Quorum Sensing Activity of Serratia fonticola Strain RB-25 Isolated from an Ex-landfill Site
Abstract
: Quorum sensing is a unique bacterial communication system which permits bacteria to synchronize their behaviour in accordance with the population density. The operation of this communication network involves the use of diffusible autoinducer molecules, termed N-acylhomoserine lactones (AHLs). Serratia spp. are well known for their use of quorum sensing to regulate the expression of various genes. In this study, we aimed to characterized the AHL production of a bacterium designated as strain RB-25 isolated from a former domestic waste landfill site. It was identified as Serratia fonticola using matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry analysis and this was confirmed by 16S ribosomal DNA sequencing. High resolution triple quadrupole liquid chromatography-mass spectrometry analysis of S. fonticola strain RB-25 spent culture supernatant indicated the existence of three AHLs namely: N-butyryl-L-homoserine lactone (C4-HSL), N-hexanoyl-L-homoserine lactone (C6-HSL) and N-(3-oxohexanoyl) homoserine-lactone (3-oxo-C6 HSL). This is the first report of the production of these AHLs in S. fonticola.1. Introduction
Serratia spp. are well known for the production of prodigiosin [1]. As part of the Enterobacteriaceae family, Serratia spp. are frequently reported to be the cause of food spoilage [1]. The ubiquitous nature of bacteria from this genus enables their survival in diverse habitats, including host digestive tracts, soil and water [2]. For example, while the majority of Serratia spp. are well-known for their role as a plant-growth promoting rhizobacterium, a large number of Serratia spp. are also repeatedly found to be isolated from respiratory tract and urinary tract clinical samples [3–6]. Due to its growing clinical significance as well as its beneficial environmental contributions, a more comprehensive investigation on the physiological properties and in particular the quorum-sensing behaviour of bacteria of this genus is crucial [7,8].
Quorum sensing (QS) is an interaction mechanism employed by unicellular bacteria to achieve cell-to-cell communication in order to regulate expression of various cell density-dependent genes [9–11]. QS is achieved by bacteria via production of and response upon detection to various QS signaling molecules termed autoinducers. These signaling mechanisms enable the bacterial population to maintain the cell density within individual species as well as among other species in the surrounding environment via detection of accumulated concentrations of exogenous signaling molecules, and subsequently allow regulation of the expression of diverse beneficial genes. Examples of QS regulated activities reported are biofilm formation and swarming activities in Pseudomonas aeruginosa [12,13], antibiotic production in Erwinia carotovora [14], and Ti plasmid conjugation in Agrobacterium tumefaciens [15]. To date, the most extensively explored QS molecules are the N-acylhomoserine lactones (AHLs), diffusible signaling molecules whose structure comprises a five-membered lactone ring that contains an amide side chain of varying length. [16]. To understand the virulence, antibiotic susceptibility, physiology and metabolism of S. fonticola one should start primarily by characterizing its AHL expression [17].
In this study, we isolated a strain RB-25 identified as S. fonticola from a site formerly used as domestic waste dumping site. Identification of the isolate was done by using matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry analysis and sequence analysis of 16S ribosomal DNA (rDNA). This isolate triggered both Chromobacterium violaeum CV026 and E. coli [pSB401] biosensors. Subsequently, its AHL profile was confirmed unequivocally by using high resolution quadrupole mass spectrometry. The former landfill site was selected as the sampling site as there is limited documentation about the microbial population with QS properties in this unique environment.
2. Experimental Section
2.1. Strain Isolation, Enrichment and Condition
A former dumping ground in Ayer Hitam (Malaysia; GPS coordinates N03′00′12.1 E101′39′33′1) was selected as the sampling site. Soil specimens were collected at a subsurface depth of 10 cm using a clean 50 mL Falcon tube. Subsequently, soil samples were inoculated into KGm medium and enrichment was done for four cycles [18,19]. In short, soil samples were mixed thoroughly with 10 mL of KGm medium. Subsequent inoculation of soil suspension into fresh KGm medium was performed every 48 h and the inoculum was incubated with agitation. The KGm medium used was supplied with 50 mM 3-oxo-C6-HSL (Sigma-Aldrich, St. Louis, MO, USA). A diluted suspension was then spread on a plate of KGm agar supplemented with 3-oxo-C6-HSL and culture was spread on Luria-Bertani (LB) agar in the last enrichment cycle. The sole growth media used throughout the entire experiment are LB broth and LB agar. All strains were cultured aerobically in 28 °C with exception of E. coli [pSB401], which was cultured in 37 °C.
