These data suggest that sil can coordinate a pathogenic program simultaneously across a genetically heterogeneous S. pyogenes population. QS in streptococci controls numerous pathogenic factors in a complex and sometimes overlapping network of regulatory pathways, suggesting that fas and sil could be fruitful targets for therapeutic intervention in GAS or GGS disease. But more research is required to fully understand how QS controls invasive infection in these species.
2.2.3. Lactobacillus Systems
Unlike agr in S. aureus, control over toxin production and modulation of adhesion is not combined into a single regulon in Lactobacillus species. As mentioned above, agr signaling increases the production of adhesins in L. plantarum, but does not appear to regulate production of the antibacterial bacteriocin peptides. Instead, both of the pln and plt operons control the production of multiple bacteriocins [158]. Of particular note, both plnC and plnD are highly homologous to agrA, and plnB is closely related to agrC [68], although it is unclear how these various 2CRS components interact. PlnA and pltA are the propetides in their two systems, but they are unlike agrD in that their processed signal peptides are linear (Table 1) and are not readily soluble in aqueous solutions [68,180]. Furthermore, these two signal peptides share features with other bacteriocin peptides [181], suggesting that these quormones are bifunctional, serving both to signal lactobacilli and thwart the growth of neighboring species. Recent findings also suggest that pln peptides cooperatively regulate behaviors across L. plantarum strains, a behavior that counters the examplar agr alleles in S. aureus [182]. These discoveries highlight the need for care in development of broad-spectrum anti-QS compounds to avoid adversely affecting beneficial species like Lactobacillus.
3. Targeting agr and Analogues to Inhibit Disease
3.1. Overview
Pharmacologic interference with pro-virulence QS processes in pathogenic Firmicutes is an attractive strategy for restricting disease progression and potentially limiting the spread of infectious organisms. Targeting virulence factors, rather than bacterial growth, could retard pathology or even prevent infection without engendering resistance to chemotherapy that accompanies antibiotic therapy [14–16]. This strategy is complementary to the efforts of antibiotic stewardship programs that attempt to address skyrocketing antibiotic resistance at a time of limited antibiotic development [27,28]. Given that soil microbiota are a reservoir for resistance genes important in human pathogens [183] and that these resistance genes are present within the normal microbiota of the human gut [184] suggests that elimination of the genetic basis of resistance is not feasible and that new approaches are required to limit selective growth pressure that results in the proliferation of resistant organisms. In this regard, anti-virulence strategies are probably best employed as a component of an antibiotic stewardship program optimized for each pathogen that includes advanced infection control measures and appropriate antibiotic therapy. Such a strategy could extend the efficacy of existing and future antibiotics. Importantly, several of the species covered in this review with agr or agr-like quorum sensing systems and with known proclivities for developing antibiotic resistance, including S. aureus, E. faecalis, and C. difficile, represent some of the most common causes of health care associated infections that result in significant morbidity and mortality as well as increased health care costs. Therefore, anti-virulence strategies for these pathogens provided either by vaccination or by drug inhibition could have a significant impact on public health.
Currently, skin and skin structure infections comprise roughly 90% of all reported S. aureus infections [185], and from Section 1.4 it is clear that inhibiting agr function early in infection could prove highly effective in limiting disease development. Furthermore, survey evidence suggests that S. aureus persistently colonizes the mucus membranes and epithelium of 30% of healthy adults, making a blockade of QS-driven pathology and transmission to limit community-acquired disease an attractive prospect. Another potential disease where inhibition of agr could be beneficial is osteomyelitis arising from traumatic injury, where bone tissue is contaminated from direct contact with S. aureus-colonized epithelial and mucosal barriers. Prophylactic antibiotic therapy against MRSA is often prescribed for orthopedic surgery and in response to traumatic injury [186–188], and animal models have demonstrated the efficacy of this therapy in reducing experimental infection rates [189–191]. Replacing antibiotic prophylaxis with anti-QS treatments could serve as a way to inhibit the earliest stages of S. aureus infection and allow host innate immune defenses to respond to injury, without promoting antibiotic resistance. Combining approaches like enhanced infection control measures with anti-agr treatment could further reduce the spread of CA-MRSA by restricting the ability of S. aureus to infect new, healthy patients.
However, anti-agr treatments will not benefit all cases of S. aureus infection. While the majority of S. aureus infections are agr+, chronically infected patients, especially many with HA-MRSA, often carry mixed populations of agr+ and agr−S. aureus [5,6,102], and in these cases blocking agr function might limit severe disease pathology but it is unlikely to allow host defenses to completely resolve infection. Vancomycin insensitivity in clinical isolates is associated with the loss of agr function and expression, often through acquiring mutations that up-regulate repressors of agr such as σB [101,108,192], and so treating VISA infections to block agr is unlikely to be efficacious. This is also likely true of targeting agr function in S. aureus biofilm infections on medical implants and in ventilator associated pneumonias, as loss of agr function leads to thicker biofilms [93,106–108], although it could be a useful strategy to treat agr-dependent S. epidermidis biofilms [193].
As we have shown here, there are numerous pathogens where inhibition of agr or agr-like pathogenic QS systems could prove effective in controlling disease or reducing infection transmission. Reducing agr-regulated production of exotoxins in L. monocytogenes and in Clostridium species could improve the ability of the infected host to respond to infection and enhance bacterial clearance. Additionally, the use of probiotic strains that produce anti-agr compounds [130] could prevent agr-dependent disease due to these gut pathogens. Targeting fas or sil in S. pyogenes, plc/pap in B. cereus, and fsr in E. faecalis could reduce tissue invasion and apoptosis or death of host cells. Whether anti-QS treatments in these species would prove effective in limiting disease remains to be ascertained. Molecular targets involved in agr and agr-like pro-virulence QS systems in Firmicutes that could be affected by chemotherapeutic intervention are shown in Figure 3 and discussed further below.
3.2. Development of Synthetic Anti-agr Compounds
Blocking agr function to reduce the development of S. aureus pathologies in vivo has previously been established. Wright et al. demonstrated in the mouse dermonecrosis model that ulcers and abscesses caused by infection with an agr I strain could be blocked by administration of inhibitory concentrations of exogenous AIP II peptide [126]. Vaccination with hapten-linked AIP-4 allowed for the production of anti-AIP-4 sera providing passive immunity that reduced the pathology of an agr IV strain [117]. These proofs of concept clearly show that targeting agr to control virulence works. However, these therapies are severely restricted in that they are agr type specific, necessitating that S. aureus infections be agr typed before treatment, unless one were to administer multiple inhibitory peptides or pooled antisera active against all four S. aureus agr types. These limitations are inherent in any therapy that specifically targets AIP production or recognition due to the hypervariability inherent in the different agr operons. But, the functionality of AgrB, C and A is the same across all agr types, which holds true for how RNAIII regulates the virulon, and it is highly likely that compounds effective against these targets in multiple S. aureus agr types, and possibly even across different agr+ species, will be discovered.
Due to its high prevalence as both a nosocomial and community-acquired pathogen, S. aureus pathogenesis has been studied extensively and numerous experimental tools have been developed to examine agr-mediated QS, and these allow for relatively easy development of screens for anti-pathogenic anti-agr therapies. For example, a first-pass high-throughput screen could be developed using a S. aureus strain with a fluorescent reporter, such as the S. aureus Newman-GFP strain developed by Xiong et al., where GFP expression is driven by the RNAIII promoter [194]. Promising compounds would specifically inhibit agr-driven fluorescence without bactericidal or bacteriostatic effects, and would be tested further to check they had no deleterious effect upon bacterial growth, membrane potential and membrane permeability. Follow-up experiments would include several in vitro assays to measure compound effects on specific agr regulon markers like α-hemolysin and capsule polysaccharides to confirm that the inhibitory molecule was not simply interfering with the fluorescence of the GFP reporter. Successful candidate drugs would at last be tested in in vivo models of S. aureus disease, such as the mouse dermonecrosis model, to investigate their bioavailability and pharmacokinetics. Then, thanks to reagents such as the suite of agr-GFP reporter strains generated in multiple agr types by Kavanaugh et al. [75], compound efficacy could be quickly assessed across the four S. aureus agr types.
The strategy outlined here for finding anti-QS effectors is one possible approach, appropriate for screening a library of small molecules, such as the NIH Molecular Library Screening Center. Computer-aided virtual screening of chemical libraries for QS inhibitors of the lasR sensor in Pseudomonas aeruginosa has identified several existing drugs with strong similarities to known anti-QS molecules [195], and a similar approach virtually identified novel compounds that partially inhibit the DNA-binding activity of S. aureus AgrA [196]. Use of these in silico methods may shorten the time required to select compounds to physically test against single targets with associated datasets, such as protein crystal structure, or the structure-activity relationships of known but clinically unfeasible inhibitors.
3.3. Inhibition of Pro-Virulence rRNAs
RNAIII may be the single most important target in the S. aureus virulence regulon, as it alters the expression of dozens of genes contributing to S. aureus pathogenesis [1] as well as many other genes controlling metabolism and biofilm production [1,2,83,96]. Reducing or inhibiting expression of RNAIII, or altering its ability to post-transcriptionally regulate virulence factor expression, should be the most efficacious way to restrict the pathogenesis program in S. aureus and possibly related species. Targeting this molecule could encompass both prophylactic and therapeutic applications. Surgical implants and grafts could be treated pre-surgery with an anti-RNAIII compound to reduce the dissemination of S. aureus from tenacious biofilms.
A direct approach would be to interfere with the action of RNAIII (Figure 3, arrow 8), which could be achieved in S. aureus by disrupting or destabilizing the secondary structure of the regulatory RNA molecule, decreasing the half-life of dsRNA species, or interfering with the mechanism of action of regulatory RNAs. Because there are multiple regulatory RNAs in S. aureus that exert control over a wide range of bacterial activities [83], broad interference with rRNA mechanisms could adversely affect bacterial survival. This is evident in the recent report by Olson et al., where chemical inhibition of the RNAse RnaP decreased mRNA turnover, leading to reduced pathogenesis in a mouse model of S. aureus sepsis. However, this treatment was also bacteriostatic which could explain the observed reduction in disease [197]. If this approach could be fine-tuned to directly target the rRNAs responsible for virulence regulation, then this method could prove effective against other pathogenic Staphylococcus species, and possibly work to limit the effects of FasX in S. pyogenes and VR-RNA in C. perfringens [33,63,145,146].
In addition to directly targeting RNAIII to limit its ability to activate virulence factor expression in S. aureus, interfering with AgrA provides another opportunity to ameliorate disease development. Preventing the phosphorylation and activation of AgrA by AgrC (Figure 3, arrows 5 and 6), or blocking the ability of AgrA to bind DNA (Figure 3, arrow 7), would prevent RNAIII expression and block the RNAIII regulon and also abolish the smaller AgrA-driven virulence program. Leonard et al. recently published their efforts to identify molecular patterns which prevent AgrA from binding DNA at the P3 site, with moderate success in vitro [196], but it remains unclear whether their target, a hydrophobic cleft in the LytR domain, provides sufficient leverage to fully block AgrA function.
Another problem is the high degree of homology between AgrAC and other LytTRS family members, several of which are vital to S. aureus growth and survival. Siamycin I, a peptide effective against HIV fusion with human T cells, attenuates the ability of GBAP to activate FsrC and gelatinase production in E. faecalis, but its effects on other LytTR family members has not yet been ascertained [198,199]. Moreover, it is toxic for Enterococcus at doses sufficient for fsr inhibition. This example demonstrates that small molecule inhibitors must be exquisitely targeted to AgrA or AgrC alone. Small molecule inhibitors of the kinase domain of AgrC or of AgrA could also prove useful in limiting or blocking disease development in L. monocytogenes, Group A streptococci, and E. faecalis. In contrast, some RNPP family members do not employ a phosphorelay like AgrC/AgrA, and instead signal through other enzymes like the Rap phosphatases [159], requiring an entirely different class of compounds to inhibit.
