Microbial Biosensors: Engineered Microorganisms as the Sensing Machinery
Abstract
: Whole-cell biosensors are a good alternative to enzyme-based biosensors since they offer the benefits of low cost and improved stability. In recent years, live cells have been employed as biosensors for a wide range of targets. In this review, we will focus on the use of microorganisms that are genetically modified with the desirable outputs in order to improve the biosensor performance. Different methodologies based on genetic/protein engineering and synthetic biology to construct microorganisms with the required signal outputs, sensitivity, and selectivity will be discussed.1. Introduction
Monitoring toxins, environmental pollutants, traces of dangerous chemicals, hormones, and/or pathogens accurately and rapidly is an important task in the field of environmental stewardship, health care and homeland security, due to their non-negligible impact on the ecosystem and human society [1–6]. Compared with conventional chromatographic methodologies such as gas chromatography (GC) or high-performance liquid chromatography (HPLC), that are tedious and not portable, the use of biosensors is gaining more attention as they offer rapid and on-site/point-of-care monitoring of even trace levels of targets [2,6,7]. A biosensor is a device that combines biological molecules as the recognition element with a physical transducer and provides quantitative or semi-quantitative analytical data corresponding to the target concentration [4,7,8]. By taking advantage of the high specificity and tight interaction between biological molecules and target compounds, numerous studies have been conducted in order to develop biosensors with higher accuracy and sensitivity. These sensing devices utilize whole cells, different configurations of antibodies, DNA, RNA, enzymes or receptor proteins as the recognition elements [1–3,9]. In the presence of target analytes, biological events are converted into an electrical and/or optical signal in proportion to the target concentration. Among these biological recognition elements, enzymes are most widely used because they are highly selective. However, the need for costly and tedious purification is one limitation of constructing enzyme-based biosensors [7]. Moreover, multienzymes or cofactors are typically required to generate a measurable signal for certain targets.
Whole cells are a good alternative to enzymes since they have the benefit of low cost and improved stability comparing to enzymes or other proteins. Tedious purification could be avoided and cells can be massively produced through a simple cell culturing step and the necessary cofactors are already present inside the cells. Moreover, microbes are easy to manipulate and have better stability under harsh environments [4,7]. Live microorganisms have been employed as biosensors using different sensing techniques such as electrochemical and optical detection [10–13]. The electrochemical biosensor is a widely used technique offering high sensitivity and rapid detection. Electroactive species produced or consumed by microbes are monitored by conductimetric, amperometric, impedimetric, or potentiometric methods. Optical techniques are based on the measurement of fluorescent, luminescent, colorimetric, or other optical outputs from microorganisms [7]. Since one of the important features of these biosensors is how to link a physical signal from the recognition element to a given transducer, methods to immobilize whole cells onto the transducer represent a crucial topic in the field of biosensors. Several excellent reviews have already been published on the use of physical and chemical immobilization methods to maintain the functionality and the accessibility of whole cells towards the targets and will not be repeated here [8,9,14]. Rather, we will focus on the use of microorganisms that are genetically modified with different types of measurable signals in order to improve the biosensor performance and/or to develop biosensors for novel targets [15–17]. Different methodologies based on genetic/protein engineering and synthetic biology to construct microorganisms with the required signal outputs, sensitivity, and selectivity will be discussed.
2. Genetically Engineered Readouts of Microbial Sensors
In whole-cell biosensing, changes in cellular metabolism, pH, and gene expression have been quantified as a response of the sensing elements to the presence of target molecules [18–21]. Microbial auxotrophy have been used to monitor growth-limiting small molecules. For example, Pfleger et al. constructed an autotrophic E. coli strain for the detection and quantification of mevalonate, an intermediate in the biosynthesis of isoprenoids, a large class of industrially important secondary metabolites that includes flavor, fragrance, anti-oxidants, steroids, and the anti-malarial drug artemisinin [18]. Since mevalonate is a key precursor whose production must be increased in order to improve the production of isoprenoids, optimizing the level of mevalonate is important in developing recombinant strains for enhanced isoprenoid production. By deleting the native pathway for the production of isopentenyl pyrophosphate (IPP) and dimethylallyl pyrophosphate (DMAPP) that are necessary for growth, and incorporating the mevalonate-utilizing pathway (exogenous mevalonate can convert into IPP and DMAPP), the mevalonate sensor was developed to measure the concentration of mevalonate in mevalonate-producing cultures through simple growth monitoring [18]. In addition, the by-products generated from the target compounds can be used as the biosensor readout. Two early examples included the conversion of α-naphthyl acetate to α-naphthol and acetate resulting in a change in pH [19], and the hydrolysis of paraoxon, an organophosphate pesticide, to a yellow colored product, p-nitrophenol [22]. By expressing the enzyme γ-hexachlorocyclohexane (HCH) dehydrochlorinase from Sphingomonas paucimobilis that degrades lindane with the concomitant generation of hydrochloric acid in E. coli immobilized on a polyaniline film, a whole-cell electrochemical biosensor was developed for detection of the persistent organochlorine pesticide lindane by monitoring the change in the conductivity using pulsed amperometry [23]. This whole-cell electrochemical biosensor provides a more suitable platform for on-site monitoring of environmental samples as purified HCH dehydrochlorinase is likely to be inhibited by several physicochemical factors such as organophosphrous pesticides [23].
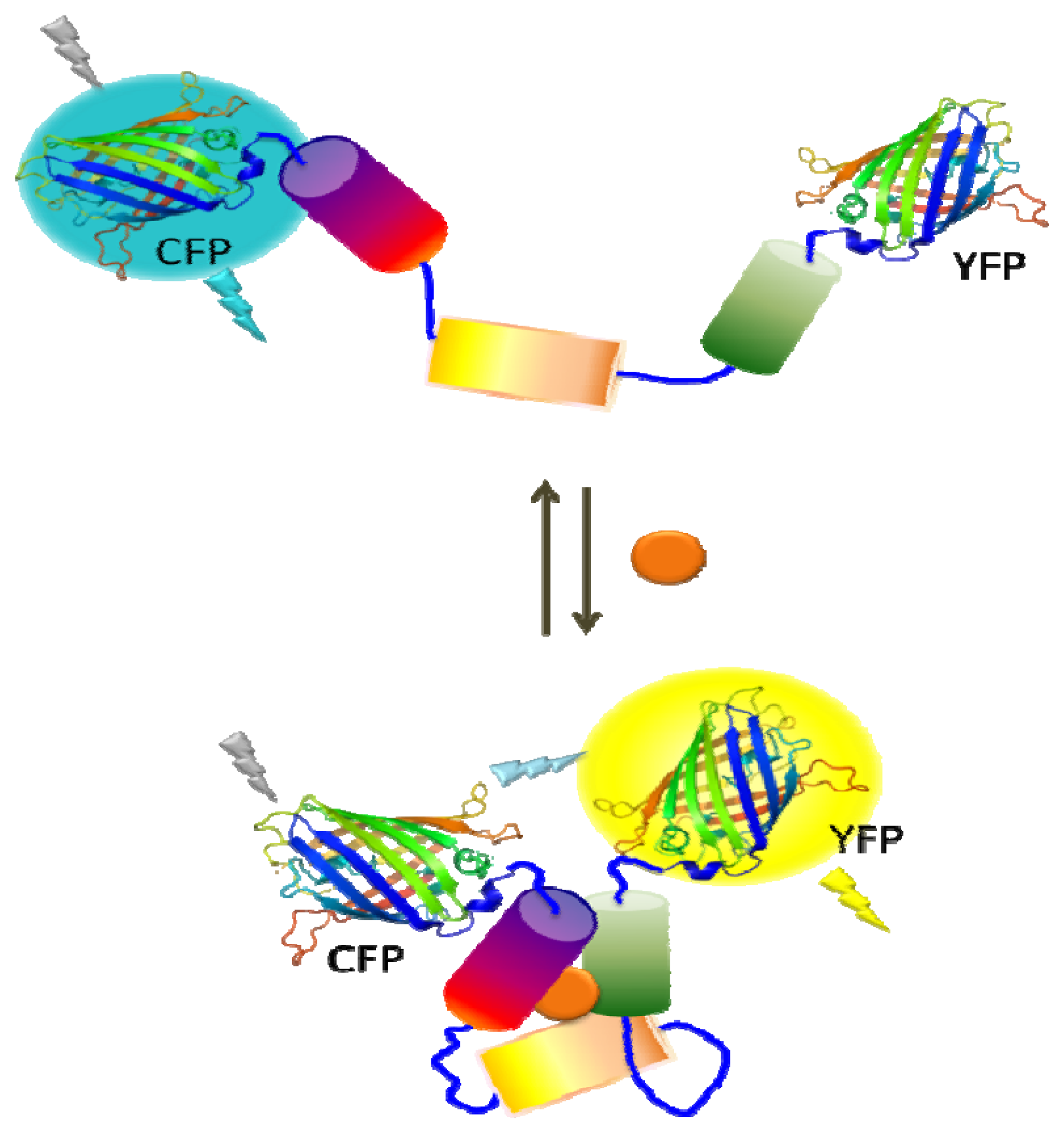
Reporter gene expression under the control of a specific regulatory network is another powerful readout as it offers not only increased sensitivity, but also provides a simple and easy sensor platform. Commonly used reporter genes are enzymes with activities for colorimetric, fluorescent, or luminescent readouts [20,24,25]. One commonly used enzyme is β-galactosidase (β-gal) which has the advantage of detection based on either colorimetric or fluorescent methods that are simple and rapid [24]. Moreover, the availability of chemiluminescent and electrochemical substrates for β-gal offers ultrahigh sensitivity offering detection as low as 2 fg and a wide dynamic range of detection [10,26] Luciferase, which catalyzes a light-emitting reaction, is another popular enzyme for whole-cell biosensors. The bacterial luciferase catalyzes the oxidation of a reduced flavin mononucleotide (FMNH2) and a long-chain fatty aldehyde in the presence of molecular oxygen and results in a blue-green light emission. The primary advantages of bacterial luciferase are its sensitivity, broad dynamic range, and simplicity. Moreover, expression of whole lux operon (luxCDABE) has benefit of not requiring the addition of a substrate in the system [20,27]. Firefly luciferase, on the other hand, utilizes luciferin as the substrate in the presence of ATP and oxygen, resulting in the visible light emission. In addition to the high sensitivity (subattomole level) and simplicity like bacterial luciferase, firefly luciferase has the advantage of a higher quantum yield than bacterial luciferase [25,28]. Fluorescent proteins are also widely utilized in microbial sensors as there is no need to supply substrates due to their autofluorescence. In addition to its simplicity, green fluorescent protein (GFP) has been used to measure gene expression and study cell-trafficking mechanisms [29]. Fluorescent proteins of different colors such as yellow fluorescent protein (YFP), cyan fluorescent protein (CFP), and mCherry have been generated to provide the ability of simultaneous detection of multiple targets and the possibility of fluorescence resonance energy transfer (FRET) between genetically fused fluorescent protein pairs. As the efficiency of fluorescence energy transfer is dependent on the distance and orientation of two fluorescent proteins, ligand binding can be used to trigger a conformational change resulting in a change in the FRET efficiency (Figure 1). Beyond exploration of reporter enzymes for better capability, these genetically modified fluorescent proteins have been used to evaluate a wide range of target compounds [29–32].