2.2. AHL Production Screening
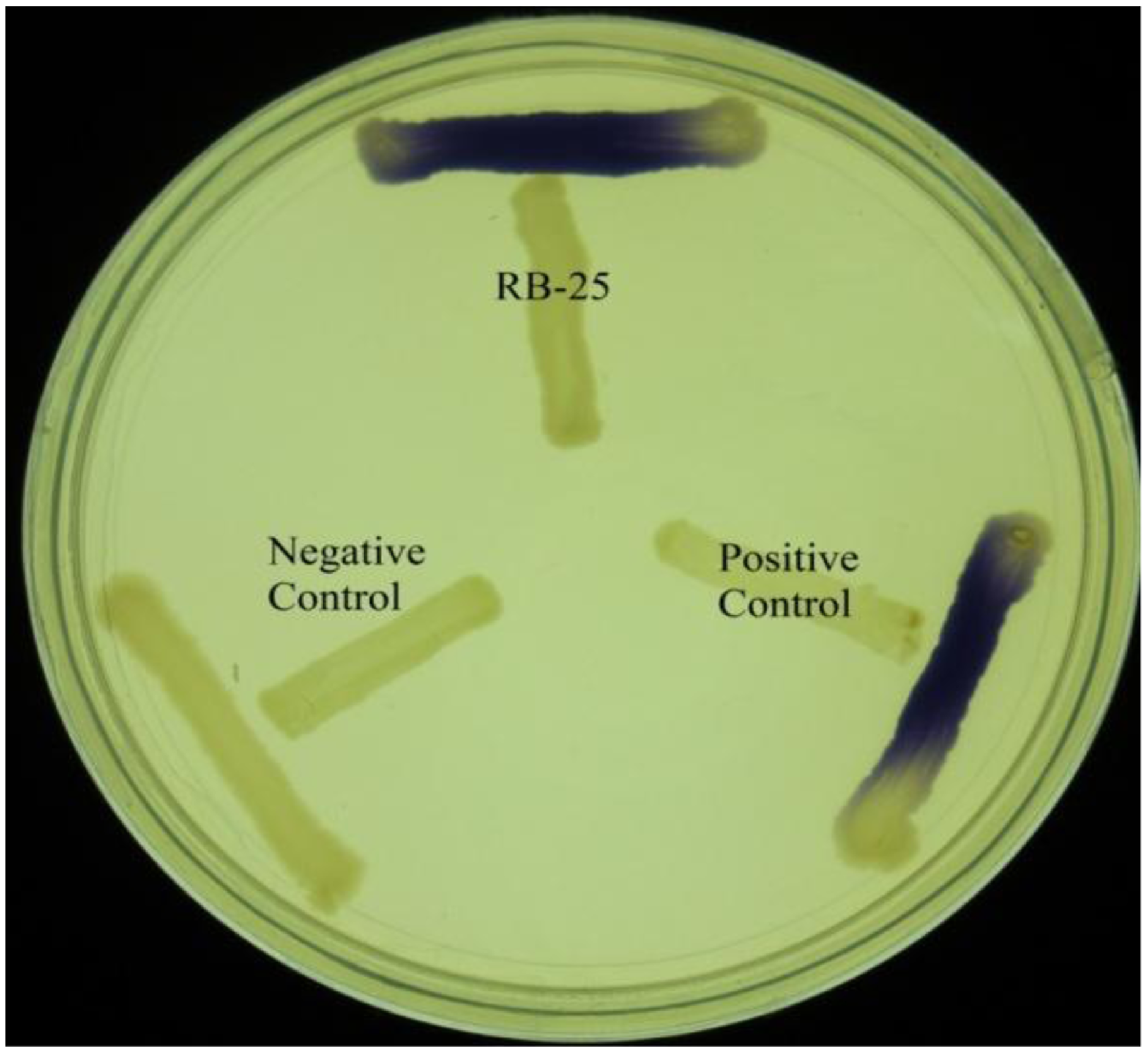
Short chain AHLs production was screened by using two different biosensors namely C. violaceum CV026 and E. coli [pSB401]. The C. violaceum CV026 biosensor responds to short chain AHLs by producing a purple pigmentation after an overnight incubation at 28 °C. AHL activity was then re-confirmed with a bioluminescence assay involving the use of the lux-based biosensor E. coli [pSB401]. Positive and negative controls for short chain AHLs detection were E. carotovora GS101 and E. carotovora PNP22, respectively. All reporter strains and both positive and negative control strains (Table 1) were gifts kindly provided by Professor Paul Williams (University of Nottingham, Nottingham, UK) (Table 1).
2.3. Extraction of AHLs
Isolates that showed QS activity were cultured in LB medium buffered to pH 5.5 with 50 mM (MOPS) in order to avoid alkaline-pH-driven deterioration of any AHLs. Log phase culture was subsequently extracted twice via the solvent extraction method. The solvent utilised in this extraction procedure was ethyl acetate supplemented with 0.1% glacial acetic acid [20]. The dried extracts were kept at −20 °C for storage purposes.