3.4. Blocking the Production and Action of AIP
As mentioned in Section 1, production and recognition of the thiolactone-containing signaling molecule AIP is a highly sequence-specific process which can be inhibited at several points by AIP molecules from other agr types or by peptide analogs (Figure 3, arrows 1–5). Current technology facilitates the design and synthesis of numerous peptides or modified peptide analogs that can inhibit agr-mediated QS in vitro. Two studies clearly demonstrate effective inhibition of multiple agr alleles at nanomolar peptide concentrations using a single, common peptide analog to AIP molecules [81,200], but the efficacy of these peptides in preventing or reducing disease in vivo has not been demonstrated. Peptide-based cross-type inhibition of agr during skin infection in vivo has been shown in a proof-of-concept paper, with a single, high dose of AIP 2 transiently limiting agr activation but without an apparent effect on pathology [126]. Given the observation that the biological lifetime of an AIP molecule in solution in vivo is about 3 hours targeting S. aureus infections with an AIP-mimetic peptides as an anti-virulence therapy suffers from severe limitations, and by extension, the rational design of inhibitory molecules based off of the structure of AIP is of limited use. However, there are several unique aspects of the agr system which may be targeted effectively by non-peptide molecules.
AgrB is unusual in that it combines several disparate functions into a single enzyme (Figure 3, arrows 1–3). While peptide analogs could be generated to irreversibly antagonize the active site which binds, cleaves, and cyclizes AgrD, the enzyme structure could also be destabilized or forced into an inactive conformation by a small molecule, an approach for which there are in vivo examples from drug studies in mammals (for example FK506 Binding Proteins, the mTOR pathway, and PPARs [201–203]). Another approach would be identification of small molecule inhibitors that bind AgrB or an associated exporter to prevent export of AgrD. Because the mechanism by which AIP is transported from the cytoplasm is unknown, the feasibility of blockading AgrD export remains to be determined. There are numerous examples from the pharmacological literature demonstrating that export channels in both prokaryotes and eukaryotes can be targeted specifically to inhibit transporter functions. Ambuic acid, an antifungal agent, weakly and partially inhibits the production of AIP 1 in S. aureus and GBAP in E. faecalis, but the mechanism of action remains unclear [204]. Either approach could serve as a prophylactic treatment for S. aureus infection and to prevent biofilm maturation and dissemination. But without identification of high affinity irreversible inhibitors such therapies could prove ineffective for extant infections in which agr-mediated QS is ongoing and producing ever more AgrB, AgrD and AIP. Additionally, peptide analogs or compounds working in these ways would need to function at the extracellular face of AgrB or first gain entry to the bacterial cell before they could block its cytoplasmic functions. However, this type of treatment should provide compounds effective against all of the species discussed in this review and represents a strategy likely to have significant commonality.
AIP itself can be targeted, as demonstrated by the numerous examples listed in Section 1.5 detailing how the innate immune system interferes with agr-mediated QS, and also by the anti-AIP-4 vaccine [117]. How current pharmacological approaches with small molecules could improve upon the existing, natural defenses against AIP and S. aureus QS is difficult to predict. However, interfering with AgrC in its ability to bind AIP and dimerize (Figure 3, arrows 4 and 5) could prove a fruitful target. Several reports detail attempts to generate synthetic peptide analogs capable of inhibiting all types of AIP simultaneously, but many of these compounds suffer from a poor affinity to at least one of the agr alleles, and these analogs remain untested in in vivo models of S. aureus disease [81,126,200,205]. But as with AgrB, small molecules that alter AgrC's conformation so that it fails to bind AIP or fails to dimerize are possible to discover through bulk screens. However, the prospect of developing compounds acting against peptide quormone production or recognition are no better in bacteria with only a single QS allele: a peptide-derived analog of the fsr quormone in E. faecalis never completely inhibits gelatinase production [206]. As this approach targets the ability of S. aureus to recognize and respond to AIP, this treatment should prove effective both as prophylaxis and post facto treatment for Staphylococcus pathogens, as well as members of the B. cereus group, L. monocytogenes, groups A, B, and G streptococci, and against E. faecalis, all of which employ signaling peptides or possess known AgrC homologues. The difficulty in targeting AgrC is similar to finding molecules that inhibit AgrA, as there are numerous 2CRS and HPK homologues to AgrC, so any anti-agr compounds that work against AgrC would need to be tested extensively to confirm they do not demonstrate broader antimicrobial effects.
4. Conclusions
Targeting non-vital, pro-pathogenic QS mechanisms in S. aureus and G- pathogens like E. coli and P. aeruginosa has been suggested as a way to curtail bacterial virulence without engendering resistance [14,15]. One of the complicating factors in targeting the AHL and AI-2 pathways in G- pathogens, as well as some G+ species, is the large and diverse array of organic small molecules and their cognate intracellular sensors employed, making it difficult to predict whether a treatment that works against these QS mechanisms in any given pathogen could work against any other. However, the marked degree of homology and distinct functional features shared between agr family members and orthologues across the Firmicutes offers the prospect of developing a small suite of therapies that can inhibit pathogenesis in numerous community-acquired and nosocomial pathogens beyond S. aureus.
References
- Queck, S.Y.; Jameson-Lee, M.; Villaruz, A.E.; Bach, T.-H.L.; Khan, B.; Sturdevant, D.E.; Ricklefs, S.M.; Li, M.; Otto, M. RNAIII-independent target gene control by the agr quorum-sensing system: insight into the evolution of virulence regulation in staphylococcus aureus. Mol.Cell 2008, 32, 150–158. [Google Scholar]
- Thoendel, M.; Kavanaugh, J.S.; Flack, C.E.; Horswill, A.R. Peptide signaling in the staphylococci. Chem. Rev. 2011, 111, 117–151. [Google Scholar]
- Loughman, J.; Fritz, S.; Storch, G.; Hunstad, D. Virulence gene expression in human community-acquired Staphylococcus aureus infection. J. Infec. Dis. 2009, 199, 294–301. [Google Scholar]
- Gagnaire, J.; Dauwalder, O.; Boisset, S.; Khau, D.; Freydière, A.-M.; Ader, F.; Bes, M.; Lina, G.; Tristan, A.; Reverdy, M.-E.; et al. Detection of staphylococcus aureus delta-toxin production by whole-cell MALDI-TOF mass spectrometry. PLoS One 2012. [Google Scholar] [CrossRef]
- Shopsin, B.; Drlica-Wagner, A.; Mathema, B.; Adhikari, R.P.; Kreiswirth, B.N.; Novick, R.P. Prevalence of agr dysfunction among colonizing Staphylococcus aureus strains. J. Infec. Dis. 2008, 198, 1171–1174. [Google Scholar]
- Smyth, D.S.; Kafer, J.M.; Wasserman, G.A.; Velickovic, L.; Mathema, B.; Holzman, R.S.; Knipe, T.A.; Becker, K.; Von Eiff, C.; Peters, G.; et al. Nasal carriage as a source of agr-defective staphylococcus aureus bacteremia. J. Infec. Dis. 2012, 206, 1168–1177. [Google Scholar]
- Shopsin, B.; Eaton, C.; Wasserman, G.A.; Mathema, B.; Adhikari, R.P.; Agolory, S.; Altman, D.R.; Holzman, R.S.; Kreiswirth, B.N.; Novick, R.P. Mutations in agr do not persist in natural populations of methicillin-resistant staphylococcus aureus. J. Infec. Dis. 2010, 202, 1593–1599. [Google Scholar]
- Cheung, G.Y.C.; Wang, R.; Khan, B.; Sturdevant, D.E.; Otto, M. Role of the accessory gene regulator agr in community-associated methicillin-resistant staphylococcus aureus pathogenesis. Infec. Immunol. 2011, 79, 1927–1935. [Google Scholar]
- Wang, R.; Braughton, K.R.; Kretschmer, D.; Bach, T.-H.L.; Queck, S.Y.; Li, M.; Kennedy, A.D.; Dorward, D.W.; Klebanoff, S.J.; Peschel, A.; et al. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat. Med. 2007, 13, 1510–1514. [Google Scholar]