3. Whole-Cell Sensor Based on Intracellular System
3.1. Protein: Transcriptional Regulator/Inducible Promoter Pairs
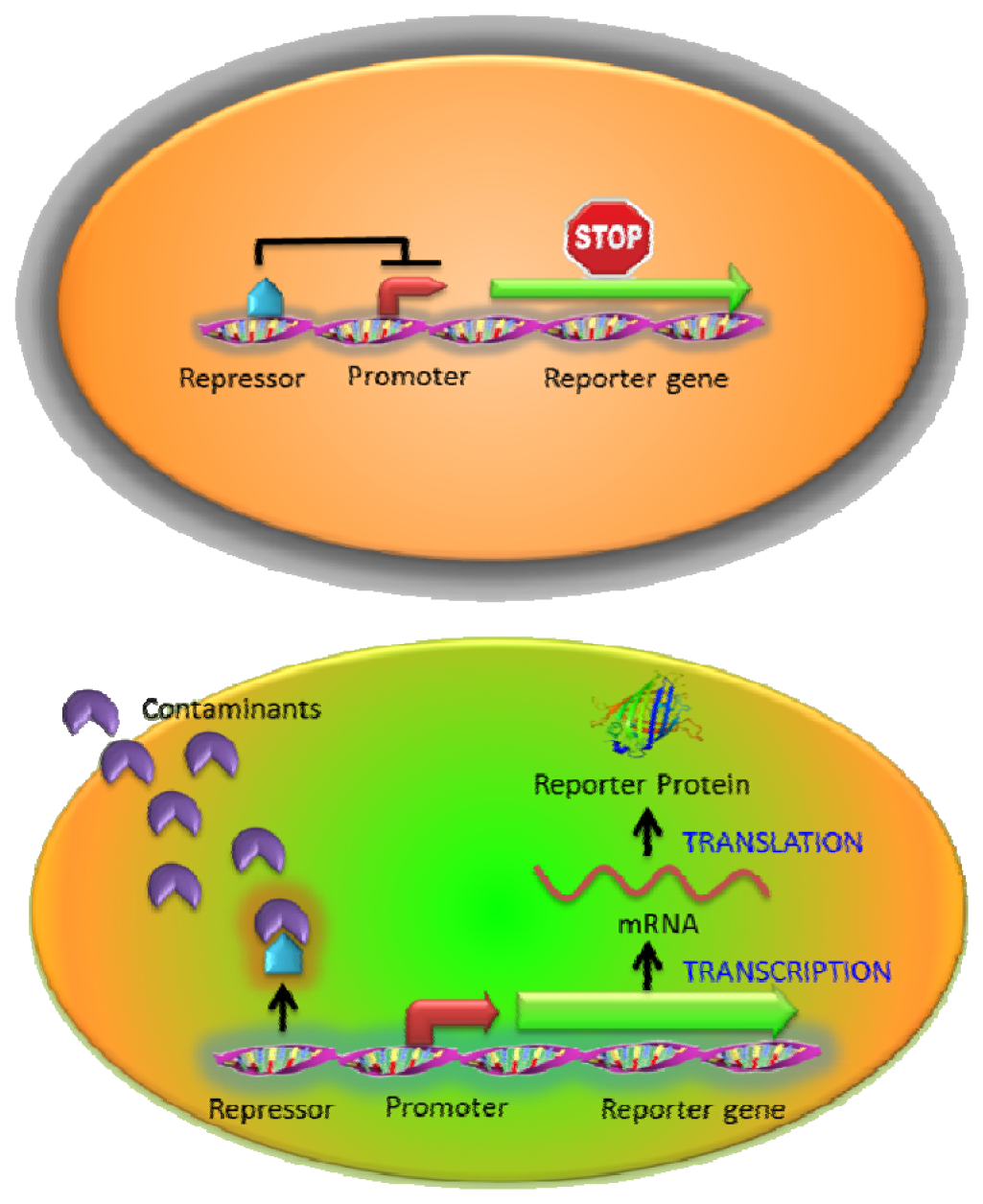
One of the most widely used intracellular sensing mechanisms is based on the coupling between a transcriptional regulator and an inducible promoter in response to different nutrient conditions, external toxicants, or communication signals [10,11]. The interaction between the target molecules and transcriptional regulators activates or represses the expression of the reporter gene resulting in a measurable signal change in a concentration-dependent manner. Most regulator protein/promoter pairs employed to detect environmental contaminants are based on the natural resistance mechanisms (such as heavy metals or antibiotics) [20,28,33] or the metabolisms of toxic compounds (such as toluene or phenol) [34,35]. In the presence of targets, native responses of the regulator and its corresponding promoter control the expression of an artificial output (reporter gene) (Figure 2). For example, a whole-cell biosensor was developed in Bacillus subtilis based on the regulatory protein CadC and the cadC promoter from Staphylococcus sp. for cadmium detection. The firefly luciferase genes (lucFF) were regulated under the cadC promoter and this luminescent bacterial sensor responded to cadmium (Cd), lead (Pb) and antimony (Sb) with nanomolar sensitivity [28] which was hundred to million folds more sensitive than earlier ones using bacterial luciferase [36,37]. Similarly, the ZntR regulatory protein and its corresponding zatAp promoter from E. coli were used to monitor Zn, Pb and Cd [20]. However, the whole-cell sensors stated above were not able to detect arsenic, which is far more toxic [38]. To address this issue, the arsenic detoxification ars operon, which is one of the most commonly exploited mechanisms for arsenic detoxification, was employed [39]. Expression from the ars operon is controlled by the regulatory protein ArsR through binding to the arsR promoter and represses the expression of reporter proteins in the absence of arsenite [40]. Upon the introduction of arsenite, the interaction between ArsR and arsenite causes the dissociation of the complex from the promoter and the reporter protein is subsequently expressed. However, earlier designs that placed the reporter genes downstream of the arsR gene resulted in a relatively high background due to leaky expression [39]. To reduce the background expression, Stocker et al. placed a transcriptional insulator, a small DNA fragment, for a second ArsR binding site downstream of the arsR gene and upstream of the reporter gene. Further binding of an ArsR dimer would block RNA polymerase from reading through and background expression of the reporter gene was reduced, resulting in a much lower detection limit [39].
While being a favorable feature in controlling arsenic detoxification for the cell, ars operon is not necessarily the most optimal for obtaining the highest sensitivity and response as a designed cellular reporter for arsenic detection. For this issue, Tani et al. [41] uncoupled the natural regulatory configuration by placing the expression of arsR in trans under control of either a T7 or lac promoter while maintaining GFP expression under the control of the arsR promoter [41]. Recently, another group led by van der Meer systematically studied the effect of promoter strength on ArsR expression and found that a stronger constitutive ArsR production decreases arsenite-dependent EGFP output from the ars promoter and vice versa [42]. These results suggested higher expression levels and sensitivities of arsenic bioreporters using uncoupled circuits may be achieved for improving field-test assays.