2.4. Bioluminescence Analysis
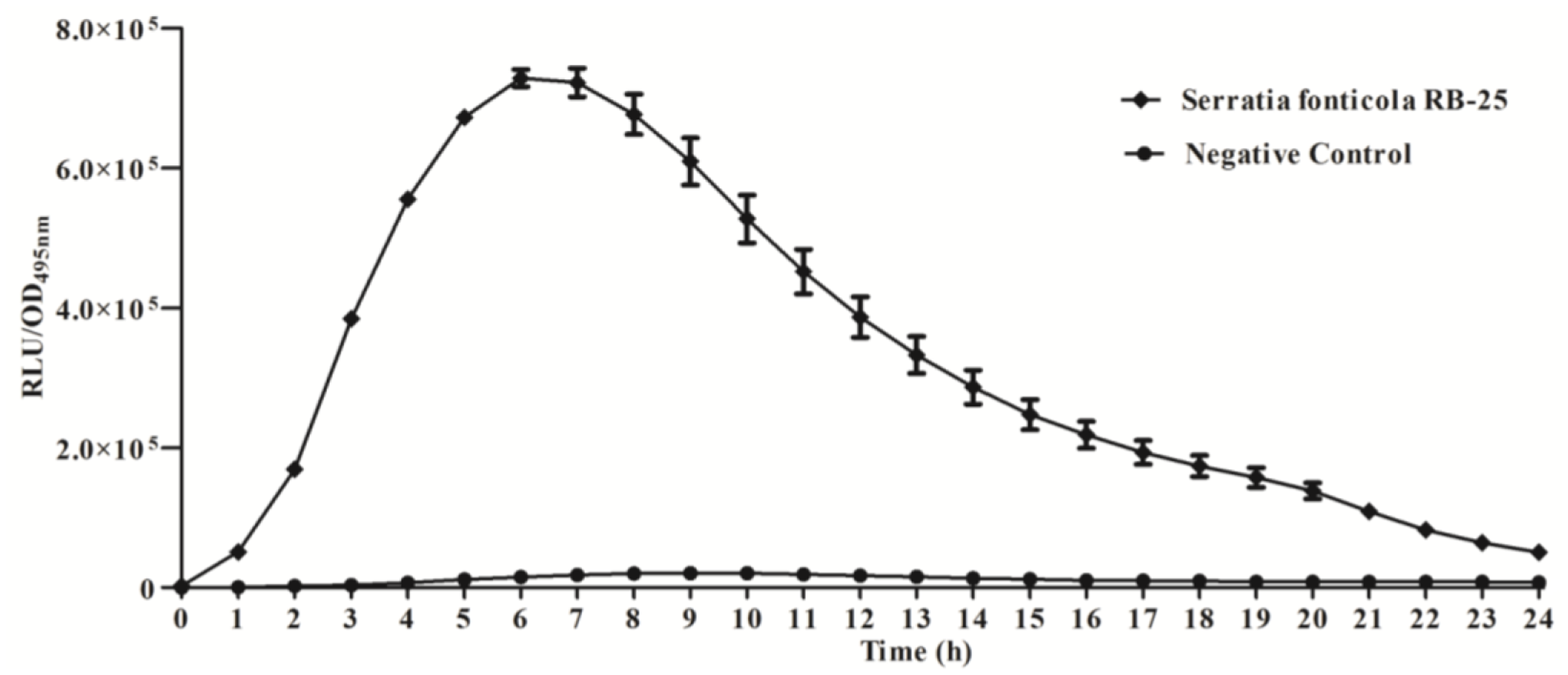
Measurement of bioluminescence were determined by using an Infinite M200 luminometer (Tecan, Männerdorf, Switzerland). Dessicated AHL extracts were resuspended with lux-based E. coli [pSB401] biosensor (OD600 of 0.1) before dispensing into each well of 96-microtitre well plates in triplicate and the experiments were independently repeated three times. Bioluminescence activities were analyzed as luminescence (relative light unit) at the excitation wavelength of 495 nm (RLU/OD495nm) over the course of 24 h. Reading of optical density and bioluminescence were automatically and simultaneously recorded at 60 min intervals [18,21].
2.5. Assay of AHLs Using Liquid Chromatography Triple Quadrupole Mass Spectrometry (LC-MS/MS)
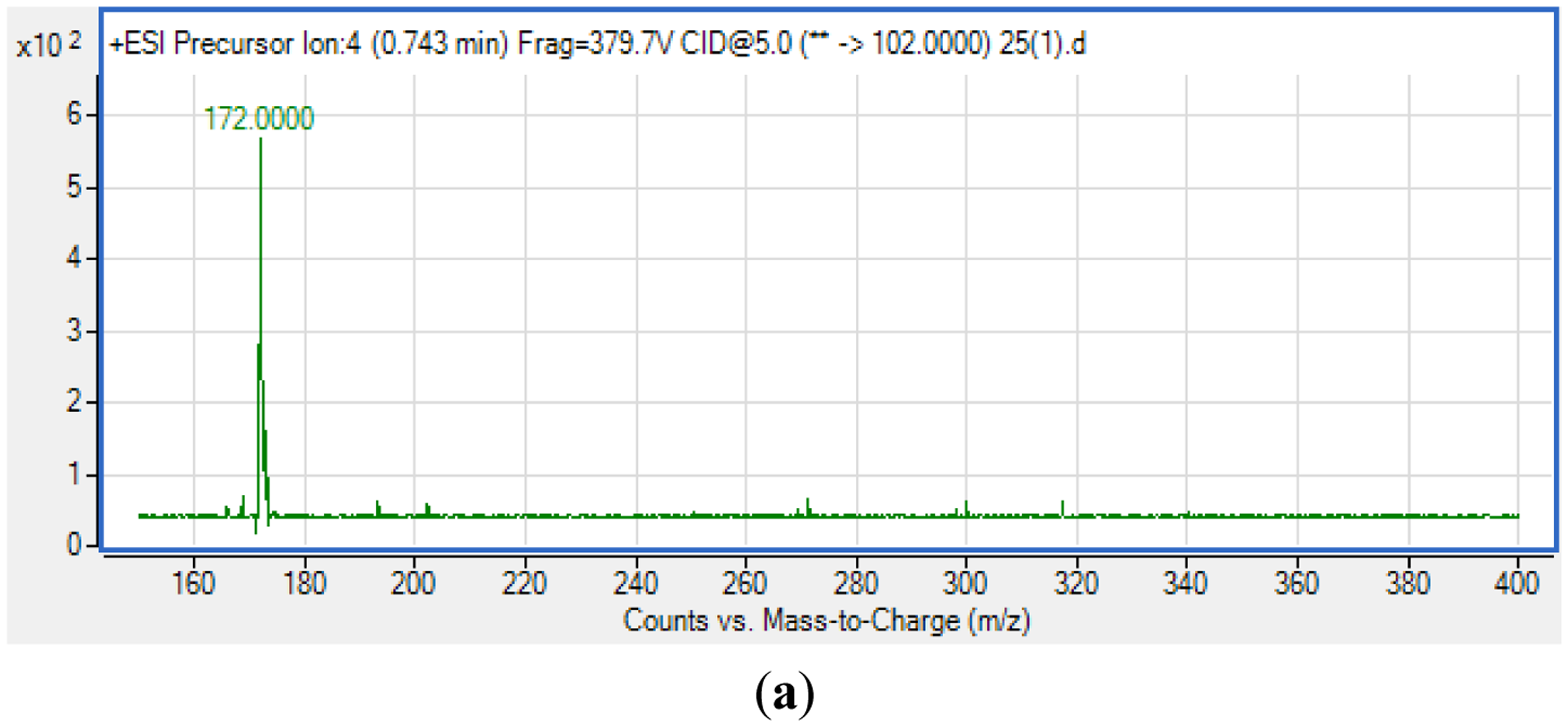
AHLs produced by the QS isolate were studied using a High Resolution Tandem Liquid Chromatography Quadrupole Mass Spectrometry (LC-MS/MS; Agilent 1290 Infinity LC and Agilent 6490 Triple Quadrupole LC/MS systems, Agilent Technologies Inc., Santa Clara, CA, USA) as described