- Montgomery, C.P.; Boyle-Vavra, S.; Daum, R.S. Importance of the global regulators Agrand SaeRS in the pathogenesis of CA-MRSA USA300 infection. PLoS One 2010. [Google Scholar] [CrossRef]
- Gillaspy, A. F.; Hickmon, S. G.; Skinner, R. A.; Thomas, J. R.; Nelson, C. L.; Smeltzer, M. S. Role of the accessory gene regulator (agr) in pathogenesis of staphylococcal osteomyelitis. Infection and Immunity 1995, 63, 3373–3380. [Google Scholar]
- Abdelnour, A.; Arvidson, S.; Bremell, T.; Rydén, C.; Tarkowski, A. The accessory gene regulator (agr) controls staphylococcus aureus virulence in a murine arthritis model. Infec. Immun. 1993, 61, 3879–3885. [Google Scholar]
- Abel, J.; Goldmann, O.; Ziegler, C.; Höltje, C.; Smeltzer, M.S.; Cheung, A.L.; Bruhn, D.; Rohde, M.; Medina, E. Staphylococcus aureus evades the extracellular antimicrobial activity of mast cells by promoting its own uptake. J. Innate Immun. 2011, 3, 495–507. [Google Scholar]
- Rasko, D.; Sperandio, V. Anti-virulence strategies to combat bacteria-mediated disease. Nat. Rev. Drug Discov. 2010, 9, 117–128. [Google Scholar]
- Cegelski, L.; Marshall, G.R.; Eldridge, G.R.; Hultgren, S.J. The biology and future prospects of antivirulence therapies. Nat. Rev. Microbiol. 2008, 6, 17–27. [Google Scholar]
- Rutherford, S.T.; Bassler, B.L. Bacterial quorum sensing: Its role in virulence and possibilities for its control. Cold Spr. Harbor Per. Med. 2012. [Google Scholar] [CrossRef]
- Gordon, C.P.; Williams, P.; Chan, W.C. Attenuating staphylococcus aureus virulence gene regulation: A medicinal chemistry perspective. J. Med. Chem. 2013, 56, 1389–1404. [Google Scholar]
- Wuster, A.; Babu, M.M. Conservation and evolutionary dynamics of the agr cell-to-cell communication system across firmicutes. J. Bacteriol. 2008, 190, 743–746. [Google Scholar]
- Yao, Y.; Vuong, C.; Kocianova, S.; Villaruz, A.E.; Lai, Y.; Sturdevant, D.E.; Otto, M. Characterization of the Staphylococcus epidermidis accessory-gene regulator response: Quorum-sensing regulation of resistance to human innate host defense. J. Infec. Dis. 2006, 193, 841–848. [Google Scholar]
- Heilbronner, S.; Holden, M.T.G.; Van Tonder, A.; Geoghegan, J.A.; Foster, T.J.; Parkhill, J.; Bentley, S.D. Genome sequence of Staphylococcus lugdunensis N920143 allows identification of putative colonization and virulence factors. FEMS Microbiol. Lett. 2011, 322, 60–67. [Google Scholar]
- Li, J.; Chen, J.; Vidal, J.E.; McClane, B.A. The agr-like quorum-sensing system regulates sporulation and production of enterotoxin and beta2 toxin by Clostridium perfringens type A non-food-borne human gastrointestinal disease strain F5603. Infec. Immun. 2011, 79, 2451–2459. [Google Scholar]
- Slamti, L.; Lereclus, D. Specificity and polymorphism of the plcr-papr quorum-sensing system in the bacillus cereus group specificity and polymorphism of the plcr-papr quorum-sensing system in the bacillus cereus group. J. Bacteriol. 2005, 187, 1182–1187. [Google Scholar]
- Garmyn, D.; Gal, L.; Briandet, R.; Guilbaud, M.; Lemaître, J.-P.; Hartmann, A.; Piveteau, P. Evidence of autoinduction heterogeneity via expression of the agr system of listeria monocytogenes at the single-cell level. Appl. Environ. Microbiol. 2011, 77, 6286–6289. [Google Scholar]
- Wright, J.S.; Traber, K.E.; Corrigan, R.; Benson, S.A.; Musser, J.M.; Novick, R.P.; Iii, J.S.W. The agr radiation: An early event in the evolution of staphylococci. J. Bacteriol. 2005, 187, 5585–5594. [Google Scholar]
- Wynendaele, E.; Bronselaer, A.; Nielandt, J.; D'Hondt, M.; Stalmans, S.; Bracke, N.; Verbeke, F.; Van De Wiele, C.; De Tré, G.; De Spiegeleer, B. Quorumpeps database: chemical space, microbial origin and functionality of quorum sensing peptides. Nucl. Acids Res. 2013, 41, D655–D659. [Google Scholar]
- Rasko, D.; Moreira, C.G.; Li, D.R.; Reading, N.C.; Ritchie, J.M.; Waldor, M.K.; Williams, N.; Taussig, R.; Wei, S.; Roth, M.; et al. Targeting QseC signaling and virulence for antibiotic development. Science 2008, 321, 1078–1080. [Google Scholar]
- Spellberg, B. The antibiotic crisis: Can we reverse 65 years of failed stewardship? Arch. Int. Med. 2011, 171, 1080–1081. [Google Scholar]
- Tamma, P.D.; Cosgrove, S.E. Antimicrobial stewardship. Infec. Dis. Clin. North Amer. 2011, 25, 245–260. [Google Scholar]
- Stryjewski, M.E.; Chambers, H.F. Skin and soft-tissue infections caused by community-acquired methicillin-resistant Staphylococcus aureus. Clin. Infec. Dis. 2008, 46, S368–S377. [Google Scholar]
- Novick, R.P. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 2003, 48, 1429–1449. [Google Scholar]
- Cassat, J.; Dunman, P.M.; Murphy, E.; Projan, S.J.; Beenken, K.E.; Palm, K.J.; Yang, S.-J.; Rice, K.C.; Bayles, K.W.; Smeltzer, M.S. Transcriptional profiling of a staphylococcus aureus clinical isolate and its isogenic agr and sarA mutants reveals global differences in comparison to the laboratory strain RN6390. Microbiology 2006, 152, 3075–3090. [Google Scholar]
- Beenken, K.E.; Dunman, P.M.; McAleese, F.; Macapagal, D.; Murphy, E.; Projan, S.J.; Blevins, J.S.; Smeltzer, M.S. Global gene expression in staphylococcus aureus biofilms. J. Bacteriol. 2004, 186, 4665–4684. [Google Scholar]
- Rowe, S.E.; Mahon, V.; Smith, S.G.; O'Gara, J.P. A novel role for SarX in staphylococcus epidermidis biofilm regulation. Microbiology 2011, 157, 1042–1049. [Google Scholar]
- Van Wamel, W.J.; Van Rossum, G.; Verhoef, J.; Vandenbroucke-Grauls, C.M.; Fluit, A.C. Cloning and characterization of an accessory gene regulator (agr)-like locus from Staphylococcus epidermidis. FEMS Microbiol. Lett. 1998, 163, 1–9. [Google Scholar]
- Otto, M.; Süssmuth, R.; Jung, G.; Götz, F. Structure of the pheromone peptide of the Staphylococcus epidermidis agr system. FEBS Lett. 1998, 424, 89–94. [Google Scholar]
- Vuong, C.; Dürr, M.; Carmody, A.B.; Peschel, A.; Klebanoff, S.J.; Otto, M. Regulated expression of pathogen-associated molecular pattern molecules in Staphylococcus epidermidis: Quorum-sensing determines pro-inflammatory capacity and production of phenol-soluble modulins. Cell. Microbiol. 2004, 6, 753–759. [Google Scholar]
- Tegmark, K.; Morfeldt, E.; Arvidson, S.; Morfeldt, E.V.A. Regulation of agr-Dependent virulence genes in staphylococcus aureus by RNAIII from coagulase-negative staphylococci. Microbiology 1998, 180, 3181–3186. [Google Scholar]
- Batzilla, C.F.; Rachid, S.; Engelmann, S.; Hecker, M.; Hacker, J.; Ziebuhr, W. Impact of the accessory gene regulatory system (agr) on extracellular proteins, codY expression and amino acid metabolism in Staphylococcus epidermidis. Proteomics 2006, 6, 3602–3613. [Google Scholar]
- Vandenesch, F.; Projan, S.J.; Kreiswirth, B.N.; Etienne, J.; Novick, R.P. Agr-related sequences in Staphylococcus lugdunensis. FEMS Microbiol. Lett. 1993, 111, 115–122. [Google Scholar]
- Sakinc, T.; Kulczak, P.; Henne, K.; Gatermann, S.G. Cloning of an agr homologue of Staphylococcus saprophyticus. FEMS Microbiol. Lett. 2004, 237, 157–161. [Google Scholar]
- Ji, G.; Pei, W.; Zhang, L.; Qiu, R.; Lin, J.; Benito, Y.; Lina, G.; Novick, R.P. Staphylococcus intermedius produces a functional agr autoinducing peptide containing a cyclic lactone. J. Bacteriol. 2005, 187, 3139–3150. [Google Scholar]
- Bannoehr, J.; Ben Zakour, N.L.; Waller, A.S.; Guardabassi, L.; Thoday, K.L.; Van den Broek, A.H.M.; Fitzgerald, J.R. Population genetic structure of the Staphylococcus intermedius group: insights into agr diversification and the emergence of methicillin-resistant strains. J. Bacteriol. 2007, 189, 8685–8692. [Google Scholar]
- Dufour, P.; Jarraud, S.; Vandenesch, F.; Novick, R.P.; Bes, M.; Lina, G.; Etienne, J. High genetic variability of the agr locus in staphylococcus species. J. Bacteriol. 2002, 184, 1180–1186. [Google Scholar]
- Cooksley, C.M.; Davis, I.J.; Winzer, K.; Chan, W.C.; Peck, M.W.; Minton, N.P. Regulation of neurotoxin production and sporulation by a Putative agrBD signaling system in proteolytic Clostridium botulinum. Appl. Environ. Microbiol 2010, 76, 4448–4460. [Google Scholar]
- Monot, M.; Boursaux-Eude, C.; Thibonnier, M.; Vallenet, D.; Moszer, I.; Medigue, C.; Martin-Verstraete, I.; Dupuy, B. Reannotation of the genome sequence of clostridium difficile strain 630. J. Med. Microbiol. 2011, 60, 1193–1199. [Google Scholar]
- Ohtani, K.; Yuan, Y.; Hassan, S.; Wang, R.; Wang, Y.; Shimizu, T. Virulence gene regulation by the agr system in clostridium perfringens. J. Bacteriol. 2009, 191, 3919–3927. [Google Scholar]