For organic contaminants, the XylR and Pu promoter pair from the xylene degradation pathway in Pseudomonas have been used to detect xylene, benzene and toluene [43], and the regulatory protein TbuT and the tbuA1p promoter derived from the toluene degradation pathway in Ralstonia pickettii were used to control luciferase expression in response to these volatile compounds [41]. The regulatory protein DmpR and the Po promoter was used to monitor phenols in Pseudomonas putida with a modest detection limit of 3 μM [35], but a mutant DmpR was generated to increase the sensitivity of the biosensor by more than 4-fold than using the wild-type DmpR [44].
An E. coli biosensor to detect L-arabinose was developed with a detection limit of 0.5 μM and a high selectivity over D-arabinose, a stereoisomer of L-arabinose, through the AraC regulatory protein and PBAD promoter pair [45]. The AraC regulatory protein of E. coli ara operon represses transcription in the absence of L-arabinose but induces transcription in the presence of L-arabinose [46]. Structurally similar pentoses and hexoses including D-arabinose, cannot activate AraC and the PBAD promoter system [45]. By engineering this regulatory protein, Tang et al. generated a mutant AraC regulatory protein with specificity only toward D-arabinose but not L-arabinose and other sugars [47]. In addition to altering the specificity of the regulatory protein to other structurally similar compounds, efforts have been made in engineering the regulatory proteins to recognize the targets of completely different chemical structures in order to expand the flexibility of the sensing repertoire. Recently, Tang et al. (2011) identified a mevalonate-responsive AraC variant and employed this newly generated variant to monitor mevalonate [48]. Mevalonate is an important intracellular intermediate of the isoprenoid biosynthesis pathway and in particular, the mevalonate sensor was used as a reporter in library screening for improved mevalonate production [18]. The novel AraC variant (AraC-mev) only responded to mevalonate while there was no response observed with other chemically similar compounds L-arabinose, triacetic acid lactone, and succinate [48]. This AraC-mev variant was coupled with a PBAD-LacZ fusion to screen mutants of hydroxymethylglutaryl-CoA reductase (HMGR), one of mevalonate-dependent isoprenoid-pathway enzymes, to enhance mevalonate production as the HMGR expression level is highly sensitive to mevalonate production [48]. We think these are powerful examples demonstrating the capability to fine tune the specificity of any regulatory protein to sense a wide range of novel targets by protein engineering.
3.2. RNA: Riboswitch and Reporter Gene Expression
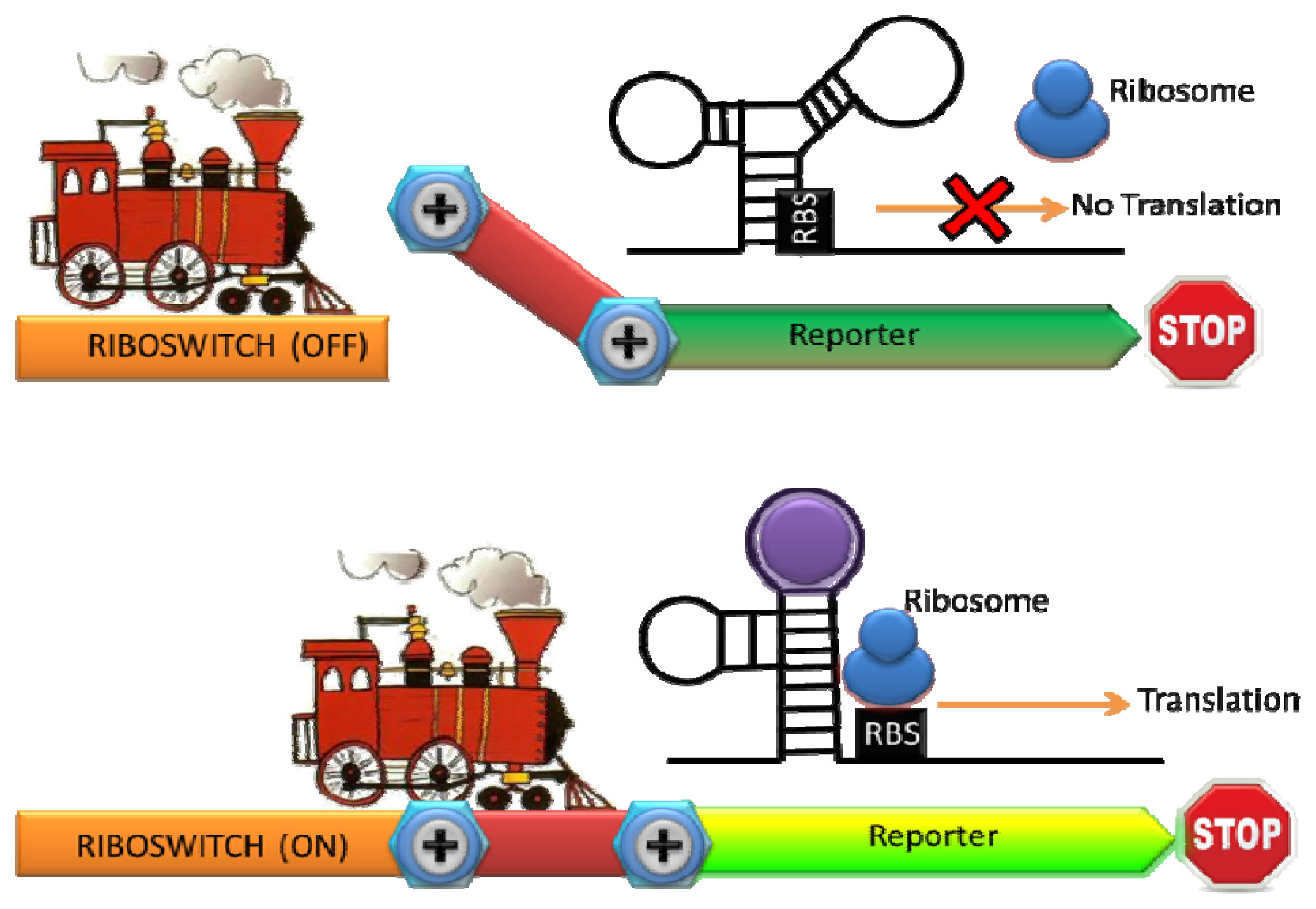
Ribosomal switches are structured RNA domains which detect molecules and regulate the expression of associated genes [49]. A riboswitch is typically consisted of an aptamer domain that undergoes a conformational change upon binding to the target molecule, resulting in the expression of a reporter gene [50] (Figure 3). Since RNA is a key regulatory molecule in natural biological system and the design and engineering of RNA are relatively easy, the interest of RNA-based detection and regulation have been significantly growing last decade [51]. Riboswitches with natural and synthetic RNA aptamers have been developed to sense temperature, metal ions, nucleic acids, small molecule, and proteins [50,52–54]. A whole-cell sensor based on an engineered riboswitch was developed to detect one of the commonly used antiasthmatic drugs, theophylline [55,56]. As an overdose of theophylline leads to serious side effects, monitoring theophylline concentration in the blood serum is clinically important [57]. Thymidylate synthase, which is essential for cell growth, was linked with an anti-theophylline aptamer and the concentration of theophylline was monitored by the theophylline-dependent growth of E. coli [56]. To simplify the sensing format, they inserted the theophylline binding aptamer before the start codon in an engineered 5′ proximal coding region of GFP. A structural change in the 5′ proximal coding region induced by theophylline resulted in reduced ribosomal accessibility. Consequently a dose-dependent repression of GFP expression by theophylline with exquisite selectivity was observed (there was no dose-dependent repression of GFP expression by a structural analogue such as caffeine) [55].
Furthermore, an artificial riboswitch has been used for in vivo monitoring of intracellular metabolites and for engineering metabolic pathways [58]. This group used the natural thiamine pyrophosphate (TPP) riboswitch found in the 5′UTR of the E. coli thiM gene to generate a TPP-activated riboswitch. The engineered TPP-activate riboswitch was fused to β-galactosidase and GFP and the expression of these reporters was shown to induce in the presence of thiamine (which is converted to TPP intracellularly). As the interaction between the analyte and riboswitch is one of the most significant factors in determining sensitivity, a TPP riboswitch library was constructed using the TPP aptamer of the B. anthracis tenA gene with a significant higher affinity for TPP (Kd = 210 pM comparing to Kd = 600 nM of the thiM) [59]. From the library, they identified several new TPP-activated riboswitches with significantly enhanced sensitivity [59]. This result again highlights our ability to fine-tune the sensitivity by engineering the natural recognition element. Even though targets are typically limited to small molecules, the whole cell biosensors with engineered riboswitch have the potential for monitoring novel targets such as drugs and metabolites in clinical and environmental samples as riboswitch can be easily integrated into the expression platform and modulate them.