previously [18,19]. The column used was Agilent Zorbax Rapid Resolution High Definition SB-C18 Threaded Column. Precursor ion scan mode was used for AHL detection where the product ion m/z value was fixed at m/z 102. Spectra analysis using the Agilent Mass Hunter software was performed as reported [18,19].
2.6. Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS) for Strain Identification
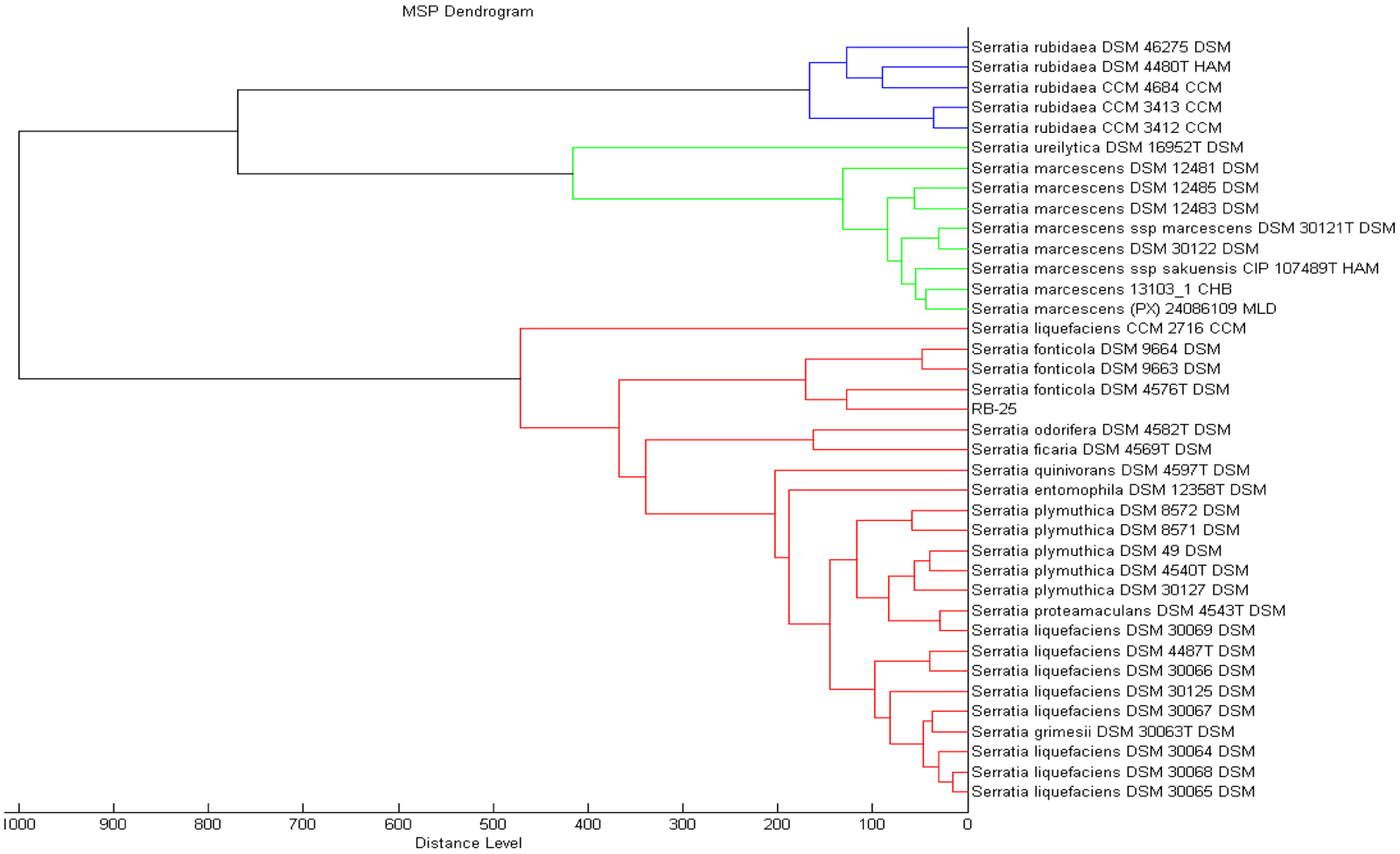
MALDI TOF MS was performed essentially as reported previously [18,19]. Bacterial samples were smeared very thinly on the MSP 96 target polished steel BC plate and MALDI matrix solution was added. A Microflex MALDI-TOF (Bruker Daltonik GmbH, Leipzig, Germany) bench-top mass spectrometer was used for identification of sample isolates [18,19,22] and for analysis of the MALDI-TOF mass spectra, we used the version 3.1 (Build 65) of the Bruker MALDI Biotyper Real Time Classification Classification software. To design the dendrogram for bacteria identification, we then used the MALDI Biotyper MSP creation method (Bruker Daltonics, Bremen, Germany) as reported previously [18,19,22].