- Chen, J.; Rood, J.I.; McClane, B.A. Epsilon-toxin production by clostridium perfringens type D strain CN3718 is dependent upon the agr operon but not the VirS/VirR two-component regulatory system. Mbio 2011. [Google Scholar] [CrossRef]
- Nakayama, J.; Chen, S.; Oyama, N.; Nishiguchi, K.; Azab, E.; Tanaka, E.; Kariyama, R.; Sonomoto, K. Revised model for enterococcus faecalis fsr quorum-sensing system: The small open reading frame fsrD encodes the gelatinase biosynthesis-activating pheromone propeptide corresponding to staphylococcal agrd. J. Bacteriol. 2006, 188, 8321–8326. [Google Scholar]
- Hancock, L.E.; Perego, M. The Enterococcus faecalis fsr two-component system controls biofilm development through production of gelatinase. J. Bacteriol. 2004, 186, 5629. [Google Scholar]
- Qin, X.; Singh, K.V.; Weinstock, G.M.; Murray, B.E. Effects of Enterococcus faecalis fsr genes on production of gelatinase and a serine protease and virulence. Infec. Immun. 2000, 68, 2579–2586. [Google Scholar]
- Autret, N.; Raynaud, C.; Dubail, I.; Charbit, A.; Berche, P. Identification of the agr locus of listeria monocytogenes: Role in bacterial virulence. Infec. Immun. 2003, 71, 4463–4471. [Google Scholar]
- Rieu, A.; Weidmann, S.; Garmyn, D.; Piveteau, P.; Guzzo, J. Agr system of Listeria monocytogenes EGD-e: Role in adherence and differential expression pattern. Appl. Environ. Microbiol. 2007, 73, 6125–6133. [Google Scholar]
- Riedel, C.U.; Monk, I.R.; Casey, P.G.; Waidmann, M.S.; Gahan, C.G.M.; Hill, C. AgrD-dependent quorum sensing affects biofilm formation, invasion, virulence and global gene expression profiles in Listeria monocytogenes. Mol. Microbiol. 2009, 71, 1177–1189. [Google Scholar]
- Sturme, M.H.J.; Nakayama, J.; Molenaar, D.; Murakami, Y.; Kunugi, R.; Fujii, T.; Elaine, E.; Kleerebezem, M.; De Vos, W.M. An agr-Like two-component regulatory system in Lactobacillus plantarum is involved in production of a novel cyclic peptide and regulation of adherence. J. Bacteriol. 2005, 187, 5224–5235. [Google Scholar]
- Slamti, L.; Lereclus, D. A cell-cell signaling peptide activates the PlcR virulence regulon in bacteria of the bacillus cereus group. EMBO J 2002, 21, 4550–4559. [Google Scholar]
- Perchat, S.; Dubois, T.; Zouhir, S.; Gominet, M.; Poncet, S.; Lemyl, C.; Aumont-Nicaise, M.; Deutscher, J.; Gohar, M.; Nessler, S.; et al. A cell–cell communication system regulates protease production during sporulation in bacteria of the Bacillus cereus group. Mol. Microbiol. 2011, 82, 619–633. [Google Scholar]
- Moyer, L.; Ramadan, R.T.; Thurman, J.; Burroughs, A.; Callegan, M.C. Bacillus cereus induces permeability of an in vitro blood-retina barrier. Infec. Immun. 2008, 76, 1358–1367. [Google Scholar]
- Huillet, E.; Tempelaars, M.H.; André-Leroux, G.; Wanapaisan, P.; Bridoux, L.; Makhzami, S.; Panbangred, W.; Martin-Verstraete, I.; Abee, T.; Lereclus, D. PlcRa, a new quorum-sensing regulator from bacillus cereus, plays a role in oxidative stress responses and cysteine metabolism in stationary phase. PLoS One 2012. [Google Scholar] [CrossRef]
- Dubois, T.; Faegri, K.; Perchat, S.; Lemy, C.; Buisson, C.; Nielsen-LeRoux, C.; Gohar, M.; Jacques, P.; Ramarao, N.; Kolstø, A.-B.; et al. Necrotrophism is a quorum-sensing-regulated lifestyle in Bacillus thuringiensis. PLoS Pathog 2012. [Google Scholar] [CrossRef]
- Lereclus, D.; Agaisse, H.; Gominet, M.; Salamitou, S.; Sanchis, V. Identification of a Bacillus thuringiensis gene that positively regulates transcription of the phosphatidylinositol-specific phospholipase C gene at the onset of the stationary phase. J. Bacteriol. 1996, 178, 2749–2756. [Google Scholar]
- Declerck, N.; Bouillaut, L.; Chaix, D.; Rugani, N.; Slamti, L.; Hoh, F.; Lereclus, D.; Arold, S.T. Structure of PlcR: Insights into virulence regulation and evolution of quorum sensing in gram-positive bacteria. Proc. Natl. Acad. Sci. USA 2007, 104, 18490–18495. [Google Scholar]
- Belotserkovsky, I.; Baruch, M.; Peer, A.; Dov, E.; Ravins, M.; Mishalian, I.; Persky, M.; Smith, Y.; Hanski, E. Functional analysis of the quorum-sensing streptococcal invasion locus (sil). PLoS Pathog 2009. [Google Scholar] [CrossRef]
- Kreikemeyer, B.; Boyle, M.D.; Buttaro, B.; Heinemann, M.; Podbielski, A. Group a streptococcal growth phase-associated virulence factor regulation by a novel operon (Fas) with homologies to two-component-type regulators requires a small RNA molecule. Mol. Microbiol. 2001, 39, 392–406. [Google Scholar]
- Cvitkovitch, D.G.; Li, Y.H.; Ellen, R.P. Quorum sensing and biofilm formation in streptococcal infections. J. Clin. Invest. 2003, 112, 1626–1632. [Google Scholar]
- Ramirez-Peña, E.; Treviño, J.; Liu, Z.; Perez, N.; Sumby, P. The group a streptococcus small regulatory RNA FasX enhances streptokinase activity by increasing the stability of the ska mRNA transcript. Mol. Microbiol. 2010, 78, 1332–1347. [Google Scholar]
- Mashburn-Warren, L.; Morrison, D.; Federle, M.J. The cryptic competence pathway in streptococcus pyogenes is controlled by a peptide pheromone. J. Bacteriol. 2012, 194, 4589–4600. [Google Scholar]
- Mashburn-Warren, L.; Morrison, D.A.; Federle, M.J. A novel double-tryptophan peptide pheromone is conserved in mutans and pyogenic streptococci and controls competence in streptococcus mutans via an rgg regulator. Mol. Microbiol. 2010, 78, 589–606. [Google Scholar]
- Diep, D.B.; Håvarstein, L.S.; Nissen-Meyer, J.; Nes, I.F. The gene encoding plantaricin A, a bacteriocin from Lactobacillus plantarum C11, is located on the same transcription unit as an agr-like regulatory system. Appl. Environ. Microbiol. 1994, 60, 160–166. [Google Scholar]
- Dandekar, A.; Chugani, S.; Greenberg, E.P. Bacterial quorum sensing and metabolic incentives to cooperate. Science 2012, 338, 264–266. [Google Scholar]
- Mellbye, B.; Schuster, M. The Sociomicrobiology of antivirulence drug resistance: A proof of concept. Mbio 2011. [Google Scholar] [CrossRef]
- Novick, R.P.; Geisinger, E. Quorum sensing in staphylococci. Ann. Rev. Genet. 2008, 42, 541–564. [Google Scholar]
- Zhang, L.; Lin, J.; Ji, G. Membrane anchoring of the AgrD N-terminal amphipathic region is required for its processing to produce a quorum-sensing pheromone in Staphylococcus aureus. J. Biol. Chem. 2004, 279, 19448–19456. [Google Scholar]
- Thoendel, M.; Horswill, A.R. Random mutagenesis and topology analysis of the autoinducing peptide biosynthesis proteins in Staphylococcus aureus. Mol. Microbiol. 2013, 87, 318–337. [Google Scholar]
- Zhang, L.; Gray, L.; Novick, R.P.; Ji, G. Transmembrane topology of AgrB, the protein involved in the post-translational modification of AgrD in staphylococcus aureus. J. Biol. Chem. 2002, 277, 34736–34742. [Google Scholar]
- Kavanaugh, J.S.; Thoendel, M.; Horswill, A.R. A role for type I signal peptidase in Staphylococcus aureus quorum sensing. Mol. Microbiol. 2007, 65, 780–798. [Google Scholar]
- Carnes, E.C.; Lopez, D.M.; Donegan, N.P.; Cheung, A.; Gresham, H.; Timmins, G.S.; Brinker, C.J. Confinement-induced quorum sensing of individual staphylococcus aureus bacteria. Nat. Chem. Biol. 2010, 6, 41–46. [Google Scholar]
- George Cisar, E.A.; Geisinger, E.; Muir, T.W.; Novick, R.P. Symmetric signalling within asymmetric dimers of the staphylococcus aureus receptor histidine kinase AgrC. Mol. Microbiol. 2009, 74, 44–57. [Google Scholar]
- Koenig, R.L.; Ray, J.L.; Maleki, S.J.; Smeltzer, M.S.; Hurlburt, B.K. Staphylococcus aureus AgrA binding to the RNAIII- agr regulatory region. J. Bacteriol. 2004, 186, 7459–7555. [Google Scholar]
- Reynolds, J.; Wigneshweraraj, S. Molecular insights into the control of transcription initiation at the staphylococcus aureus agr operon. J. Mol. biol. 2011, 412, 862–881. [Google Scholar]
- Geisinger, E.; Chen, J.; Novick, R.P. Allele-dependent differences in quorum-sensing dynamics result in variant expression of virulence genes in staphylococcus aureus. J. Bacteriol. 2012, 194, 2854–2864. [Google Scholar]
- Lyon, G.J.; Wright, J.S.; Muir, T.W.; Novick, R.P. Key determinants of receptor activation in the agr autoinducing peptides of staphylococcus aureus. Biochemistry 2002, 41, 10095–10104. [Google Scholar]
- Ji, G.; Beavis, R.; Novick, R.P. Bacterial interference caused by autoinducing peptide variants. Science 1997, 276, 2027–2030. [Google Scholar]
- Felden, B.; Vandenesch, F.; Bouloc, P.; Romby, P. The staphylococcus aureus RNome and its commitment to virulence. PLoS Pathog 2011. [Google Scholar] [CrossRef]