3.3. Quorum Sensing
Microorganisms have different cell-cell communication methods based on their lifestyles. Quorum sensing (QS), one of the most commonly used methods, is based on the use of diffusible small molecules for communication [60]. These small molecules, also known as autoinducers, are produced, secreted, and recognized via the internal biochemical pathways of the specific microorganisms. Conventionally, autoinducers have been divided into two generic groups: the acyl homoserine lactones (AHLs), used by Gram-negative bacteria and the autoinducing polypeptides (AIPs), used by Gram-positive bacteria [61]. Recently, a universal autoinducer (AI-2) was found to be used by both Gram-negative and Gram-positive bacteria [62]. When the concentration of autoinducers is beyond a defined threshold, various features such as virulence, biofilm formation, sporulation, genetic competence and bioluminescence are regulated [63]. This unique machinery has been employed in many sensing applications. Pathogens such as Pseudomonas aeruginosa and Burkholderia cepacia employ diffusible AHL signal molecules to regulate the production of a number of virulence determinants and can cause destructive lung diseases in cystic fibrosis patients [64]. Middleton and coworkers extracted sputum samples from cystic fibrosis patients colonized by either P. aeruginosa or B. cepacia, and demonstrated the clinical detection of short- and long-chain AHLs using an E. coli strain engineered to upregulate the expression of a reporter in response to these signal molecules [65,66]. Although this approach is capable of performing well in the laboratory, an integrated analytical device capable of on-site monitoring of pathogens would be much more practical. To adapt this sensing system into a portable format, a whole cell biosensor employing components of the AHL-mediated QS regulatory system as the recognition element and β-galactosidase as the reporter protein was liquid-dried on filter paper strips [67]. After reconstitution in LB broth for 1.5 h, the disposable biosensor was successfully applied to detect AHLs in physiological samples such as saliva at concentrations down to 1 × 10−8 M to correlate the presence of pathogens.
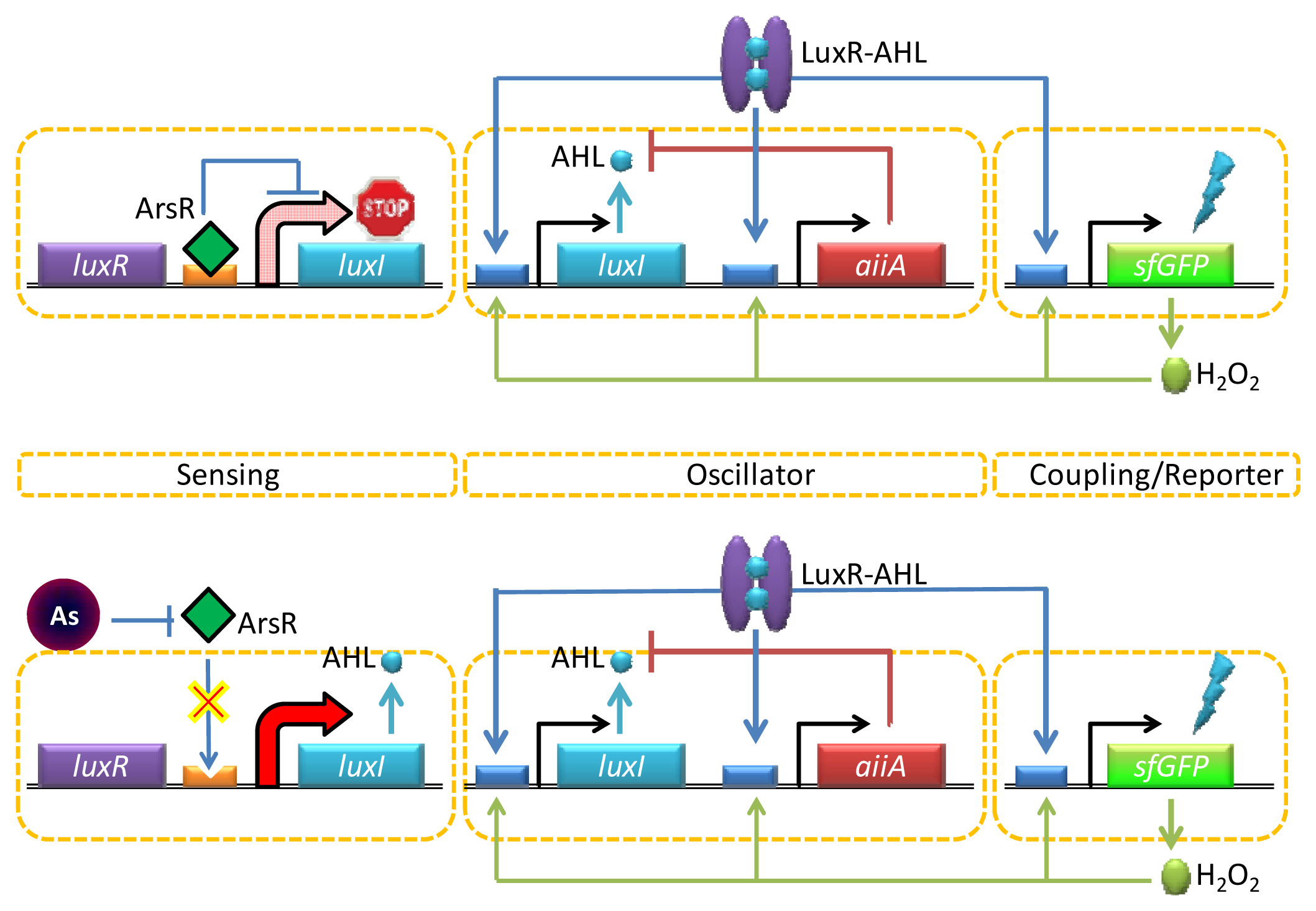
One interesting way to enable both strong and long-range target coupling is to synchronize the response of millions of cells, and the AHL-mediated quorum sensing has recently been incorporated into this format for enhanced arsenic sensing. Quorum sensing typically involves a strong intercellular coupling over tens of micrometers [68], yet the relatively slow diffusion time of molecular communication leads to signal delays over the millimeter length scale. To overcome this challenge, Prindle et al. used a gene circuitry that produces an oscillating amount of GFP in an E. coli reporter strain maintained inside microfluidic cavities (so called ‘biopixels’) [69]. In this design, the AHL synthase LuxI is under the control of a native arsenite-responsive promoter, but the reporter gene (gfp) is regulated by the AHL QS system (luxR or luxI) (Figure 4). In the presence of arsenite, LuxI was expressed, and AHL was diffused across the cell membrane and mediated intercellular coupling. Interestingly, rather than taking the absolute level of GFP expression as a measure for arsenite-dependent depression of the system, this group used the duration of the oscillation period. The fast gaseous diffusion of hydrogen peroxide that is produced spontaneously as a result of oxygen radical formation at high GFP fluorescence is used to increase the production of AiiA. This in turn down-regulates the lux promoter controlling gfp expression resulting in an increase in the oscillatory amplitude and duration period depending on the arsenite concentrations. Since the oscillation period is independent of the absolute fluorescence intensity, the biopixel system output can avoid the errors coming from detector/light source fluctuations.
3.4. Others
Apart from the sensing mechanisms stated above, there are still several other mechanisms for the development of microbial sensors. For example, conformational changes as a result of protein-protein interactions are another way to construct live cell biosensors. Marvin et al. described the generation of high-signal-to-noise single wavelength biosensor for maltose [70]. By fusing the circularly permuted fluorescent proteins into a bacterial periplasmic binding protein (PBP), a maltodextrin-binding protein, the designed sensor was able to observe rapid maltose transport across the plasma membrane and the addition of maltose to extracellular media. Similarly, E. coli mutants with a genetically engineered glucose/galactose-binding protein (GBP) have been also used to serve as biosensors for glucose [71]. Interestingly, while the E. coli GBP has a high-binding affinity of 0.2 mM for glucose [72], they targeted mutants with new binding affinities within the physiological range for blood glucose monitoring [73].
While low sensitivity of live cell biosensors is always questioned by others and hinders their use, a system for developing live cell biosensors via the use of biological nanopores as sensing platforms provides the possibility toward single-molecule detection [74]. In this format, biological pores and channels that open in response to binding of individual ligands stand out as relatively simple components of single-molecule sensors [75]. In nature, single-molecule detection is routinely achieved through ligand-gated ion channel proteins [76]. A critical feature of this signaling mechanism is the strong molecular amplification that it entails. The binding of a single molecule can cause 104–107 ions to pass through the membrane to generate a significant change in the transmembrane potential. Using genetically engineered hemolysin pores, Bayley's group developed sensors for the detection of ionic species and organic molecules in solution [77–79].
While the systems described above are based on a known interaction between the target molecules and the bio-components, the yeast two-hybrid assay using recombinant DNA technology can be used as a biosensor to provide an important first hint for the identification of interaction partners. For the two-hybrid screening, the transcription factor is split into two parts, a binding domain and an activating domain, and the activation of downstream reporter gene(s) is achieved via the binding of a transcription factor to an upstream activating sequence by the desired interacting partners [80]. Using the two hybrid assay in yeast, interactions between the estrogen receptor and its coactivators were detected and shown to be dependent on the presence of estrogen [81]. The specificity of this assay was assessed by determining the effects of steroids, known estrogen receptor agonists, and phytoestrogens. The pattern of response to chemicals was consistent with estrogenic activity measured by other assay systems, indicating that this assay system is reliable in measuring estrogenic activity. Lee et al. also constructed an efficient and reliable yeast two-hybrid detection system to evaluate the estrogenic activity of potential endocrine disruptors. This system employed the interaction between the human estrogen receptor β (hERβ) ligand binding domain and the co-activator SRC1 [82].