2.7. Molecular Identification by 16S Ribosomal DNA (16S rDNA) Sequencing
To amplify the 16S rDNA genes, we used the reported method [23] with the following universal primers: 27F forward primers, 515F forward primers and 1525R reverse primers. Purification of genomic DNA, 16S rDNA PCR template and PCR condition was performed as reported elsewehere [23] and a phylognetic tree was conducted using MEGA5 [24].
3. Results and Discussion
3.1. Ioslation of Soil Bacteriaand Screening of Their AHLs Production
KGm medium is a minimal medium which facilitates growth of non-fastidious bacteria and quorum quenching bacteria [18,19]. Soil samples were inoculated into sterile KGm medium and four cycles of enrichment was conducted. It was noted that KGm medium became turbid after two days. This indicated growth of microorganisms during each enrichment cycle. No growth was detected under our present experimental condition in the control KGm medium where AHL was not supplemented (data not shown). Bacterial isolates were streaked in several passages in order to obtain single, pure colonies and the bacteria were examined for both AHL production and degradation using the two types of biosensors. For screening of AHL activities, a number of AHL biosensors have been constructed, for instance C. violaceum CV026 was used for screening of short chain AHL production [21,25,26]. This AHL biosensor relies on the Cvi-based QS system whereby violacein is produced as a result of the presence of short chain AHLs. This makes CV026 a very convenient biosensor for initial and rapid screening of AHLs based on pigment formation [25]. Based on this preliminary screening, we detected that strain RB-25 produced short chain AHLs (Figure 1). However no significant degradation of AHLs was observed, suggesting strain RB-25 is a non-fastidous bacteria capable of thriving under nutrient limiting conditions.
We further studied the production of AHLs using bioluminescence analysis with E. coli [pSB401] biosensor cells which detect short chain AHLs [21, 26], confirming that strain RB-25 produced short chain AHLs (Figure 2). Production of short chain AHLs was seen as the increment of RLU/OD495nm value observed over 24 h.
3.2. Molecular and MALDI-TOF Identification of Strain RB-25
MALDI-TOF has been proven to be a rapid and reliable technique useful for bacteria biotyping [27–29]. In the present work, preliminary identification was done using MALDI-TOF where we could identified isolate RB-25 as S. fonticola with a value of 2.361, which indicated a reliable detection at both the genus and species level. To obtain confident identification at the species level, the MALDI-TOF scoring system value should be higher than 2.3 (Figure 3) [18,19,22].
To further confirm the identity of strain RB-25, we then used a molecular approach by analyzing its 16S rDNA gene nucleotide sequences using phylogenetics. Web-based comparison against the NCBI database and phylogenetic analysis of 16S rDNA sequences of strain RB-25 confirmed that this isolate is indeed S. fonticola (Figure 4). In this work, the Neighbor-Joining method [30] with bootstrap value from 1,000 replicates [31] and Maximum Composite Likelihood method [32] were used.
3.3. Identification of AHLs
AHLs production has been documented in some species of Serratia where the most frequently detected AHL is C6-HSL [1]. Production of C4-HSL and C6-HSL had also been documented in Serratia sp. ATCC 39006 [33] and S. marcescens MG1 [34,35], whereas some Serratia spp. like S. plymuthica RVH1, S. marcescens SS-1 and S. proteamaculans B5a have been reported to produce other types of AHLs [8,36,37].