- Cheung, A.L.; Koomey, J.M.; Butler, C.A.; Projan, S.J.; Fischetti, V.A. Regulation of exoprotein expression in staphylococcus aureus by a locus (sar) distinct from. agr. Proc. Natl. Acad. Sci. USA 1992, 89, 6462–6466. [Google Scholar]
- Cheung, A.L.; Projan, S.J. Cloning and sequencing of sarA of Staphylococcus aureus, a gene required for the expression of. agr. J. Bacteriol. 1994, 176, 4168–4172. [Google Scholar]
- Reyes, D.; Andrey, D.O.; Monod, A.; Kelley, W.L.; Zhang, G.; Cheung, A.L. Coordinated Regulation by AgrA, SarA, and SarR to control agr expression in staphylococcus aureus. J. Bacteriol. 2011, 193, 6020–6031. [Google Scholar]
- Luong, T.T.; Dunman, P.M.; Murphy, E.; Steven, J.; Lee, C.Y. Transcription profiling of the mgrA regulon in staphylococcus aureus. J. Bacteriol. 2006, 188, 1899–1910. [Google Scholar]
- Manna, A.C.; Cheung, A.L. Expression of SarX, a negative regulator of agr and exoprotein synthesis, is activated by MgrA in staphylococcus aureus. J. Bacteriol. 2006, 188, 4288–4299. [Google Scholar]
- Manna, A.C.; Cheung, A.L. SarU, a sarA Homolog, is repressed by SarT and regulates virulence genes in staphylococcus aureus. Infec. Immun. 2003, 71, 343–353. [Google Scholar]
- Majerczyk, C.D.; Sadykov, M.R.; Luong, T.T.; Lee, C.; Somerville, G.; Sonenshein, A.L. Staphylococcus aureus CodY negatively regulates virulence gene expression. J. Bacteriol. 2008, 190, 2257–2265. [Google Scholar]
- Pohl, K.; Francois, P.; Stenz, L.; Schlink, F.; Geiger, T.; Herbert, S.; Goerke, C.; Schrenzel, J.; Wolz, C. CodY in staphylococcus aureus: A regulatory link between metabolism and virulence gene expression. J. Bacteriol. 2009, 191, 2953–2963. [Google Scholar]
- Shaw, L.N.; Aish, J.; Davenport, J.E.; Brown, M.C.; Lithgow, J.K.; Simmonite, K.; Crossley, H.; Travis, J.; Potempa, J.; Foster, S.J. Investigations into sigmaB-modulated regulatory pathways governing extracellular virulence determinant production in staphylococcus aureus. J. Bacteriol. 2006, 188, 6070–6080. [Google Scholar]
- Lauderdale, K.J.; Boles, B.R.; Cheung, A.L.; Horswill, A.R. Interconnections between Sigma B, agr, and proteolytic activity in Staphylococcus aureus biofilm maturation. Infec. Immun. 2009, 77, 1623–1635. [Google Scholar]
- Sun, F.; Liang, H.; Kong, X.; Xie, S.; Cho, H.; Deng, X.; Ji, Q.; Zhang, H.; Alvarez, S.; Hicks, L.M.; et al. Quorum-sensing agr mediates bacterial oxidation response via an intramolecular disulfide redox switch in the response regulator AgrA. Proc. Natl. Acad. Sci. USA 2012, 109, 9095–9100. [Google Scholar]
- Balaban, N.; Novick, R.P. Translation of RNAIII, the staphylococcus aureus agr regulatory RNA molecule, can be activated by a 3′-end deletion. FEMS Microbiol. Lett. 1995, 133, 155–161. [Google Scholar]
- Cheung, A.L.; Bayer, A.S.; Zhang, G.; Gresham, H.; Xiong, Y.-Q. Regulation of virulence determinants in vitro and in vivo in staphylococcus aureus. FEMS Immunol. Med. Microbiol. 2004, 40, 1–9. [Google Scholar]
- Kennedy, A.D.; Bubeck Wardenburg, J.; Gardner, D.J.; Long, D.; Whitney, A.R.; Braughton, K.R.; Schneewind, O.; DeLeo, F.R. Targeting of alpha-hemolysin by active or passive immunization decreases severity of USA300 skin infection in a mouse model. J. Infec. Dis. 2010, 202, 1050–1058. [Google Scholar]
- Heyer, G.; Saba, S.; Adamo, R.; Soong, G.; Cheung, A.; Prince, A. Staphylococcus aureus agr and sarA functions are required for invasive infection but not inflammatory responses in the lung. Infec. Immun. 2002, 70, 127–133. [Google Scholar]
- Bubeck Wardenburg, J.; Patel, R.J.; Schneewind, O. Surface proteins and exotoxins are required for the pathogenesis of staphylococcus aureus pneumonia. Infec. Immun. 2007, 75, 1040–1044. [Google Scholar]
- Cheung, A.L.; Eberhardt, K.J.; Chung, E.; Yeaman, M.R.; Sullam, P.M.; Ramos, M.; Bayer, A.S. Diminished virulence of a sar-/agr- Mutant of staphylococcus aureus in the rabbit model of endocarditis. J. Clin. Invest. 1994, 94, 1815–1822. [Google Scholar]
- Fowler, V.G.; Sakoulas, G.; Mcintyre, L.M.; Meka, V.G.; Arbeit, R.D.; Cabell, C.H.; Stryjewski, M.E.; Eliopoulos, G.M.; Reller, L.B.; Corey, G.R.; et al. Persistent bacteremia due to methicillin-resistant staphylococcus aureus infection is associated with agr dysfunction and low-level in vitro resistance to thrombin-induced platelet microbicidal protein. J. Infec. Dis. 2004, 190, 1140–1149. [Google Scholar]
- Raber, K.E.; Lee, E.; Benson, S.; Corrigan, R.; Cantera, M.; Shopsin, B.; Novick, R.P. Agr function in clinical Staphylococcus aureus isolates. Microbiology 2008, 154, 2265–2274. [Google Scholar]
- Rudkin, J.K.; Edwards, A.M.; Bowden, M.G.; Brown, E.L.; Pozzi, C.; Waters, E.M.; Chan, W.C.; Williams, P.; O'Gara, J.P.; Massey, R.C. Methicillin resistance reduces the virulence of healthcare-associated methicillin-resistant Staphylococcus aureus by interfering with the agr quorum sensing system. J. Infec. Dis. 2012, 205, 798–806. [Google Scholar]
- Yarwood, J.M.; Schlievert, P.M. Quorum sensing in Staphylococcus infections. J. Clin. Invest. 2003, 112, 1620–1625. [Google Scholar]
- Schwan, W.R.; Langhorne, M.H.; Ritchie, H.D.; Stover, C.K. Loss of hemolysin expression in Staphylococcus aureus agr mutants correlates with selective survival during mixed infections in murine abscesses and wounds. FEMS Immunol. Med. Microbiol. 2003, 38, 23–28. [Google Scholar]
- Boles, B.R.; Horswill, A.R. Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog 2008, 4, e1000052. [Google Scholar]
- Beenken, K.E.; Mrak, L.N.; Griffin, L.M.; Zielinska, A.K.; Shaw, L.N.; Rice, K.C.; Horswill, A.R.; Bayles, K.W.; Smeltzer, M.S. Epistatic relationships between sarA and agr in Staphylococcus aureus biofilm formation. PLoS One 2010, 5, e10790. [Google Scholar]
- Sakoulas, G.; Moellering, R.C.; Eliopoulos, G.M. Adaptation of methicillin-resistant staphylococcus aureus in the face of vancomycin therapy. Clin. Infec. Dis. 2006, 42, S40–S50. [Google Scholar]
- Periasamy, S.; Joo, H.-S.; Duong, A.C.; Bach, T.-H.L.; Tan, V.Y.; Chatterjee, S.S.; Cheung, G.Y.C.; Otto, M. How staphylococcus aureus biofilms develop their characteristic structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1281–1286. [Google Scholar]
- Del Pozo, J.L.; Patel, R. The challenge of treating biofilm-associated bacterial infections. Clin. Pharmacol. Therapeut. 2007, 82, 204–209. [Google Scholar]
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial bioflims: A common cause of persistent infections. Science 1999, 284, 1318–1322. [Google Scholar]
- Van Belkum, A.; Melles, D.C.; Nouwen, J.; Van Leeuwen, W.B.; Van Wamel, W.; Vos, M.C.; Wertheim, H.F.L.; Verbrugh, H.A. Co-evolutionary aspects of human colonisation and infection by staphylococcus aureus. Infec. Genet. Evolution. 2009, 9, 32–47. [Google Scholar]
- Goerke, C.; Wirtz, C.; Flückiger, U.; Wolz, C. Extensive phage dynamics in Staphylococcus aureus contributes to adaptation to the human host during infection. Mol. Microbiol. 2006, 61, 1673–1685. [Google Scholar]
- Daum, R.S.; Spellberg, B. Progress toward a staphylococcus aureus vaccine. Clin. Infec. Dis. 2011, 54, 560–567. [Google Scholar]
- Ohlsen, K.; Lorenz, U. Immunotherapeutic strategies to combat staphylococcal infections. Int. J. Med. Microbiol. 2010, 300, 402–410. [Google Scholar]
- Schaffer, A.C.; Lee, J.C. Staphylococcal vaccines and immunotherapies. Infec. Dis. Clin. North Amer. 2009, 23, 153–171. [Google Scholar]
- Park, J.; Jagasia, R.; Kaufmann, G.F.; Mathison, J.C.; Ruiz, D.L.; Moss, J.A.; Meijler, M.M.; Ulevitch, R.J.; Janda, K.D. Infection control by antibody disruption of bacterial quorum sensing signaling. Chem. Biol. 2007, 14, 1119–1127. [Google Scholar]
- Chan, P.F.; Foster, S.J.; Ingham, E.; Clements, M.O. The staphylococcus aureus alternative sigma factor ς B controls the environmental stress response but not starvation survival or pathogenicity in a mouse abscess model. J. Bacteriol. 1998, 180, 6082–6089. [Google Scholar]
- Regassa, L.B.; Novick, R.P.; Betley, M.J. Glucose and nonmaintained pH decrease expression of the accessory gene regulator (agr) in staphylococcus aureus. Infec. Immun. 1992, 60, 3381–3388. [Google Scholar]
- Bore, E.; Langsrud, S.; Langsrud, Ø.; Rode, T.M.; Holck, A. Acid-shock responses in Staphylococcus aureus investigated by global gene expression analysis. Microbiology 2007, 153, 2289–2303. [Google Scholar]