Another elegant way for estrogen detection is via the use of chimeric proteins. These biosensors are typically constructed by fusing a target ligand-binding domain to an easily assayed reporter protein. Properly designed fusions allow ligand-induced conformational changes in the target ligand-binding domain to be transmitted to the reporter and allosterically modulate its properties. Skretas et al. combined the ligand-binding domains of estrogen receptors with a highly sensitive thymidylate synthase reporter for identifying diverse estrogenic compounds [83]. Expression of the reporter protein creates a hormone dependent growth phenotype in thymidylate synthase deficient E. coli strains. By using this sensor, they were able to rapidly screen a small chemical library and identify structurally novel estrogen receptors modulators, while predicting their agonistic/antagonistic bio-characteristics in human cell assays. This system was further employed to assay for estrogenic behavior in complex mixtures found in consumer products, including perfumes, hand and body washes, deodorants, and herbal supplements [84].
4. Whole-Cell Sensor Based on Extracellular System
4.1. Surface Display System
Surface display of peptides, proteins and epitopes on living cells has the advantage for target molecules or substrates that are not accessible to the intracellular environment. Even for targets that can be readily uptake, the overall kinetics could be still significantly improved by bypassing membrane transport. Moreover, surface display provides stabilization of enzymes and proteins through attachment to the cell wall and easy purification comparing to free proteins [19,85]. Many outer membrane proteins, lipoproteins, S-layer proteins, cell-surface appendages and autotransporters have been utilized for cell surface display. The ice nucleation protein (INP), one of outer membrane proteins of plant pathogenic bacteria, has been widely used for bacteria cell surface display [15,86–88].
Since the utilization of enzyme based biosensors is limited by the cost and the tedious work for the purification of enzymes [4,7], whole cell biosensors expressing anticipated enzymes can overcome these limitations. It has been shown that the E. coli cells expressing organophosphorus hydrolase (OPH) were more stable and robust than purified OPH, retaining 100% activity over a period of one month [89]. In particular, E. coli cells expressing OPH on the cell surface degrade parathion and paraoxon sevenfold faster than whole cells expressing OPH intracellularly [89], suggesting that reduction of the mass-transport limitation of substrates and products across the cell membrane results in consequent improvement of the overall kinetics. Our group has developed several engineered strains displaying organophosphorus hydrolase (OPH) on the cell surface for the simultaneous hydrolysis of organophosphorous pesticides into p-nitrophenol and its subsequent conversion into electrochemically active intermediates [15,86]. By immobilizing these engineered cells onto a carbon paste electrode, different organophosphorus pesticides can be detected by measuring the corresponding electrooxidation current generated from these intermediates [90]. Similarly, Liu et al. displayed xylose dehydrogenase (XDH) on the cell surface using the INP anchor for the detection of D-xylose, a compound of significant interest in genetics, human nutrition, food technology and pharmacology [87]. In that study, XDH converts D-xylose to D-xylonolactone using NAD+ as the cofactor and the production of NADH determined at 340 nm was used to correlate with the initial xylose concentration. The XDH-displayed bacteria showed a detection limit of 2 μM of D-xylose and a broad linear range of detection up to 900 μM. Furthermore, the XDH surface displaying system selectively detected D-xylose in real samples such as food and degraded products of lignocelluloses.
In addition to using outer membrane proteins as the anchor, proteins have been successfully presented on the cell surface through the autotransporter protein system. Autotransporter proteins are displayed on the surface of Gram-negative bacteria through secretion to the periplasm by an N-terminal signal peptide and anchored on the outer membrane by the C-terminal β-domain. Both OPH and GFP were displayed onto the surface through the E. coli AIDA-I (adhesion involved in diffuse adherence I) autotransporter protein and used for monitoring organophosphate compounds [91].
In addition to enzymes, human antigens were displayed on the yeast surface to detect monoclonal antibodies since the direct preparation of these mammalian proteins remains challenging and laborious [92,93]. More importantly, it is more likely to produce soluble and functional mammalian proteins in yeast with the appropriate post-translational glycosylation or proteolytic modification [94]. Tang et al. presented human antigens on the yeast surface via the glucoamylase secretion signal sequence and the glycosylphosphatidylinositol-anchor attachment signal sequence of the native yeast glycoprotein, α-agglutinin. The antigen-displaying yeast was used to detect monoclonal antibodies using both immunofluorescence and the enzyme-linked immunosorbent assay (ELISA). An added benefit to direct sensing is the potential application in screening positive antibody-producing hybridoma cell lines for large-scale applications [94].
4.2. G-Protein Coupled Receptors (GPCRs)
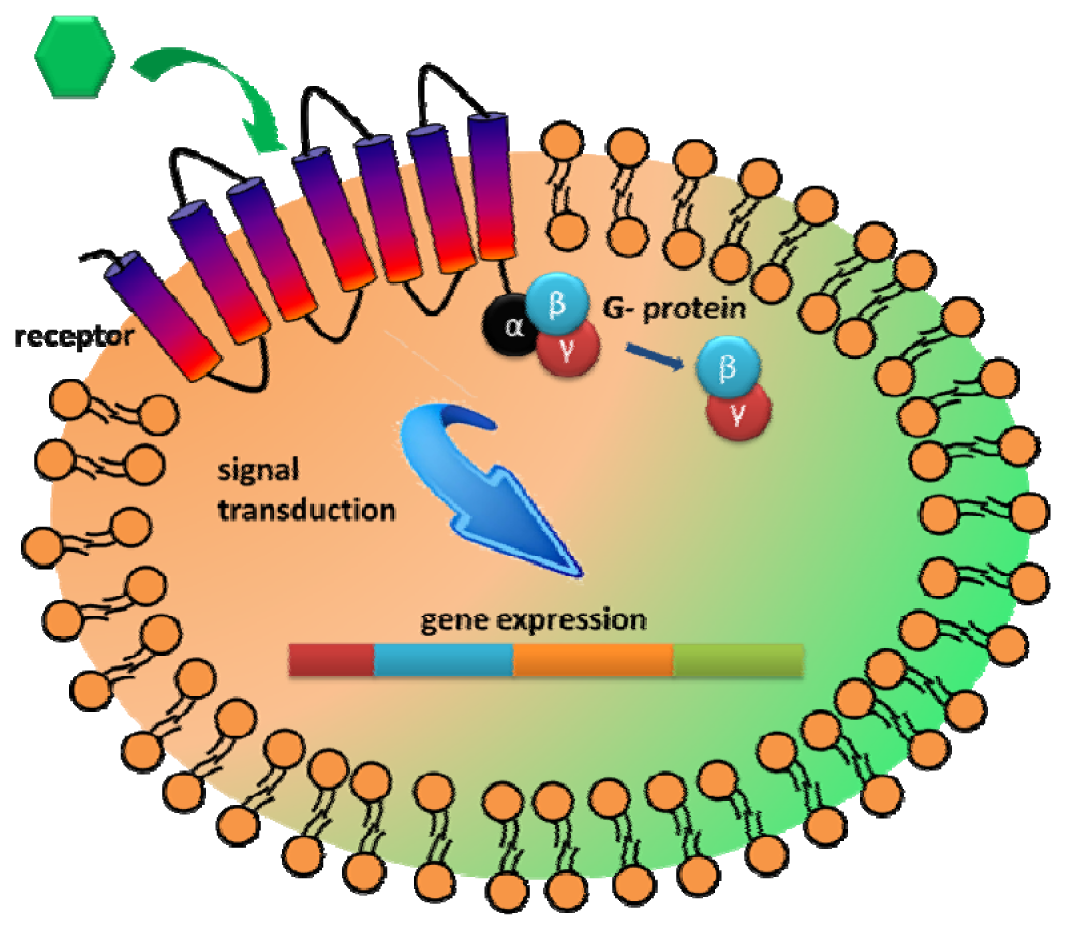
Another interesting extracellular component that has received attention as a recognition element is the G-protein coupled receptors (GPCRs). GPCRs represent the largest family of integral membrane receptors and are involved in many intracellular communications in response to diverse external stimuli [95,96]. The primary benefit to explore GPCR is the large range of natural binding repertoire ranging from small molecules to peptides and glycoproteins [14,95,96]. As GPCRs allow transmission of a wide range of stimulants, including hormones, neurotransmitters, taste, and chemicals, they are highly modular in nature and can be customized to sense either a wide range of targets [17,96,97]. Upon ligand binding to GPCR, a signal is initiated intracellularly via interaction with the GTP-binding protein (G protein) to further transmit cellular responses [95,97] (Figure 5). Thus, the main advantage of utilizing GPCR for whole cell biosensing is that various cellular processes could be used as the sensor readout.
