Rhodotorula Mucilaginosa, a Quorum Quenching Yeast Exhibiting Lactonase Activity Isolated from a Tropical Shoreline
Abstract
: Two microbial isolates from a Malaysian shoreline were found to be capable of degrading N-acylhomoserine lactones. Both Matrix Assisted Laser Desorption Ionization-Time of Flight-Mass Spectrometry and 18S rDNA phylogenetic analyses confirmed that these isolates are Rhodotorula mucilaginosa. Quorum quenching activities were detected by a series of bioassays and rapid resolution liquid chromatography analysis. The isolates were able to degrade various quorum sensing molecules namely N-hexanoyl-L-homoserine lactone (C6-HSL), N-(3-oxo-hexanoyl)-L-homoserine lactone (3-oxo-C6-HSL) and N-(3-hydroxyhexanoyl)-L-homoserine lactone (3-hydroxy-C6-HSL). Using a relactonisation assay to verify the quorum quenching mechanism, it is confirmed that Rh. mucilaginosa degrades the quorum sensing molecules via lactonase activity. To the best of our knowledge, this is the first documentation of the fact that Rh. mucilaginosa has activity against a broad range of AHLs namely C6-HSL, 3-oxo-C6-HSL and 3-hydroxy-C6-HSL.1. Introduction
Quorum sensing (QS) enables microorganisms to communicate via secreted signaling molecules called autoinducers and contributes to the regulation of gene expression in response to bacterial population density [1]. Many Gram-negative bacteria use the N-acyl homoserine lactones (AHLs) as autoinducers [2] in which AHLs consist of a 4- to 18-carbon N-acyl side chain linked to a lactone ring [3]. AHLs are synthesized by the activity of LuxI synthase using S-adenosylmethionine and acylated acyl carrier protein as substrates [4]. The stability of AHLs is pH-dependent, whereby the lactone ring will hydrolyze under alkaline conditions, resulting in a homoserine structure with an opened ring. Such a process is reversible with a switch of pH to acidic conditions [5,6]. Gram-negative bacteria employ AHL as QS signals in their communication circuits to coordinate various physiological activities. These signals lead to an activation of many processes including symbiosis, virulence, competence, conjugation, antibiotic production, motility, sporulation, and biofilm formation [2].
On the other hand, quorum quenching (QQ) is known as a process in which disrupts the QS signals by using several ways which includes enzymatic destruction of the signal molecules, development of antibodies or through QS signalling molecules blocking agents [7–9]. The first documented QQ enzyme was produced by a soil bacterium from the genus Bacillus in which was encoded by the aiiA gene. It was later characterized as an AHL-lactonase [8]. Since then, these QQ systems have been found in various microorganisms that use them to prevent the benefits of QS in an attempt to gain competitive advantage in polymicrobial environment [10]. The QQ properties of the genera Bacillus and Pseudomonas have been well established [11]. Examples are the AiiA lactonase homologs of Bacillus and PvdQ and also QuiP of Pseudomonas [12,13]. These reactions are produced enzymatically by lactonases such as AiiA, AttM, AiiB [14,15] and AhlD [16]. The disruption of QS signaling molecules is considered as a potential way of preventing and treating infections besides preventing plant diseases [17].
In view of this, we have isolated Rhodotorula mucilaginosa from a Malaysian shoreline which possesses QQ properties. The genus Rhodotorula is known as a saprophytic yeast that can be obtained from environmental sources [18] and has been described as a pathogen antagonist [19]. This study is the first documentation of AHL-degrading activity produced by a marine yeast member from the genus Rhodotorula.
2. Experimental Section
2.1. Sample Collection and Microbial Isolation
A water sample (50 mL) was collected in a sterile container from the shoreline of Batu Maung Penang, Malaysia. The sample was immediately processed upon returning to the laboratory. Water sample was diluted ten-fold and plated onto Luria Bertani (LB) agar. Pure colony was obtained by repeated streaking on LB agar grown at 28 °C.
2.2. Isolation and Identification of Microbial Strains
Microbial isolates of interest were identified using a Matrix Assisted Laser Desorption Ionization-Time of Flight-Mass Spectrometer (MALDI-TOF-MS, Bruker, München, Germany) [20] extraction method with a UV laser wavelength of 337 nm and acceleration voltage of 20 kV. Each spot on the target plate was measured by the MBT-autoX.axe autoExecute method. The bacterial spectra were then analyzed in the Bruker MALDI Biotyper Real Time Classification (RTC) Version 3.1 (Build 65) software. In order to verify the isolates identification, phylogenetic analysis was done via 18S rDNA gene nucleotide sequences.
The genomic DNA of the yeast isolates were extracted using the QIAamp® DNA Mini Kit (Qiagen, Frankfurt, Germany) and used as template for PCR. The 18S rDNA gene was PCR-amplified using the following primers namely ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) as described previously [21,22]. Nucleotide sequences were compared with GenBank databases using the BLASTN program followed by sequence alignment [23,24]. A phylogenetic tree was generated using the Molecular Evolutionary Genetic Analysis (MEGA) version 5.2 with Neighbour-Joining algorithm and 1,000 re-samplings [25,26].