- Rothfork, J.M.; Timmins, G.S.; Harris, M.N.; Chen, X.; Lusis, A.J.; Otto, M.; Cheung, A.L.; Gresham, H.D. Inactivation of a bacterial virulence pheromone by phagocyte-derived oxidants: New role for the NADPH oxidase in host defense. Proc. Natl. Acad. Sci. USA 2004, 101, 13867–13872. [Google Scholar]
- Schlievert, P.M.; Case, L.C.; Nemeth, K.A.; Davis, C.C.; Sun, Y.; Qin, W.; Wang, F.; Brosnahan, A.J.; Mleziva, J.A.; Peterson, M.L.; et al. Alpha and beta chains of hemoglobin inhibit production of staphylococcus aureus exotoxins. Biochemistry 2007, 46, 14349–14358. [Google Scholar]
- Peterson, M.M.; Mack, J.L.; Hall, P.R.; Alsup, A.A.; Alexander, S.M.; Sully, E.K.; Sawires, Y.S.; Cheung, A.L.; Otto, M.; Gresham, H.D. Apolipoprotein B is an innate barrier against invasive staphylococcus aureus infection. Cell Host Micro 2008, 4, 555–566. [Google Scholar]
- Pynnonen, M.; Stephenson, R.E.; Schwartz, K.; Hernandez, M.; Boles, B.R. Hemoglobin promotes staphylococcus aureus nasal colonization. PLoS Pathog 2011. [Google Scholar] [CrossRef]
- Attia, A.S.; Benson, M.A.; Stauff, D.L.; Torres, V.J.; Skaar, E.P. Membrane damage elicits an immunomodulatory program in staphylococcus aureus. PLoS Pathog 2010. [Google Scholar] [CrossRef]
- Wright, J.S.; Jin, R.; Novick, R.P. Transient interference with staphylococcal quorum sensing blocks abscess formation. Proc. Natl. Acad. Sci. USA 2005, 102, 1691–1696. [Google Scholar]
- Otto, M.; Echner, H.; Voelter, W.; Gotz, F. Pheromone cross-inhibition between staphylococcus aureus and staphylococcus epidermidis. Infec. Immun. 2001, 69, 1957–1960. [Google Scholar]
- Otto, M.; Süssmuth, R.; Vuong, C.; Jung, G.; Götz, F. Inhibition of virulence factor expression in staphylococcus aureus by the staphylococcus epidermidis agr pheromone and derivatives. FEBS Lett 1999, 450, 257–262. [Google Scholar]
- Lina, G.; Boutite, F.; Tristan, A.; Bes, M.; Etienne, J.; Vandenesch, F. Bacterial competition for human nasal cavity colonization: role of staphylococcal agr alleles. Appl. Environ. Microbiol. 2003, 69, 18–23. [Google Scholar]
- Li, J.; Wang, W.; Xu, S.X.; Magarvey, N.A.; McCormick, J.K. Lactobacillus reuteri-produced cyclic dipeptides quench agr-mediated expression of toxic shock syndrome toxin-1 in staphylococci. Proc. Natl. Acad. Sci. USA 2011, 108, 3360–3365. [Google Scholar]
- Qazi, S.; Middleton, B.; Muharram, S.H.; Cockayne, A.; Hill, P.; Shea, P.O.; Chhabra, S.R.; Ca, M.; Williams, P. N-acylhomoserine lactones antagonize virulence gene expression and quorum sensing in staphylococcus aureus. Infec. Immun. 2006, 74, 910–919. [Google Scholar]
- Mitchell, G.; Séguin, D.L.; Asselin, A.-E.; Déziel, E.; Cantin, A.M.; Frost, E.H.; Michaud, S.; Malouin, F. Staphylococcus aureus sigma B-dependent emergence of small-colony variants and biofilm production following exposure to Pseudomonas aeruginosa 4-hydroxy-2-heptylquinoline-N-oxide. BMC Microbiol 2010, 10, 33. [Google Scholar]
- Vuong, C.; Otto, M. Staphylococcus epidermidis infections. Micro. Infec. 2002, 4, 481–489. [Google Scholar]
- Wang, R.; Khan, B.A.; Cheung, G.Y.C.; Bach, T.L.; Jameson-lee, M.; Kong, K.; Queck, S.Y.; Otto, M. Staphylococcus epidermidis surfactant peptides promote biofilm maturation and dissemination of biofilm-associated infection in mice. J. Clin. Invest. 2011, 121, 238–248. [Google Scholar]
- Dai, L.; Yang, L.; Parsons, C.; Findlay, V.J.; Molin, S.; Qin, Z. Staphylococcus epidermidis recovered from indwelling catheters exhibit enhanced biofilm dispersal and “self-renewal” through downregulation of agr. BMC Microbiol 2012, 12, 102. [Google Scholar]
- Lambe, D., Jr; Ferguson, K.; Keplinger, J.; Gemmell, C.; Kalbfleisch, J. Pathogenicity of staphylococcus lugdunensis, staphylococcus schleiferi, and three other coagulase-negative staphylococci in a mouse model and possible virulence factors. Can. Microbiol. 1990, 36, 455–463. [Google Scholar]
- Vandenesch, F.; Storrs, M.; Poitevin-Later, F.; Etienne, J.; Courvalin, P.; Fleurette, J. Delta-like haemolysin produced by Staphylococcus lugdunensis. FEMS Microbiol. Lett. 1991, 62, 65–68. [Google Scholar]
- Donvito, B.; Etienne, J.; Greenland, T.; Mouren, C.; Delorme, V.; Vandenesch, F. Distribution of the synergistic haemolysin genes hld and slush with respect to agr in human staphylococci. FEMS Microbiol. Lett. 1997, 151, 139–144. [Google Scholar]
- Fitzgerald, J.R. The staphylococcus intermedius group of bacterial pathogens: species re-classification, pathogenesis and the emergence of meticillin resistance. Veterin. Dermatol. 2009, 20, 490–495. [Google Scholar]
- Ben Zakour, N.L.; Bannoehr, J.; Van den Broek, A.H.M.; Thoday, K.L.; Fitzgerald, J.R. Complete genome sequence of the canine pathogen staphylococcus pseudintermedius. J. Bacteriol. 2011, 193, 2363–2364. [Google Scholar]
- Kalkum, M.; Lyon, G.J.; Chait, B.T. Detection of secreted peptides by using hypothesis-driven multistage mass spectrometry. Proc. Natl. Acad. Sci. USA 2003, 100, 2795–2800. [Google Scholar]
- Stabler, R.A.; He, M.; Dawson, L.; Martin, M.; Valiente, E.; Corton, C.; Lawley, T.D.; Sebaihia, M.; Quail, M.A.; Rose, G.; et al. Comparative genome and phenotypic analysis of clostridium difficile 027 strains provides insight into the evolution of a hypervirulent bacterium. Genome Biol 2009, 10, R102. [Google Scholar]
- Vidal, J.E.; Ma, M.; Saputo, J.; Garcia, J.; Uzal, F.A.; McClane, B.A. Evidence that the Agr-like quorum sensing system regulates the toxin production, cytotoxicity and pathogenicity of Clostridium perfringens type C isolate CN3685. Mol. Microbiol. 2012, 83, 179–194. [Google Scholar]
- Chen, J.; McClane, B. A role of the agr-like quorum-sensing system in regulating toxin production by clostridium perfringens type B strains CN1793 and CN1795. Infec. Immun. 2012, 80, 3008–3017. [Google Scholar]
- Ohtani, K.; Hirakawa, H.; Tashiro, K.; Yoshizawa, S.; Kuhara, S.; Shimizu, T. Identification of a two-component VirR/VirS regulon in clostridium perfringens. Anaerobe 2010, 16, 258–264. [Google Scholar]
- Shimizu, T.; Yaguchi, H.; Ohtani, K.; Banu, S.; Hayashi, H. Clostridial VirR/VirS regulon involves a regulatory RNA molecule for expression of toxins. Mol. Microbiol. 2002, 43, 257–265. [Google Scholar]
- Pinkston, K.L.; Gao, P.; Diaz-Garcia, D.; Sillanpää, J.; Nallapareddy, S.R.; Murray, B.E.; Harvey, B.R. The fsr quorum-sensing system of enterococcus faecalis modulates surface display of the collagen-binding MSCRAMM ace through regulation of gelE. J. Bacteriol. 2011, 193, 4317–4325. [Google Scholar]
- Engelbert, M.; Mylonakis, E.; Ausubel, F.M.; Calderwood, S.B.; Gilmore, M.S. Contribution of gelatinase, serine protease, and fsr to the pathogenesis of enterococcus faecalis endophthalmitis. Infec. Immun. 2004, 72, 3628–3633. [Google Scholar]
- Bourgogne, A.; Hilsenbeck, S.G.; Dunny, G.M.; Murray, B.E. Comparison of OG1RF and an isogenic fsrB deletion mutant by transcriptional analysis: the fsr system of enterococcus faecalis is more than the activator of gelatinase and serine protease. J. Bacteriol. 2006, 188, 2875–2884. [Google Scholar]
- Teixeira, N.; Santos, S.; Marujo, P.; Yokohata, R.; Iyer, V.S.; Nakayama, J.; Hancock, L.E.; Serror, P.; Silva Lopes, M.D.F. The incongruent gelatinase genotype and phenotype in Enterococcus faecalis are due to shutting off the ability to respond to the gelatinase biosynthesis-activating pheromone (GBAP) quorum-sensing signal. Microbiology 2012, 158, 519–528. [Google Scholar]
- Bourgogne, A.; Thomson, L.C.; Murray, B.E. Bicarbonate enhances expression of the endocarditis and biofilm associated pilus locus, ebpR-ebpABC, in enterococcus faecalis. BMC Microbiol 2010, 10, 17. [Google Scholar]
- Nakayama, J.; Kariyama, R.; Kumon, H. Description of a 23.9-kilobase chromosomal deletion containing a region encoding fsr genes which mainly determines the gelatinase-negative phenotype of clinical isolates of enterococcus faecalis in urine. Appl. Environ. Microbiol. 2002, 68, 3152–3155. [Google Scholar]
- Shankar, J.; Walker, R.G.; Ward, D.; Horsburgh, M.J. The enterococcus faecalis exoproteome: identification and temporal regulation by Fsr. PLoS One 2012. [Google Scholar] [CrossRef]
- Williams, T.; Bauer, S.; Beier, D.; Kuhn, M. Construction and characterization of listeria monocytogenes mutants with in-frame deletions in the response regulator genes identified in the genome sequence. Infec. Immun. 2005, 73, 3152–3159. [Google Scholar]
- Montalban-Lopez, M.; Sanchez-Hidalgo, M.; Valdivia, E.; Martinez-Bueno, M.; Maqueda, M. Are bacteriocins underexploited? Novel applications for old antimicrobials. Curr. Pharm. Biotechnol. 2011, 12, 1205–1220. [Google Scholar]