Although GPCR-based biosensors have been proposed for drug screening by measuring the level of secondary messengers such as GTPγS, cAMP and Ca2+ in mammalian cell, GPCR-based sensors have been developed by coupling with the yeast mating response pathway. Different mammalian GPCRs targeting opioids, neurotransmitters, adenosines, somatostatin or sugar nucleotides have been successfully expressed and displayed the ability of ligand binding in yeast [14,17,95,96,98]. By controlling imidazoleglycerol-phosphate dehydratase, an enzyme involved in histidine biosynthesis, under the GPCR-signaling responsive promoter, cell growth was used to correlate with the external concentration of target molecules [14,95,96,98]. Furthermore, the sensitivity and selectivity can be fine-tuned by engineering the receptors via direct evolution [17,97] and the mating responsive pathway [14,99]. For example, Broach et al. created a new UDP-glucose receptor by random mutagenesis and demonstrated a thirty fold lower detection limit for UDP-glucose comparing to the original receptor [17]. These sugar nucleotides are significant reagents in the biological or chemoenzymatic synthesis of carbohydrates. Since sugar nucleotides are structurally diverse and have similar physicochemical properties, they are challenging targets for inexpensive and simple high-throughput detection [17]. In addition to increasing the sensitivity by 30-fold, mutant UDP-glucose receptors with altered specificity for other sugar nucleotides have been created to extend the range of targets that can be detected [17].
5. Conclusions
In this review, we have highlighted recent approaches based on protein/cellular engineering and synthetic biology as an emerging tool in the field of whole-cell biosensors. In addition to engineering the sensor readouts to enhance signals, there have been continuous efforts in manipulating and generating recognition elements such as RNA, enzyme and non-enzymatic proteins with improved affinity and selectivity. Many of the approaches currently employed for improving product synthesis can easily be adapted for whole-cell biosensor design in order to optimize the sensitivity, selectivity and robustness. To use these whole-cell biosensors for rapid, point-of-care applications, developing better materials to interface with the biological elements would be of great importance.
Acknowledgments
This work is partially supported by NSF (CBET1129012).
Conflicts of Interests
The authors declare no conflict of interest.
References
- Cella, L.N.; Sanchez, P.; Zhong, W.W.; Myung, N.V.; Chen, W.; Mulchandani, A. Nano aptasensor for protective antigen toxin of anthrax. Anal. Chem. 2010, 82, 2042–2047. [Google Scholar]
- Wu, C.H.; Le, D.; Mulchandani, A.; Chen, W. Optimization of a whole-cell cadmium sensor with a toggle gene circuit. Biotechnol. Prog. 2009, 25, 898–903. [Google Scholar]
- Park, M.; Cella, L.N.; Chen, W.; Myung, N.V.; Mulchandani, A. Carbon nanotubes-based chemiresistive immunosensor for small molecules: Detection of nitroaromatic explosives. Biosens. Bioelectron. 2010, 26, 1297–1301. [Google Scholar]
- Lagarde, F.; Jaffrezic-Renault, N. Cell-based electrochemical biosensors for water quality assessment. Anal. Bioanal. Chem. 2011, 400, 947–964. [Google Scholar]
- Abbaszadegan, M.; Alum, A.; Abbaszadegan, H.; Stout, V. Cell surface display of poliovirus receptor on Escherichia coli, a novel method for concentrating viral particles in water. Appl. Environ. Microb. 2011, 77, 5141–5148. [Google Scholar]
- Liu, C.; Yong, D.; Yu, D.; Dong, S. Cell-based biosensor for measurement of phenol and nitrophenols toxicity. Talanta 2011, 84, 766–770. [Google Scholar]
- Su, L.; Jia, W.; Hou, C.; Lei, Y. Microbial biosensors: A review. Biosens. Bioelectron. 2011, 26, 1788–1799. [Google Scholar]
- Eltzov, E.; Marks, R. Whole-cell aquatic biosensors. Anal. Bioanal. Chem. 2011, 400, 895–913. [Google Scholar]
- Liu, Q.; Cai, H.; Xu, Y.; Li, Y.; Li, R.; Wang, P. Olfactory cell-based biosensor: A first step towards a neurochip of bioelectronic nose. Biosens. Bioelectron. 2006, 22, 318–322. [Google Scholar]
- Daunert, S.; Barrett, G.; Feliciano, J.S.; Shetty, R.S.; Shrestha, S.; Smith-Spencer, W. Genetically engineered whole-cell sensing systems: Coupling biological recognition with reporter genes. Chem. Rev. 2000, 100, 2705–2738. [Google Scholar]
- Van der Meer, J.R.; Belkin, S. Where microbiology meets microengineering: Design and applications of reporter bacteria. Nat. Rev. Microbiol. 2010, 8, 511–522. [Google Scholar]
- Belkin, S. Microbial whole-cell sensing systems of environmental pollutants. Curr. Opin. Microbiol. 2003, 6, 206–212. [Google Scholar]
- Elad, T.; Lee, J.H.; Belkin, S.; Gu, M.B. Microbial whole-cell arrays. Microb. Biotechnol. 2008, 1, 137–148. [Google Scholar]
- Fukuda, N.; Ishii, J.; Kaishima, M.; Kondo, A. Amplification of agonist stimulation of human G-protein-coupled receptor signaling in yeast. Anal. Chem. 2011, 417, 182–187. [Google Scholar]
- Shimazu, M.; Mulchandani, A.; Chen, W. Simultaneous degradation of organophosphorus pesticides and p-nitrophenol by a genetically engineered Moraxella sp with surface-expressed organophosphorus hydrolase. Biotechnol. Bioeng. 2001, 76, 318–324. [Google Scholar]
- Turner, K.; Xu, S.; Pasini, P.; Deo, S.; Bachas, L.; Daunert, S. Hydroxylated polychlorinated biphenyl detection based on a genetically engineered bioluminescent whole-cell sensing system. Anal. Chem. 2007, 79, 5740–5745. [Google Scholar]
- Ault, A.D.; Broach, J.R. Creation of GPCR-based chemical sensors by directed evolution in yeast. Protein Eng. Des. Sel. 2006, 19, 1–8. [Google Scholar]
- Pfleger, B.F.; Pitera, D.J.; Newman, J.D.; Martin, V.J.J.; Keasling, J.D. Microbial sensors for small molecules: Development of a mevalonate biosensor. Metab. Eng. 2007, 9, 30–38. [Google Scholar]
- Schultheiss, E.; Weiss, S.; Winterer, E.; Maas, R.; Heinzle, E.; Jose, J. Esterase autodisplay: Enzyme engineering and whole-cell activity determination in microplates with pH sensors. Appl. Environ. Microb. 2008, 74, 4782–4791. [Google Scholar]
- Ivask, A.; Rolova, T.; Kahru, A. A suite of recombinant luminescent bacterial strains for the quantification of bioavailable heavy metals and toxicity testing. BMC Biotechnol. 2009, 9, 41–55. [Google Scholar]
- Zheng, J.; Sagar, V.; Smolinsky, A.; Bourke, C.; LaRonde-LeBlanc, N.; Cropp, T.A. Structure and function of the macrolide biosensor protein, MphR(A), with and without Erythromycin. J. Mol. Biol. 2009, 387, 1250–1260. [Google Scholar]
- Rainina, E.I.; Efremenco, E.N.; Varfolomeyev, S.D.; Simonian, A.L.; Wild, J.R. The development of a new biosensor based on recombinant E. coli for the direct detection of organophosphorus neurotoxins. Biosens. Bioelectron. 1996, 11, 991–1000. [Google Scholar]
- Anu, P.M.U.; Chaurasia, A.K.; Sawant, S.N.; Apte, S.K. Polyaniline-based highly sensitive microbial biosensor for selective detection of lindane. Anal. Chem. 2012, 84, 6672–6678. [Google Scholar]
- Jain, V.K.; Magrath, I.T. A chemiluminescent assay for quantitation of β-galactosidase in the femtogram range: Application to quantitation of β-galactosidase in lacZ-transfected cells. Anal. Chem. 1991, 199, 119–124. [Google Scholar]
- Steghens, J.P.; Min, K.L.; Bernengo, J.C. Firefly luciferase has two nucleotide binding sites: Effect of nucleoside monophosphate and CoA on the light-emission spectra. Biochem. J. 1998, 336, 109–113. [Google Scholar]
- Biran, I.; Klimentiy, L.; Hengge-Aronis, R.; Ron, E.Z.; Rishpon, J. On-line monitoring of gene expression. Microbiology 1999, 145, 2129–2133. [Google Scholar]
- Baldwin, T.O.; Christopher, J.A.; Raushel, F.M.; Sinclair, J.F.; Ziegler, M.M.; Fisher, A.J.; Rayment, I. Structure of bacterial luciferase. Curr. Opin. Struct. Biol. 1995, 5, 798–809. [Google Scholar]