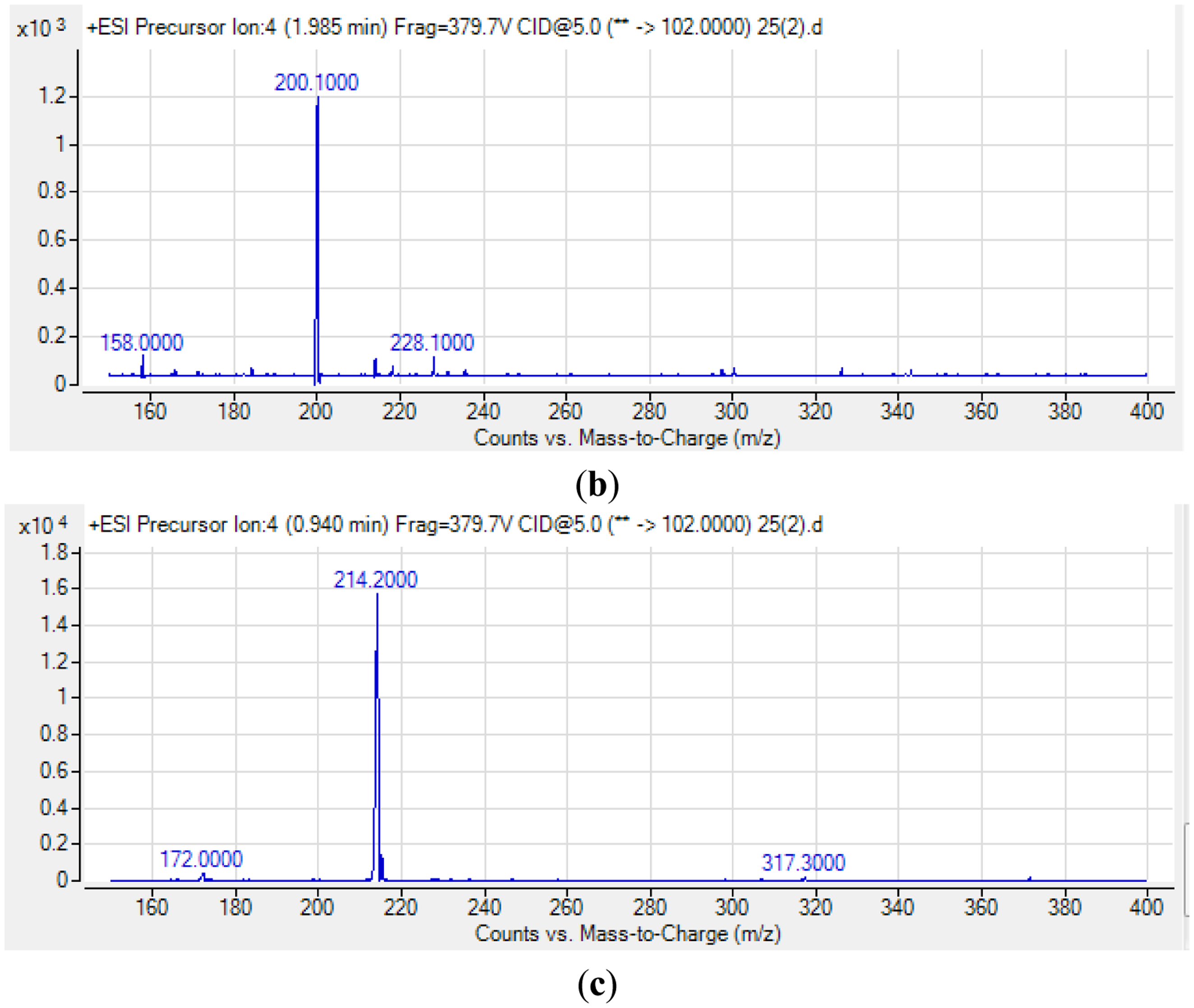
For S. fonticola, although its QS genes had been documented for rapid identification purposes [38], little is known about its AHL profile. Hence, in this work, we strived to identify if this isolate produced any detectable AHLs. Characterization of its AHLs profile is the first step in understanding the QS properties of S. fonticola and its AHL extract was assayed via tandem mass spectrometry platform that confirmed the presence of three AHLs, namely C4-HSL, C6-HSL and 3-oxo-C6 HSL (Figure 5).
4. Conclusions
In this work, we studied the AHL profile of S. fonticola strain RB-25 isolated from a former landfill site. In this paper we confirmed for the first time the presence in the spent supernatant of this strain of three short chain AHLs, namely C4-HSL, C6-HSL and 3-oxo-C6 HSL, thus providing evidence of quorum sensing activity in this isolate.
Acknowledgments
This research was financed by the University of Malaya HIR Grant (UM.C/625/1/HIR/MOHE/CHAN/01, Grant No. A-000001-50001) to Kok-Gan Chan which is gratefully acknowledged.