- Eijsink, V.G.H.; Axelsson, L.; Diep, D.B.; Håvarstein, L.S.; Holo, H.; Nes, I.F. Production of class II bacteriocins by lactic acid bacteria; an example of biological warfare and communication. Anton. Van Leeuwenhoek 2002, 81, 639–654. [Google Scholar]
- Sit, C.S.; Vederas, J.C. Approaches to the discovery of new antibacterial agents based on bacteriocins. Biochem. Cell Biol. 2008, 86, 116–123. [Google Scholar]
- Sturme, M.H.J.; Francke, C.; Siezen, R.J.; De Vos, W.M.; Kleerebezem, M. Making sense of quorum sensing in lactobacilli: a special focus on Lactobacillus plantarum WCFS1. Microbiology 2007, 153, 3939–3947. [Google Scholar]
- Rocha-Estrada, J.; Aceves-Diez, A.E.; Guarneros, G.; De La Torre, M. The RNPP family of quorum-sensing proteins in Gram-positive bacteria. Appl. Microbiol. Biotechnol. 2010, 87, 913–923. [Google Scholar]
- Bozue, J.; Powell, B.S.; Cote, C.K.; Moody, K.L.; Gelhaus, H.C.; Vietri, N.J.; Rozak, D.A. Disrupting the luxS quorum sensing gene does not significantly affect Bacillus anthracis virulence in mice or guinea pigs. Virulence 2012, 3, 504–509. [Google Scholar]
- Jones, M.B.; Peterson, S.N.; Benn, R.; Braisted, J.C.; Jarrahi, B.; Shatzkes, K.; Ren, D.; Wood, T.K.; Blaser, M.J. Role of luxS in Bacillus anthracis growth and virulence factor expression. Virulence 2010, 1, 72–83. [Google Scholar]
- Jones, M.B.; Jani, R.; Ren, D.; Wood, T.K.; Blaser, M.J. Inhibition of bacillus anthracis growth and virulence-gene expression by inhibitors of quorum-sensing. J. Infec. Dis. 2005, 191, 1881–1888. [Google Scholar]
- Rocha, J.; Flores, V.; Cabrera, R.; Soto-Guzmán, A.; Granados, G.; Juaristi, E.; Guarneros, G.; De La Torre, M. Evolution and some functions of the NprR-NprRB quorum-sensing system in the Bacillus cereus group. Appl. Microbiol. Biotechnol. 2011, 94, 1069–1078. [Google Scholar]
- Bouillaut, L.; Perchat, S.; Arold, S.; Zorrilla, S.; Slamti, L.; Henry, C.; Gohar, M.; Declerck, N.; Lereclus, D. Molecular basis for group-specific activation of the virulence regulator PlcR by PapR heptapeptides. Nucl. Acids Res. 2008, 36, 3791–3801. [Google Scholar]
- Pomerantsev, A.P.; Pomerantseva, O.M.; Camp, A.S.; Mukkamala, R.; Goldman, S.; Leppla, S.H. PapR peptide maturation: Role of the NprB protease in Bacillus cereus 569 PlcR/PapR global gene regulation. FEMS Immunol. Med. Microbiol. 2010, 55, 361–377. [Google Scholar]
- Pakula, R. Factors regulating competence in transformation of streptococci. J. Bacteriol. 1965, 90, 1320–1324. [Google Scholar]
- Martin, B.; Quentin, Y.; Fichant, G.; Claverys, J.-P. Independent evolution of competence regulatory cascade in streptococci? Trends Microbiol 2006, 14, 339–345. [Google Scholar]
- Kreth, J.; Merritt, J.; Zhu, L.; Shi, W.; Qi, F. Cell density- and come-dependent expression of a group of mutacin and mutacin-like genes in Streptococcus mutans. FEMS Microbiol. Lett. 2006, 265, 11–17. [Google Scholar]
- Kreth, J.; Hung, D.C.I.; Merritt, J.; Perry, J.; Zhu, L.; Goodman, S.D.; Cvitkovitch, D.G.; Shi, W.; Qi, F. The response regulator come in streptococcus mutans functions both as a transcription activator of mutacin production and repressor of CSP biosynthesis. Microbiology 2007, 153, 1799–1807. [Google Scholar]
- Tamura, S.; Yonezawa, H.; Motegi, M.; Nakao, R.; Yoneda, S.; Watanabe, H.; Yamazaki, T. Inhibiting effects of streptococcus salivarius on dependent biofilm formation by streptococcus mutans. Oral Microbiol. Immunol. 2009, 24, 152–161. [Google Scholar]
- Desai, K.; Mashburn-Warren, L.; Federle, M.J.; Morrison, D. Development of competence for genetic transformation of streptococcus mutans in a chemically defined medium. J. Bacteriol. 2012, 194, 3774–3780. [Google Scholar]
- Wenderska, I.B.; Lukenda, N.; Cordova, M.; Magarvey, N.; Cvitkovitch, D.G.; Senadheera, D.B. A novel function for the competence inducing peptide, XIP, as a cell death effector of Streptococcus mutans. FEMS Microbiol. Lett. 2012, 336, 104–112. [Google Scholar]
- Yano, M.; Gohil, S.; Coleman, J.R.; Manix, C.; Pirofski, L. Antibodies to streptococcus pneumoniae capsular polysaccharide. Mbio 2011, 2, 1–10. [Google Scholar]
- Klenk, M.; Koczan, D.; Guthke, R.; Nakata, M.; Thiesen, H.-J.; Podbielski, A.; Kreikemeyer, B. Global epithelial cell transcriptional responses reveal streptococcus pyogenes fas regulator activity association with bacterial aggressiveness. Cell. Microbiol. 2005, 7, 1237–1250. [Google Scholar]
- Malke, H.; Ferretti, J.J. CodY-affected transcriptional gene expression of streptococcus pyogenes during growth in human blood. J. Med. Microbiol. 2007, 56, 707–714. [Google Scholar]
- Hidalgo-Grass, C.; Ravins, M.; Dan-Goor, M.; Jaffe, J.; Moses, A.E.; Hanski, E. A locus of group a streptococcus involved in invasive disease and DNA transfer. Mol. Microbiol. 2002, 46, 87–99. [Google Scholar]
- Eran, Y.; Getter, Y.; Baruch, M.; Belotserkovsky, I.; Padalon, G.; Mishalian, I.; Podbielski, A.; Kreikemeyer, B.; Hanski, E. Transcriptional regulation of the sil locus by the SilCR signalling peptide and its implications on group A streptococcus virulence. Mol. Microbiol. 2007, 63, 1209–22. [Google Scholar]
- Salim, K.Y.; De Azavedo, J.C.; Bast, D.J.; Cvitkovitch, D.G. Regulation of sagA, siaA and scpC by SilCR, a putative signaling peptide of streptococcus pyogenes. FEMS Microbiol. Lett. 2008, 289, 119–125. [Google Scholar]
- Hidalgo-Grass, C.; Dan-Goor, M.; Maly, A.; Eran, Y.; Kwinn, L.A.; Nizet, V.; Ravins, M.; Jaffe, J.; Peyser, A.; Moses, A.E.; Hanski, E. Effect of a bacterial pheromone peptide on host chemokine degradation in group a streptococcal necrotising soft-tissue infections. Lancet 2004, 363, 696–703. [Google Scholar]
- Nakayama, J.; Akkermans, A.D.L.; De Vos, W.M. High-throughput PCR screening of genes for three-component regulatory system putatively involved in quorum sensing from low-G + C gram-positive bacteria. Biosci. Biotechnol. Biochem. 2003, 67, 480–489. [Google Scholar]
- Anderssen, E.E.L.; Diep, D.B.; Nes, I.F.; Eijsink, V.G.H.; Nissen-Meyer, J. Antagonistic activity of Lactobacillus plantarum C11: Two new two-peptide bacteriocins, plantaricins EF and JK, and the induction factor plantaricin A. Appl. Environ. Microbiol. 1998, 64, 2269–2272. [Google Scholar]
- Calasso, M.; Di Cagno, R.; De Angelis, M.; Campanella, D.; Minervini, F.; Gobbetti, M. Effects of the peptide pheromone plantaricin A and cocultivation with lactobacillus sanfranciscensis DPPMA174 on the exoproteome and the adhesion capacity of lactobacillus plantarum DC400. Appl. Environ. Microbiol. 2013, 79, 2657–2669. [Google Scholar]
- Forsberg, K.J.; Reyes, A.; Wang, B.; Selleck, E.M.; Sommer, M.O.A.; Dantas, G. The shared antibiotic resistome of soil bacteria and human pathogens. Science 2012, 337, 1107–1111. [Google Scholar]
- Sommer, M.O.A.; Dantas, G.; Church, G.M. Functional characterization of the antibiotic resistance reservoir in the human microflora. Science 2009, 325, 1128–1131. [Google Scholar]
- Tong, S.Y.C.; Chen, L.F.; Fowler, V.G. Colonization, pathogenicity, host susceptibility, and therapeutics for Staphylococcus aureus: what is the clinical relevance? Semin. Immunopathol. 2012, 34, 185–200. [Google Scholar]
- Soriano, A.; Popescu, D.; Garcia, S.; Bori, G.; Martinez, J.A.; Balasso, V.; Marco, F.; Almela, M.; Mensa, J. Usefulness of teicoplanin for preventing methicillin-resistant staphylococcus aureus infections in orthopedic surgery. Eur. J. Clin. Microbiol. Infec. Dis. 2006, 25, 35–38. [Google Scholar]
- Gemmell, C.G.; Edwards, D.I.; Fraise, A.P.; Gould, F.K.; Ridgway, G.L.; Warren, R.E. Guidelines for the prophylaxis and treatment of methicillin-resistant Staphylococcus aureus (MRSA) infections in the UK. J. Antimicrob. Chemother. 2006, 57, 589–608. [Google Scholar]
- Giannoudis, P.V.; Parker, J.; Wilcox, M.H. Methicillin-resistant Staphylococcus in trauma and orthopaedic practice. J. Bone Joint Surg. 2005, 87, 749–754. [Google Scholar]
- Baldoni, D.; Haschke, M.; Rajacic, Z.; Zimmerli, W.; Trampuz, A. Linezolid alone or combined with rifampin against methicillin-resistant Staphylococcus aureus in experimental foreign-body infection. Antimicrob. Agent. Chemother. 2009, 53, 1142–1148. [Google Scholar]
- Kowalski, R.P.; Romanowski, E.G.; Mah, F.S.; Sasaki, H.; Fukuda, M.; Gordon, Y.J. A comparison of moxifloxacin and levfloxacin topical prophylaxis in a fluoroquinolone-resistant Staphylococcus aureus rabbit model. Jpn. J. Opthalmol. 2008, 53, 211–216. [Google Scholar]