- Tauriainen, S.; Karp, M.; Chang, W.; Virta, M. Luminescent bacterial sensor for cadmium and lead. Biosens. Bioelectron. 1998, 13, 931–938. [Google Scholar]
- Kremers, G.-J.; Gilbert, S.G.; Cranfill, P.J.; Davidson, M.W.; Piston, D.W. Fluorescent proteins at a glance. J. Cell Sci. 2011, 124, 157–160. [Google Scholar]
- Ha, J.S.; Song, J.J.; Lee, Y.M.; Kim, S.J.; Sohn, J.H.; Shin, C.S.; Lee, S.G. Design and application of highly responsive fluorescence resonance energy transfer biosensors for detection of sugar in living Saccharomyces cerevisiae cells. Appl. Environ. Microb. 2007, 73, 7408–7414. [Google Scholar]
- Campbell, R.E. Fluorescent-protein-based biosensors: Modulation of energy transfer as a design principle. Anal. Chem. 2009, 81, 5972–5979. [Google Scholar]
- Ewald, J.C.; Reich, S.; Baumann, S.; Frommer, W.B.; Zamboni, N. Engineering genetically encoded nanosensors for real-time in vivo measurements of citrate concentrations. PLoS One 2011, 6. [Google Scholar] [CrossRef]
- Urban, A.; Eckermann, S.; Fast, B.; Metzger, S.; Gehling, M.; Ziegelbauer, K.; Rubsamen-Waigmann, H.; Freiberg, C. Novel whole-cell antibiotic biosensors for compound discovery. Appl. Environ. Microb. 2007, 73, 6436–6443. [Google Scholar]
- Tecon, R.; Beggah, S.; Czechowska, K.; Sentchilo, V.; Chronopoulou, P.-M.; McGenity, T.J.; van der Meer, J.R. Development of a multistrain bacterial bioreporter platform for the monitoring of hydrocarbon contaminants in marine environments. Environ. Sci. Technol. 2009, 44, 1049–1055. [Google Scholar]
- Shingler, V.; Moore, T. Sensing of aromatic compounds by the DmpR transcriptional activator of phenol-catabolizing Pseudomonas sp. Strain CF600. J. Bacteriol. 1994, 176, 1555–1560. [Google Scholar]
- Tauriainen, S.; Karp, M.; Chang, W.; Virta, M. Recombinant luminescent bacteria for measuring bioavailable arsenite and antimonite. Appl. Environ. Microb. 1997, 63, 4456–4461. [Google Scholar]
- Virta, M.; Lampinen, J.; Karp, M. A luminescence-based mercury biosensor. Anal. Chem. 1995, 6, 667–669. [Google Scholar]
- Tsai, S.-L.; Singh, S.; Chen, W. Arsenic metabolism by microbes in nature and the impact on arsenic remediation. Curr. Opin. Biotechnol. 2009, 20, 659–667. [Google Scholar]
- Stocker, J.; Balluch, D.; Gsell, M.; Harms, H.; Feliciano, J.; Daunert, S.; Malik, K.A.; van der Meer, J.R. Development of a set of simple bacterial biosensors for quantitative and rapid measurements of arsenite and arsenate in potable water. Environ. Sci. Technol. 2003, 37, 4743–4750. [Google Scholar]
- Wu, J.; Rosen, B.P. Metalloregulated expression of the ars operon. J. Biol. Chem. 1993, 268, 52–58. [Google Scholar]
- Tani, C.; Inoue, K.; Tani, Y.; Harun-ur-Rashid, M.; Azuma, N.; Ueda, S.; Yoshida, K.; Maeda, I. Sensitive fluorescent microplate bioassay using recombinant Escherichia coli with multiple promoter-reporter units in tandem for detection of arsenic. J. Biosci. Bioeng. 2009, 108, 414–420. [Google Scholar]
- Merulla, D.; Hatzimanikatis, V.; van der Meer, J.R. Tunable reporter signal production in feedback-uncoupled arsenic bioreporters. Microb. Biotechnol. 2013, 6. [Google Scholar] [CrossRef]
- Willardson, B.M.; Wilkins, J.F.; Rand, T.A.; Schupp, J.M.; Hill, K.K.; Keim, P.; Jackson, P.J. Development and testing of a bacterial biosensor for toluene-based environmental contaminants. Appl. Environ. Microb. 1998, 64, 1006–1012. [Google Scholar]
- Wise, A.A.; Kuske, C.R. Generation of novel bacterial regulatory proteins that detect priority pollutant phenols. Appl. Environ. Microb. 2000, 66, 163–169. [Google Scholar]
- Shetty, R.S.; Ramanathan, S.; Badr, I.H.A.; Wolford, J.L.; Daunert, S. Green fluorescent protein in the design of a living biosensing system for L-arabinose. Anal. Chem. 1999, 71, 763–768. [Google Scholar]
- Schleif, R. AraC protein, regulation of the L-arabinose operon in Escherichia coli, and the light switch mechanism of AraC action. FEMS Microbiol. Rev. C 2010, 34, 779–796. [Google Scholar]
- Tang, S.-Y.; Fazelinia, H.; Cirino, P.C. AraC regulatory protein mutants with altered effector specificity. J. Am. Chem. Soc. 2008, 130, 5267–5271. [Google Scholar]
- Tang, S.-Y.; Cirino, P.C. Design and application of a mevalonate-responsive regulatory protein. Angew. Chem. Int. Ed. 2011, 50, 1084–1086. [Google Scholar]
- Topp, S.; Gallivan, J.P. Emerging applications of riboswitches in chemical biology. ACS Chem. Biol. 2010, 5, 139–148. [Google Scholar]
- Win, M.N.; Smolke, C.D. A modular and extensible RNA-based gene-regulatory platform for engineering cellular function. Proc. Natl. Acad. Sci. USA 2007, 104, 14283–14288. [Google Scholar]
- Liang, J.C.; Bloom, R.J.; Smolke, C.D. Engineering biological systems with synthetic RNA molecules. Mol. Cell. 2011, 43, 915–926. [Google Scholar]
- Sinha, J.; Reyes, S.J.; Gallivan, J.P. Reprogramming bacteria to seek and destroy an herbicide. Nat. Chem. Biol. 2010, 6, 464–470. [Google Scholar]
- Michener, J.K.; Thodey, K.; Liang, J.C.; Smolke, C.D. Applications of genetically-encoded biosensors for the construction and control of biosynthetic pathways. Metab. Eng. 2012, 14, 212–222. [Google Scholar]
- Baker, J.L.; Sudarsan, N.; Weinberg, Z.; Roth, A.; Stockbridge, R.B.; Breaker, R.R. Widespread genetic switches and toxicity resistance proteins for fluoride. Science 2012, 335, 233–235. [Google Scholar]
- Jo, J.-J.; Shin, J.-S. Construction of intragenic synthetic riboswitches for detection of a small molecule. Biotechnol. Lett. 2009, 31, 1577–1581. [Google Scholar]
- Kristin, M.; Thompson, H.A.S.; Knudsen, Scott, M.; Ellington, A.D. Group I aptazymes as genetic regulatory switches. BMC Biotechnol. 2002, 2, 21–32. [Google Scholar]
- Pernites, R.B.; Ponnapati, R.R.; Advincula, R.C. Surface plasmon resonance (SPR) detection of theophylline via electropolymerized molecularly imprinted polythiophenes. Macromolecules 2010, 43, 9724–9735. [Google Scholar]
- Nomura, Y.; Yokobayashi, Y. Reengineering a natural riboswitch by dual genetic selection. J. Am. Chem. Soc. 2007, 129, 13814–13815. [Google Scholar]
- Muranaka, N.; Sharma, V.; Nomura, Y.; Yokobayashi, Y. Efficient design strategy for whole-cell and cell-free biosensors based on engineered riboswitches. Anal. Lett. 2009, 42, 108–122. [Google Scholar]
- Wang, B.; Buck, M. Customizing cell signaling using engineered genetic logic circuits. Trends Microbiol. 2012, 20, 376–384. [Google Scholar]
- Galloway, W.R.J.D.; Hodgkinson, J.T.; Bowden, S.D.; Welch, M.; Spring, D.R. Quorum sensing in gram-negative bacteria: Small-molecule modulation of AHL and AI-2 quorum sensing pathways. Chem. Rev. 2010, 111, 28–67. [Google Scholar]
- Roy, V.; Adams, B.L.; Bentley, W.E. Developing next generation antimicrobials by intercepting AI-2 mediated quorum sensing. Enzym. Microb. Technol. 2011, 49, 113–123. [Google Scholar]
- Mangwani, N.; Dash, H.R.; Chauhan, A.; Das, S. Bacterial quorum sensing: functional features and potential applications in biotechnology. J. Mol. Microb. Biotechnol. 2012, 22, 215–227. [Google Scholar]
- Govan, J.R.W.; Deretic, V. Microbial pathogenesis in cystic fibrosis: Mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 1996, 60, 539–574. [Google Scholar]
- Middleton, B.; Rodgers, H.C.; Cámara, M.; Knox, A.J.; Williams, P.; Hardman, A. Direct detection of N-acylhomoserine lactones in cystic fibrosis sputum. FEMS Microbiol. Lett. 2002, 207, 1–7. [Google Scholar]
- Kumari, A.; Pasini, P.; Deo, S.K.; Flomenhoft, D.; Shashidhar, H.; Daunert, S. Biosensing systems for the detection of bacterial quorum signaling molecules. Anal. Chem. 2006, 78, 7603–7609. [Google Scholar]