Author Contributions
Robson Ee contributed in methodology design, execution of experimental tests, data analysis and interpretation, as well as manuscript writing and revision. Yan-Lue Lim conducted the experimental tests, processed and analyzed the data and co-contributed in manuscript writing and revision. Kok-Keng Tee contributed in bacterial typing analysis and advised in manuscript revision. Wai-Fong Yin coordinated and co-supervised the research activities, as well as contributed in proofreading and editing of the manuscript. Kok-Gan Chan proposed the concept of the research approach and design, supervised the research activities, and contributed in editing and revision of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Van Houdt, R.; Givskov, M.; Michiels, C.W. Quorum sensing in Serratia. FEMS Microbiol. Rev. 2007, 31, 407–424. [Google Scholar]
- Grimont, P.A.D.; Grimont, F. The Genus. Serratia. Ann. Rev. Microbiol. 1978, 32, 221–248. [Google Scholar]
- Yokota, M.; Okazawa, A.; Tanaka, T. Serratia marcescens as an opportunistic human pathogen. Nihon Saikingaku Zasshi 2001, 56, 527–535. [Google Scholar]
- Bollet, C.; Gainnier, M.; Sainty, J.M.; Orhesser, P.; De Micco, P. Serratia fonticola isolated from a leg abscess. J. Clin. Microbiol. 1991, 29, 834–835. [Google Scholar]
- Devi, U.; Khatri, I.; Kumar, N.; Sharma, D.; Subramanian, S.; Saini, A.K. Draft genome sequence of plant-growth-promoting rhizobacterium Serratia fonticola strain AU-AP2C, isolated from the pea rhizosphere. Genome Announc. 2013, 1. [Google Scholar] [CrossRef]
- Henriques, I.; Juca Ramos, R.T.; Barauna, R.A.; de Sa, P.H.; Marinho Almeida, D.; Carneiro, A.R.; Barbosa, S.; Pereira, A.; Alves, A.; Saavedra, M.J.; et al. Draft genome sequence of Serratia fonticola UTAD54, a carbapenem-resistant strain isolated from drinking water. Genome Announc. 2013, 1. [Google Scholar] [CrossRef]
- Stock, I.; Burak, S.; Sherwood, K.J.; Gruger, T.; Wiedemann, B. Natural antimicrobial susceptibilities of strains of “unusual” Serratia species: S. ficaria, S. fonticola, S. odorifera, S. plymuthica and S. rubidaea. J. Antimicrob. Chemother. 2003, 51, 865–885. [Google Scholar]
- Van Houdt, R.; Moons, P.; Aertsen, A.; Jansen, A.; Vanoirbeek, K.; Daykin, M.; Williams, P.; Michiels, C.W. Characterization of a luxI/luxR-type quorum sensing system and N-acyl-homoserine lactone-dependent regulation of exo-enzyme and antibacterial component production in Serratia plymuthica RVH1. Res. Microbiol. 2007, 158, 150–158. [Google Scholar]
- Fuqua, C.; Parsek, M.R.; Greenberg, E.P. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Ann. Rev. Genet. 2001, 35, 439–468. [Google Scholar]
- Miller, M.B.; Bassler, B.L. Quorum sensing in bacteria. Ann. Rev. Microbiol. 2001, 55, 165–199. [Google Scholar]
- Schauder, S.; Bassler, B.L. The languages of bacteria. Genes Dev. 2001, 15, 1468–1480. [Google Scholar]
- Davies, D.G.; Parsek, M.R.; Pearson, J.P.; Iglewski, B.H.; Costerton, J.W.; Greenberg, E.P. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 1998, 280, 295–298. [Google Scholar]
- Shrout, J.D.; Chopp, D.L.; Just, C.L.; Hentzer, M.; Givskov, M.; Parsek, M.R. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol. Microbiol. 2006, 62, 1264–1277. [Google Scholar]
- Bainton, N.J.; Bycroft, B.W.; Chhabra, S.R.; Stead, P.; Gledhill, L.; Hill, P.J.; Rees, C.E.; Winson, M.K.; Salmond, G.P.; Stewart, G.S.; et al. A general role for the lux autoinducer in bacterial cell signalling: Control of antibiotic biosynthesis. Erwinia. Gene 1992, 116, 87–91. [Google Scholar]
- Zhang, L.; Murphy, P.J.; Kerr, A.; Tate, M.E. Agrobacterium conjugation and gene regulation by N-acyl-L-homoserine lactones. Nature 1993, 362, 446–448. [Google Scholar]
- Chong, T.M.; Koh, C.L.; Sam, C.K.; Choo, Y.M.; Yin, W.F.; Chan, K.G. Characterization of quorum sensing and quorum quenching soil bacteria isolated from Malaysian tropical montane forest. Sensors 2012, 12, 4846–4859. [Google Scholar]
- Hong, K.W.; Koh, C.L.; Sam, C.K.; Yin, W.F.; Chan, K.G. Quorum quenching revisited-from signal decays to signalling confusion. Sensors 2012, 12, 4661–4696. [Google Scholar]
- Han-Jen, R.E.; Wai-Fong, Y.; Kok-Gan, C. Pandoraea sp. RB-44, a novel quorum sensing soil bacterium. Sensors 2013, 13, 14121–14132. [Google Scholar]
- Chen, J.W.; Koh, C.L.; Sam, C.K.; Yin, W.F.; Chan, K.G. Short Chain N-acyl homoserine lactone production by soil isolate Burkholderia sp. strain A9. Sensors 2013, 13, 13217–13227. [Google Scholar]
- Ortori, C.A.; Dubern, J.F.; Chhabra, S.R.; Camara, M.; Hardie, K.; Williams, P.; Barrett, D.A. Simultaneous quantitative profiling of N-acyl-L-homoserine lactone and 2-alkyl-4(1H)-quinolone families of quorum-sensing signaling molecules using LC-MS/MS. Anal. Bioanal. Chem. 2011, 399, 839–850. [Google Scholar]