- Niska, J.A.; Shahbazian, J.H.; Ramos, R.I.; Pribaz, J.R.; Billi, F.; Francis, K.P.; Miller, L.S. Daptomycin and tigecycline have a broader effective dose range than vancomycin as prophylaxis against a surgical implant Staphylococcus aureus infection in mice. Antimicrob. Agent. Chemother. 2012, 56, 2590–2597. [Google Scholar]
- Howden, B.P.; Davies, J.K.; Johnson, P.D.R.; Stinear, T.P.; Grayson, M.L. Reduced vancomycin susceptibility in Staphylococcus aureus, including vancomycin-intermediate and heterogeneous vancomycin-intermediate strains: Resistance mechanisms, laboratory detection, and clinical implications. Clin. Microbiol. Rev. 2010, 23, 99–139. [Google Scholar]
- Vuong, C.; Gerke, C.; Somerville, G.A.; Fischer, E.R.; Otto, M. Quorum-sensing control of biofilm factors in staphylococcus epidermidis. J. Infec. Dis. 2003, 188, 706–718. [Google Scholar]
- Xiong, Y.-Q.; Van Wamel, W.; Nast, C.C.; Yeaman, M.R.; Cheung, A.L.; Bayer, A.S. Activation and transcriptional interaction between agr RNAII and RNAIII in Staphylococcus aureus in vitro and in an experimental endocarditis model. J. Infec. Dis 2002, 186, 668–677. [Google Scholar]
- Yang, L.; Rybtke, M.T.; Jakobsen, T.H.; Hentzer, M.; Bjarnsholt, T.; Givskov, M.; Tolker-Nielsen, T. Computer-aided identification of recognized drugs as Pseudomonas aeruginosa quorum-sensing inhibitors. Antimicrob. Agent. Chemother. 2009, 53, 2432–2443. [Google Scholar]
- Leonard, P.G.; Bezar, I.F.; Sidote, D.J.; Stock, A.M. Identification of a hydrophobic cleft in the LytTR domain of AgrA as a locus for small molecule interactions that inhibit DNA binding. Biochemistry 2012, 51, 10035–10043. [Google Scholar]
- Olson, P.D.; Kuechenmeister, L.J.; Anderson, K.L.; Daily, S.; Beenken, K.E.; Roux, C.M.; Reniere, M.L.; Lewis, T.L.; Weiss, W.J.; Pulse, M.; et al. Small molecule inhibitors of Staphylococcus aureus RnpA alter cellular mRNA turnover, exhibit antimicrobial activity, and attenuate pathogenesis. PLoS Pathog 2011. [Google Scholar] [CrossRef]
- Nakayama, J.; Tanaka, E.; Kariyama, R.; Nagata, K.; Nishiguchi, K.; Mitsuhata, R.; Uemura, Y.; Tanokura, M.; Kumon, H.; Sonomoto, K. Siamycin attenuates fsr quorum sensing mediated by a gelatinase biosynthesis-activating pheromone in enterococcus faecalis. J. Bacteriol. 2007, 189, 1358–1365. [Google Scholar]
- Ma, P.; Nishiguchi, K.; Yuille, H.M.; Davis, L.M.; Nakayama, J.; Phillips-Jones, M.K. Anti-HIV siamycin I directly inhibits autophosphorylation activity of the bacterial FsrC quorum sensor and other ATP-dependent enzyme activities. FEBS Lett 2011, 585, 2660–2664. [Google Scholar]
- Scott, R.J.; Lian, L.-Y.; Muharram, S.H.; Cockayne, A.; Wood, S.J.; Bycroft, B.W.; Williams, P.; Chan, W.C. Side-chain-to-tail thiolactone peptide inhibitors of the staphylococcal quorum-sensing system. Bioorg. Med. Chem. Lett. 2003, 13, 2449–2453. [Google Scholar]
- Blackburn, E.A.; Walkinshaw, M.D. Targeting FKBP isoforms with small-molecule ligands. Curr. Opin. Pharmacol. 2011, 11, 365–371. [Google Scholar]
- Benjamin, D.; Colombi, M.; Moroni, C.; Nall, M.H. Rapamycin passes the torch: A new generation of mTOR inhibitors. Nat. Rev. Drug Discov. 2011, 10, 868–880. [Google Scholar]
- Parrill, A.L.; Sardar, V.M.; Yuan, H. Sphingosine 1-phosphate and lysophosphatidic acid receptors: agonist and antagonist binding and progress toward development of receptor-specific ligands. Semin. Cell Dev. Biol. 2004, 15, 467–476. [Google Scholar]
- Nakayama, J.; Uemura, Y.; Nishiguchi, K.; Yoshimura, N.; Igarashi, Y.; Sonomoto, K. Ambuic acid inhibits the biosynthesis of cyclic peptide quormones in gram-positive bacteria. Antimicrob. Agent. Chemother. 2009, 53, 580–586. [Google Scholar]
- Lyon, G.J.; Mayville, P.; Muir, T.W.; Novick, R.P. Rational design of a global inhibitor of the virulence response in staphylococcus aureus, based in part on localization of the site of inhibition to the receptor-histidine kinase, AgrC. Proc. Natl. Acad. Sci. USA 2000, 97, 13330–13335. [Google Scholar]
- Nakayama, J.; Yokohata, R.; Sato, M.; Suzuki, T.; Matsufuji, T.; Nishiguchi, K.; Kawai, T.; Yamanaka, Y.; Nagata, K.; Tanokura, M.; et al. Development of a peptide antagonist against fsr quorum sensing of enterococcus faecalis. ACS Chem. Biol. 2013. [Google Scholar] [CrossRef]
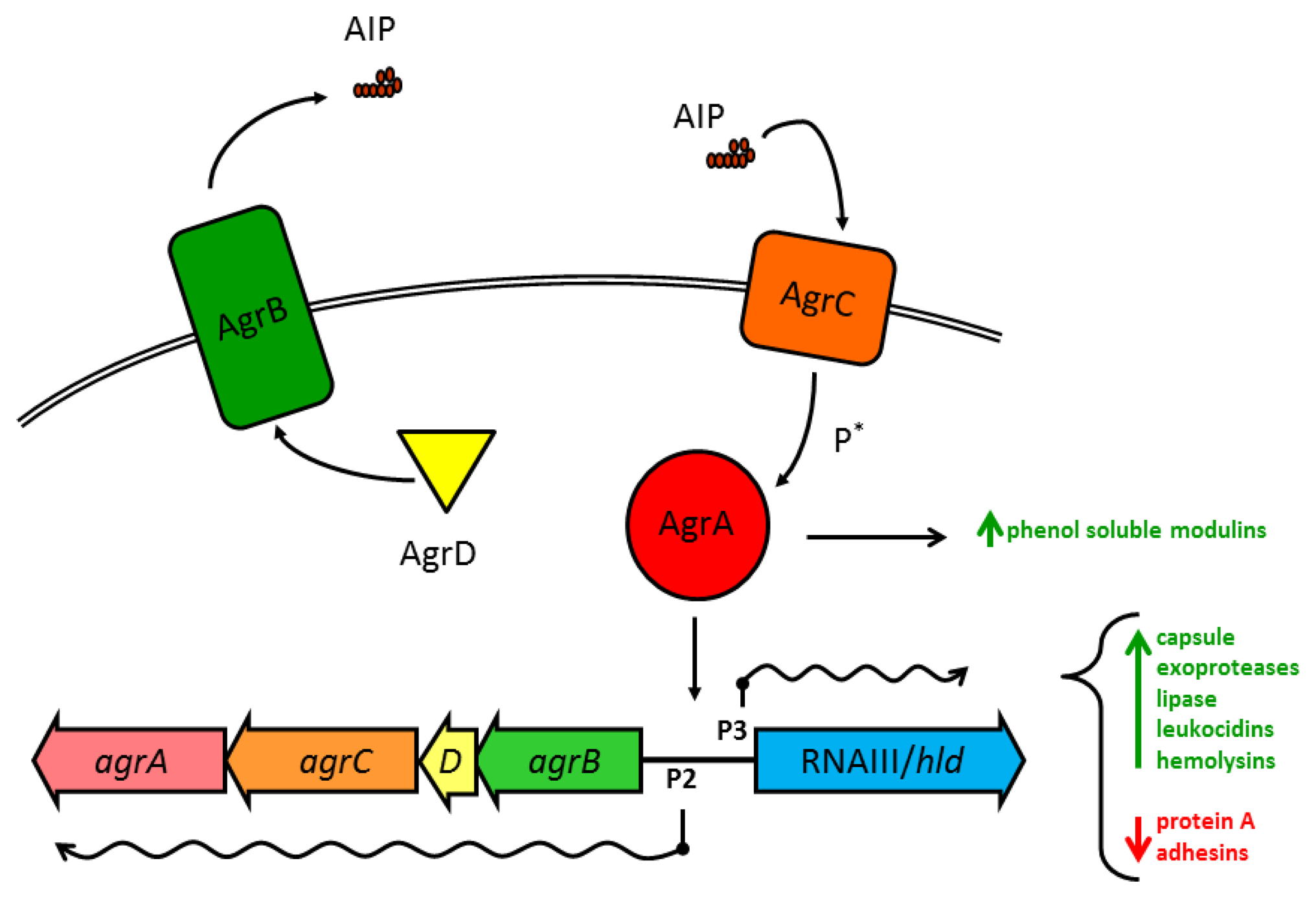
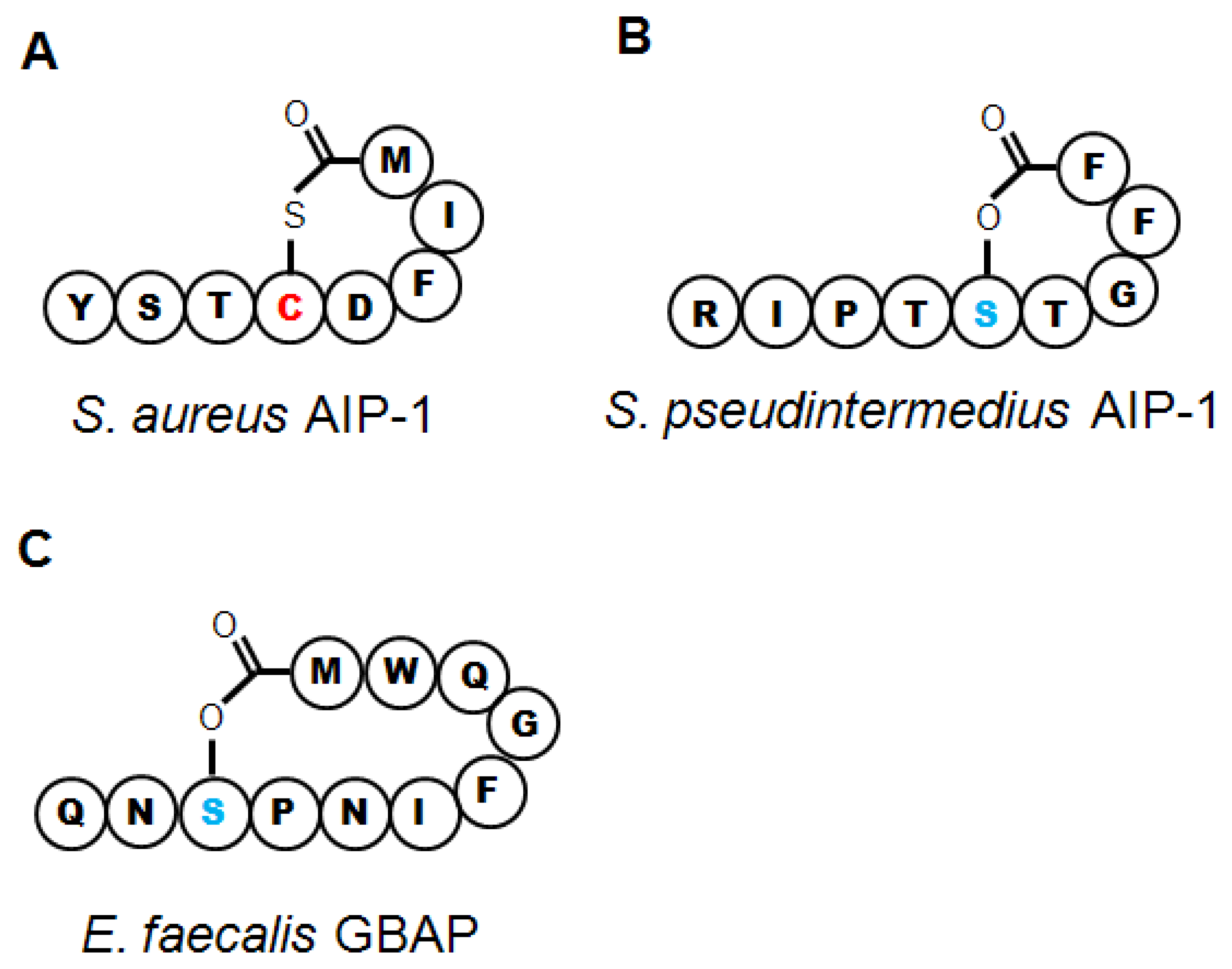
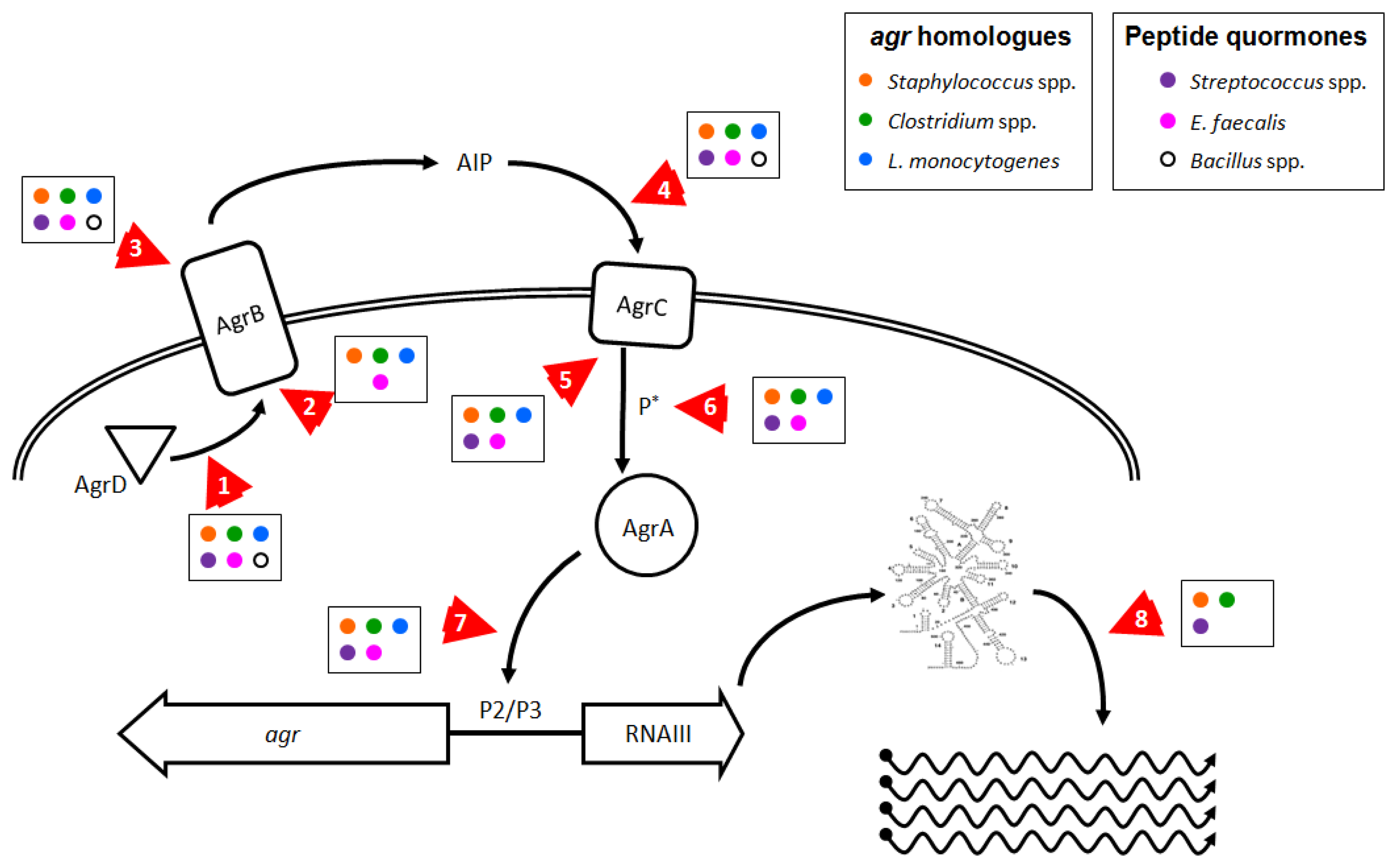
| Species | agrHomologues/Analogues | Discovery Method | Up-Regulated Behavior | Signal Peptide Name and Sequence1 | ||
|---|---|---|---|---|---|---|
| Staphylococcus aureus | agr (agrBDCA) | Mutants [2,30] | Virulence (toxins, capsule) [1,31,32] | Sa AIP 1 | YST
 DFIM DFIM | |
| Sa AIP 2 | GVNA
 SSLF SSLF | |||||
| Sa AIP 3 | IN
 DFLL DFLL | |||||
| Sa AIP 4 | YST
 YFIM YFIM | |||||
| S. epidermidis | agr | Mutants, genomics [33–35] | Virulence (toxins), biofilm [34,36–38] | AIP-Se 1 | DSV
 ASYF ASYF | |
| AIP-Se 2 | YNP
 NSYL NSYL | |||||
| AIP-Se 3 | YNP
 SAYL SAYL | |||||
| S. lugdunensis | agr | Genomics [20,39] | HKU09-01 | DI
 NAYF NAYF | ||
| N920217 | DMN
 NGYF NGYF | |||||
| S. saprophyticus | agr | Genomics [40] | agr | TINP
 FGYT FGYT | ||
| S. pseudintermedius | agr | Genomics, proteomics [41,42] | Si AIP 1 | RIPT
 TGFF TGFF | ||
| agr II | RIPI
 TGFF TGFF | |||||
| agr III | KIPT
 TGFF TGFF | |||||
| agr IV | KYPT
 TGFF TGFF | |||||
| Staphylococcus spp. | agr | Mutants, genomics, proteomics [18,37,43] | Virulence (hemolysins, surface proteins) [20,37,40,43] | |||
| Clostridium botulinum | agrD1, agrD2 | Mutants [44] | Sporulation (agrD1), virulence (toxins) (agrD2) [44] | agrD1 | ACYW
 VYQP VYQP | |
| agrD2 | ADSA
 HLGI HLGI | |||||
| C. difficile | agr, agrD2 | Genomics [44,45] | agrD | ANST
 PWII PWII | ||
| agr2 | NSA
 SWVA SWVA | |||||
| C. perfringens | agrBD | Mutants, genomics [18,21,46, 47] | Virulence (toxins, proteases), sporulation [21,46,47] | ATSA
 IWFT IWFT | ||
| Enterococcus faecalis | fsr (fsrBDCA) | Mutants, genomics [18,48–50] | Virulence (proteases), biofilms [49,50] | GBAP | QN
 PNIFGQWM PNIFGQWM | |
| Listeria monocytogenes | agr | Mutants, genomics [18,51,52] | Biofilm, virulence (internalization, toxins) [51–53] | 4b agr | MSKA
 FMFV FMFV | |
| DG119D | RLAS
 LYTQ LYTQ | |||||
| agr | ||||||
| Lactobacillus plantarum | lam (lamBDCA) | Mutants [54] | Adhesion [54] | LamD558 | LVMC
 VGIW VGIW | |
| Peptide Quormone Analogues | ||||||
| Bacillus cereus | pap, plc | Mutants, genomics [55,56] | Sporulation, virulence (cytotoxicity, toxins), stress responses [55–58] | PapR I | ADLPFEF | |
| PapR II | SDMPFEF | |||||
| PapR III | NEVPFEF | |||||
| PapR IV | SDLPFEH | |||||
| PapRa | CSIPYEY | |||||
| B. thuringiensis | pap | Mutants [59] | Virulence (toxins) [59,60] | |||
| Bacillus spp. | plc, pap | Mutants, genomics [18,55,61] | Sporulation, virulence (toxins) [55,56] | |||
| Streptococcus pyogenes | comC, comRS, fas, sil | Mutants, genomics [62–64] | Virulence (invasion, toxins), competence, biofilms [62–66] | CSP | MRLSKFFRDFILQRKK | |
| XIP | SAVDWWRL | |||||
| SilCR | DIFKLVIDHISMKARKK | |||||
| Streptococcus spp. | comC, comRS, sil | Mutants, genomics [62,64,67] | Competence, biofilms [64,67] | XIP | GLDWWSL | |
| Lactobacillus spp. | pln, plt | Mutants, genomics [18,54] | Adhesion, bacteriocin production [54,68] | PlnA | KSSAYSLQMGATAIKQVKKLFKKWGW | |
| PltA | EQLSFTSIGILQLLTIGTRSCWFFYCRY | |||||
1Sequences in italics denote peptide sequences identified through genomics methods but not yet confirmed through mass spectrometry or other chemical methods.
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Gray, B.; Hall, P.; Gresham, H. Targeting agr- and agr-Like Quorum Sensing Systems for Development of Common Therapeutics to Treat Multiple Gram-Positive Bacterial Infections. Sensors 2013, 13, 5130-5166. https://doi.org/10.3390/s130405130
Gray B, Hall P, Gresham H. Targeting agr- and agr-Like Quorum Sensing Systems for Development of Common Therapeutics to Treat Multiple Gram-Positive Bacterial Infections. Sensors. 2013; 13(4):5130-5166. https://doi.org/10.3390/s130405130
Chicago/Turabian StyleGray, Brian, Pamela Hall, and Hattie Gresham. 2013. "Targeting agr- and agr-Like Quorum Sensing Systems for Development of Common Therapeutics to Treat Multiple Gram-Positive Bacterial Infections" Sensors 13, no. 4: 5130-5166. https://doi.org/10.3390/s130405130