- Struss, A.; Pasini, P.; Ensor, C.M.; Raut, N.; Daunert, S. Paper strip whole cell biosensors: A portable test for the semiquantitative detection of bacterial quorum signaling molecules. Anal. Chem. 2010, 82, 4457–4463. [Google Scholar]
- Waters, C.M.; Bassler, B.L. Quorum sensing: Cell-to-cell communication in bacteria. Annu. Rev. Cell. Dev. Biol. 2005, 21, 319–346. [Google Scholar]
- Prindle, A.; Samayoa, P.; Razinkov, I.; Danino, T.; Tsimring, L.S.; Hasty, J. A sensing array of radically coupled genetic ‘biopixels’. Nature 2012, 481, 39–44. [Google Scholar]
- Marvin, J.S.; Schreiter, E.R.; Echevarría, I.M.; Looger, L.L. A genetically encoded, high-signal-to-noise maltose sensor. Proteins 2011, 79, 3025–3036. [Google Scholar]
- Jeffery, C.J. Engineering periplasmic ligand binding proteins as glucose nanosensors. Nano Rev. 2011, 2, 5743–5746. [Google Scholar]
- Vyas, N.K.; Vyas, M.N.; Quiocho, F.A. Sugar and signal-transducer binding sites of the Escherichia coli galactose chemoreceptor protein. Science 1988, 242, 1290–1295. [Google Scholar]
- Khan, F.; Saxl, T.E.; Pickup, J.C. Fluorescence intensity- and lifetime-based glucose sensing using an engineered high-Kd mutant of glucose/galactose-binding protein. Anal. Biochem. 2010, 399, 39–43. [Google Scholar]
- Majd, S.; Yusko, E.C.; Billeh, Y.N.; Macrae, M.X.; Yang, J.; Mayer, M. Applications of biological pores in nanomedicine, sensing, and nanoelectronics. Curr. Opin. Biotechnol. 2010, 21, 439–476. [Google Scholar]
- Hagan, B.; Paul, S.C. Stochastic sensors inspired by biology. Nature 2001, 413, 226–230. [Google Scholar]
- Cornell, B.A.; Braach-Maksvytis, V.L.B.; King, L.G.; Osman, P.D.J.; Raguse, B.; Wieczorek, L.; Pace, R.J. A biosensor that uses ion-channel switches. Nature 1997, 387, 580–583. [Google Scholar]
- Wu, H.-C.; Bayley, H. Single-molecule detection of nitrogen mustards by covalent reaction within a protein nanopore. J. Am. Chem. Soc. 2008, 130, 6813–6819. [Google Scholar]
- Heron, A.J.; Thompson, J.R.; Cronin, B.; Bayley, H.; Wallace, M.I. Simultaneous measurement of ionic current and fluorescence from single protein pores. J. Am. Chem. Soc. 2009, 131, 1652–1653. [Google Scholar]
- Soskine, M.; Biesemans, A.; Moeyaert, B.; Cheley, S.; Bayley, H.; Maglia, G. An engineered clya nanopore detects folded target proteins by selective external association and pore entry. Nano Lett. 2012, 12, 4895–4900. [Google Scholar]
- Young, K.H. Yeast two-hybrid: So many interactions, (in) so little time. Biol. Reprod. 1998, 58, 302–311. [Google Scholar]
- Nishikawa, J.-I.; Saito, K.; Goto, J.; Dakeyama, F.; Matsuo, M.; Nishihara, T. New screening methods for chemicals with hormonal activities using interaction of nuclear hormone receptor with coactivator. Toxicol. Appl. Pharm. 1999, 154, 76–83. [Google Scholar]
- Lee, H.-S.; Cho, E.-M.; Jung, J.; Ohta, A. Evaluation on antagonist activities of polycyclic aromatic hydrocarbons using the yeast two-hybrid detection system for endocrine disruptors. Environ. Monit. Assess. 2007, 129, 87–95. [Google Scholar]
- Skretas, G.; Meligova, A.K.; Villalonga-Barber, C.; Mitsiou, D.J.; Alexis, M.N.; Micha-Screttas, M.; Steele, B.R.; Screttas, C.G.; Wood, D.W. Engineered chimeric enzymes as tools for drug discovery: Generating reliable bacterial screens for the detection, discovery, and assessment of estrogen receptor modulators. J. Am. Chem. Soc. 2007, 129, 8443–8457. [Google Scholar]
- Gawrys, M.D.; Hartman, I.; Landweber, L.F.; Wood, D.W. Use of engineered Escherichia coli cells to detect estrogenicity in everyday consumer products. J. Chem. Technol. Biotechnol. 2009, 84, 1834–1840. [Google Scholar]
- Wu, C.H.; Mulchandani, A.; Chen, W. Versatile microbial surface-display for environmental remediation and biofuels production. Trends Microbiol. 2008, 16, 181–188. [Google Scholar]
- Shimazu, M.; Nguyen, A.; Mulchandani, A.; Chen, W. Cell surface display of organophosphorus hydrolase in Pseudomonas putida using an ice-nucleation protein anchor. Biotechnol. Progr. 2003, 19, 1612–1614. [Google Scholar]
- Liang, B.; Li, L.; Mascin, M.; Liu, A. Construction of Xylose dehydrogenase displayed on the surface of bacteria using ice nucleation protein for sensitive d-Xylose detection. Anal. Chem. 2011, 84, 275–282. [Google Scholar]
- Van Bloois, E.; Winter, R.T.; Kolmar, H.; Fraaije, M.W. Decorating microbes: Surface display of proteins on Escherichia coli. Trends Biotechnol. 2011, 29, 79–86. [Google Scholar]
- Richins, R.D.; Kaneva, I.; Mulchandani, A.; Chen, W. Biodegradation of organophosphorus pesticides by surface-expressed organophosphorus hydrolase. Nat. Biotechnol. 1997, 15, 984–987. [Google Scholar]
- Lei, Y.; Mulchandani, P.; Chen, W.; Mulchandani, A. Biosensor for direct determination of fenitrothion and EPN using recombinant Pseudomonas putida JS444 with surface-expressed organophosphorous hydrolase. 2. Modified carbon paste electrode. Appl. Biochem. Biotechnol. 2007, 136, 243–250. [Google Scholar]
- Li, C.; Zhu, Y.; Benz, I.; Schmidt, M.A.; Chen, W.; Mulchandani, A.; Qiao, C. Presentation of functional organophosphorus hydrolase fusions on the surface of Escherichia coli by the AIDA-I autotransporter pathway. Biotechnol. Bioeng. 2008, 99, 485–490. [Google Scholar]
- Gai, S.A.; Wittrup, K.D. Yeast surface display for protein engineering and characterization. Curr. Opin. Struct. Biol. 2007, 17, 467–473. [Google Scholar]
- Wadle, A.; Mischo, A.; Imig, J.; Wüllner, B.; Hensel, D.; Wätzig, K.; Neumann, F.; Kubuschok, B.; Schmidt, W.; Old, L.J.; et al. Serological identification of breast cancer-related antigens from a Saccharomyces cerevisiae surface display library. Int. J. Cancer 2005, 117, 104–113. [Google Scholar]
- Tang, Y.Q.; Han, S.Y.; Zheng, H.; Wu, L.; Ueda, M.; Wang, X.N.; Lin, Y. Construction of cell surface-engineered yeasts displaying antigen to detect antibodies by immunofluorescence and yeast-ELISA. Appl. Microbiol. Biotechnol. 2008, 79, 1019–1026. [Google Scholar]
- Erlenbach, I.; Kostenis, E.; Schmidt, C.; Hamdan, F.F.; Pausch, M.H.; Wess, J. Functional expression of M-1, M-3 and M-5 muscarinic acetylcholine receptors in yeast. J. Neurochem. 2001, 77, 1327–1337. [Google Scholar]
- Sarramegna, V.; Demange, P.; Milon, A.; Talmont, F. Optimizing functional versus total expression of the human μ-opioid receptor in pichia pastoris. Protein Expres. Purif. 2002, 24, 212–220. [Google Scholar]
- Schmidt, C.; Li, B.; Bloodworth, L.; Erlenbach, I.; Zeng, F.Y.; Wess, J. Random mutagenesis of the M-3 muscarinic acetylcholine receptor expressed in yeast—Identification of point mutations that “silence” a constitutively active mutant M-3 receptor and greatly impair receptor/G protein coupling. J. Biol. Chem. 2003, 278, 30248–30260. [Google Scholar]
- O'Malley, M.A.; Mancini, J.D.; Young, C.L.; McCusker, E.C.; Raden, D.; Robinson, A.S. Progress toward heterologous expression of active G-protein-coupled receptors in Saccharomyces cerevisiae: Linking cellular stress response with translocation and trafficking. Protein Sci. 2009, 18, 2356–2370. [Google Scholar]
- Paliwal, S.; Iglesias, P.A.; Campbell, K.; Hilioti, Z.; Groisman, A.; Levchenko, A. MAPK-mediated bimodal gene expression and adaptive gradient sensing in yeast. Nature 2007, 446, 46–51. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/
Share and Cite
Park, M.; Tsai, S.-L.; Chen, W. Microbial Biosensors: Engineered Microorganisms as the Sensing Machinery. Sensors 2013, 13, 5777-5795. https://doi.org/10.3390/s130505777
Park M, Tsai S-L, Chen W. Microbial Biosensors: Engineered Microorganisms as the Sensing Machinery. Sensors. 2013; 13(5):5777-5795. https://doi.org/10.3390/s130505777
Chicago/Turabian StylePark, Miso, Shen-Long Tsai, and Wilfred Chen. 2013. "Microbial Biosensors: Engineered Microorganisms as the Sensing Machinery" Sensors 13, no. 5: 5777-5795. https://doi.org/10.3390/s130505777