- Winson, M.K.; Swift, S.; Fish, L.; Throup, J.P.; Jorgensen, F.; Chhabra, S.R.; Bycroft, B.W.; Williams, P.; Stewart, G.S. Construction and analysis of luxCDABE-based plasmid sensors for investigating N-acyl homoserine lactone-mediated quorum sensing. FEMS Microbiol. Lett. 1998, 163, 185–192. [Google Scholar]
- Ngeow, Y.F.; Cheng, H.J.; Chen, J.W.; Yin, W.F.; Chan, K.G. Short chain N-acylhomoserine lactone production by clinical multidrug resistant Klebsiella pneumoniae strain CSG20. Sensors 2013, 13, 15242–15251. [Google Scholar]
- Chan, K.G.; Yin, W.F.; Sam, C.K.; Koh, C.L. A novel medium for the isolation of N-acylhomoserine lactone-degrading bacteria. J. Ind. Microbiol. Biotechnol. 2009, 36, 247–251. [Google Scholar]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar]
- McClean, K.H.; Winson, M.K.; Fish, L.; Taylor, A.; Chhabra, S.R.; Camara, M.; Daykin, M.; Lamb, J.H.; Swift, S.; Bycroft, B.W.; et al. Quorum sensing and Chromobacterium violaceum: Exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology 1997, 143, 3703–3711. [Google Scholar]
- Chen, J.-W.; Chin, S.; Tee, K.K.; Yin, W.-F.; Choo, Y.M.; Chan, K.-G. N-acyl homoserine lactone-producing Pseudomonas putida strain T2-2 from human tongue surface. Sensors 2013, 13, 13192–13203. [Google Scholar]
- Dai, Y.; Li, L.; Roser, D.C.; Long, S.R. Detection and identification of low-mass peptides and proteins from solvent suspensions of Escherichia coli by high performance liquid chromatography fractionation and matrix-assisted laser desorption/ionization mass spectrometry. Rapid. Commun. Mass Spectrom. 1999, 13, 73–78. [Google Scholar]
- Liang, X.; Zheng, K.; Qian, M.G.; Lubman, D.M. Determination of bacterial protein profiles by matrix-assisted laser desorption/ionization mass spectrometry with high-performance liquid chromatography. Rapid. Commun. Mass Spectrom. 1996, 10, 1219–1226. [Google Scholar]
- Chong, B.E.; Wall, D.B.; Lubman, D.M.; Flynn, S.J. Rapid profiling of E. coli proteins up to 500 kDa from whole cell lysates using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid. Commun. Mass Spectrom. 1997, 11, 1900–1908. [Google Scholar]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar]
- Thomson, N.R.; Crow, M.A.; McGowan, S.J.; Cox, A.; Salmond, G.P. Biosynthesis of carbapenem antibiotic and prodigiosin pigment in Serratia is under quorum sensing control. Mol. Microbiol. 2000, 36, 539–556. [Google Scholar]
- Eberl, L.; Christiansen, G.; Molin, S.; Givskov, M. Differentiation of Serratia liquefaciens into swarm cells is controlled by the expression of the flhD master operon. J. Bacteriol. 1996, 178, 554–559. [Google Scholar]
- Eberl, L.; Winson, M.K.; Sternberg, C.; Stewart, G.S.; Christiansen, G.; Chhabra, S.R.; Bycroft, B.; Williams, P.; Molin, S.; Givskov, M. Involvement of N-acyl-L-homoserine lactone autoinducers in controlling the multicellular behaviour of Serratia liquefaciens. Mol. Microbiol. 1996, 20, 127–136. [Google Scholar]
- Horng, Y.T.; Deng, S.C.; Daykin, M.; Soo, P.C.; Wei, J.R.; Luh, K.T.; Ho, S.W.; Swift, S.; Lai, H.C.; Williams, P. The LuxR family protein SpnR functions as a negative regulator of N-acylhomoserine lactone-dependent quorum sensing in Serratia marcescens. Mol. Microbiol. 2002, 45, 1655–1671. [Google Scholar]
- Christensen, A.B.; Riedel, K.; Eberl, L.; Flodgaard, L.R.; Molin, S.; Gram, L.; Givskov, M. Quorum-sensing-directed protein expression in Serratia proteamaculans B5a. Microbiology 2003, 149, 471–483. [Google Scholar]
- Zhu, H.; Sun, S.J.; Dang, H.Y. PCR detection of Serratia spp. using primers targeting pfs and luxS genes involved in AI-2-dependent quorum sensing. Curr. Microbiol. 2008, 57, 326–330. [Google Scholar]

| Strain | Description |
|---|---|
| C. violaceum CV026 | Biosensor that produces purple coloured violacein in response to short chain AHLs. |
| E. coli [pSB401] | Biosensor that produces bioluminescence in exposure to short chain AHLs. |
| E. carotovora GS101 | Positive control for biosensors. Produces short chain AHLs to activate C. violaceum CV026 and E. coli [pSB401]. |
| E. carotovora PNP22 | Negative control for biosensors. |
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Ee, R.; Lim, Y.-L.; Tee, K.-K.; Yin, W.-F.; Chan, K.-G. Quorum Sensing Activity of Serratia fonticola Strain RB-25 Isolated from an Ex-landfill Site. Sensors 2014, 14, 5136-5146. https://doi.org/10.3390/s140305136
Ee R, Lim Y-L, Tee K-K, Yin W-F, Chan K-G. Quorum Sensing Activity of Serratia fonticola Strain RB-25 Isolated from an Ex-landfill Site. Sensors. 2014; 14(3):5136-5146. https://doi.org/10.3390/s140305136
Chicago/Turabian StyleEe, Robson, Yan-Lue Lim, Kok-Keng Tee, Wai-Fong Yin, and Kok-Gan Chan. 2014. "Quorum Sensing Activity of Serratia fonticola Strain RB-25 Isolated from an Ex-landfill Site" Sensors 14, no. 3: 5136-5146. https://doi.org/10.3390/s140305136