2.3. Rapid Resolution Liquid Chromatography (RRLC) Analysis
Sample preparation for RRLC analysis was performed similar to the whole-cell AHL inactivation assay as described above, with the exception that the final concentration of AHL was 50 μM after rehydration with cell suspensions. AHLs were extracted twice using ethyl acetate, followed by drying by evaporation of the extraction. Acetonitrile (100 μL) was added to the extracted AHL and subjected to RRLC analysis using an Agilent Technologies 1200 series RRLC system [27] (Agilent, Santa Clara, CA, USA). AHL samples were separated in a Agilent Poroshell 120 EC-C18 column (4.6 mm × 100 mm, 2.7 μm particle size) with an elution procedure consisting of an isocratic profile of acetonitrile/water (35:65, v/v) for short chain AHLs with a constant flow rate of 0.7 mL/min and detection wavelength at 210 nm with 0.2. 0.4, 0.6, 0.8 and 1.0 μg/μL of synthetic AHLs were loaded as standards. AHL incubated with Escherichia coli TOP10 cells and PBS buffer served as negative controls, while AHL incubated with Bacillus cereus acted as positive control. Chromatographs were expressed as milli-absorbance units (mAUs, vertical axis) and time (minutes, horizontal axis), respectively.
2.4. Identification of AHL Lactonase Activity
Overnight cultures of QQ yeast cells were harvested by centrifugation. Cell pellets were washed twice and re-suspended in phosphate buffered saline (PBS) (100 mM, pH 6.5). Selected known concentrations of synthetic AHLs (C6-HSL, 3-oxo-C6-HSL and 3-hydroxy-C6-HSL, Sigma-Aldrich, St. Louis, MO, USA) were dispensed into sterile micro-centrifuge tubes and dried by evaporation. Yeast cell suspensions were then added to rehydrate the AHLs to final concentrations of 0.5 μM. The mixtures were then incubated at 28 °C with shaking (220 rpm) for 0 h and 24 h. All reactions were stopped by heat inactivation at 95 °C. For the detection of AHL degradation, 10 μL of reaction mixture was spotted onto sterile paper discs placed on a Chromobacterium violaceum CV026 lawn and incubated overnight at 28 °C. AHL inactivation assays involved incubation of E. coli TOP10 and PBS buffer as negative controls. Re-lactonisation with acidification using hydrochloric acid (HCl, 0.2 M) was performed as reported [11].
3. Results and Discussion
3.1. Isolation and Identification of Isolated Strains
A total of two yeast colonies (isolates B2 and B3) were purified from the tropical Malaysian shoreline. In order to identify the isolates, MALDI-TOF-MS was performed and later verified through 18S rDNA phylogenetic analysis. Both of the MALDI-TOF-MS (Figure 1) and 18S rDNA phylogenetic analyses (Figure 2) identified both isolates belonged to Rh. mucilaginosa.
3.2. Degradation of AHLs by Rh. Mucilaginosa Isolates B2 and B3
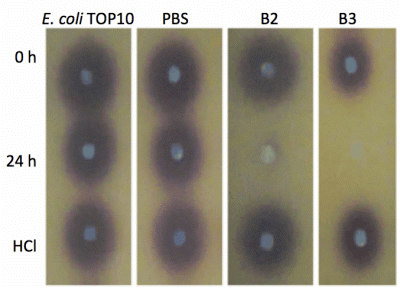
Both yeast isolates B2 and B3 showed degradation of C6-HSL (Figure 3), 3-oxo-C6-HSL (Figure 4) and 3-hydroxy-C6-HSL (Figure 5) as depicted by the reduction of the respective peaks in RRLC chromatograms after 24 h of incubation of these AHLs with Rh. mucilaginosa cells. To confirm whether both Rh. mucilaginosa isolates B2 and B3 degraded AHLs via lactonase activity, we acidified the AHL degradation mixture to promote relactonisation of the opened lactone rings [6]. Formation of purple color pigmentation after the addition of 0.2 M hydrochloric acid indicated lactonase production [28] (Figure 6).
Based on the whole-cell AHL inactivation assays and RRLC analyses on the degradation of various AHL, strong QQ activities were observed among the isolated Rh. mucilaginosa. In order to determine whether the Rh. mucilaginosa strain B2 and B3 inactivated AHLs through both the cleavage of the acyl chain or via lactonolysis, the B2 and B3 strains were incubated with different AHLs for 24 h. The cells were removed and the supernatant was collected and acidified to pH 2 and incubated for further 24 h. This resulted in the pH-mediated recyclization of any opened lactone ring compounds present [6], which would be subsequently detected using the C. violaceum CV026 AHL biosensor [29]. Degradation of C6-HSL, 3-oxo-C6-HSL and 3-hydroxy-C6-HSL was detected in the supernatant after 24 h incubation and recovered by acidification indicated that both B2 and B3 possess lactonase activity.
Rh. mucilaginosa has been reported as an emerging pathogen, especially in immunocompromised patients [30,31]. Furthermore, Rh. mucilaginosa has also been reported previously to reduce the development of natural decay of apples [32] while it inhibits the growth of Penicillium expansum and Botrytis cinerea which cause blue and gray mold decay, respectively [33]. We speculate that the QQ activity may play a part in restricting the QS activity of surrounding microorganisms, thus reducing QS-mediated phenotypes responsible for food spoilage. However, further analysis of its QQ mechanism should be conducted to confirm this speculation. Both Rh. mucilaginosa and Pseudomonas species produce a wide range of antimicrobials which help to confer a high competitive ability and to ensure dominance within their respective microbial communities [34]. It may be that QQ activity also plays a key role in enabling Rh. mucilaginosa and other microbial weed species to dominate their respective habitats. Further studies of the biology of QQ are needed to fully characterize the ecophysiological consequences [34]. Recently, QQ yeast Trichosporon loubieri has been reported to be isolated from tropical wetland waters and capable to grow on N-3-oxo-hexanoyl homoserine as carbon and nitrogen source for growth [35]. This work may indicate eukaryotic cells possesses QQ activity that could compete with the prokaryotic cells that rely on QS for coordinated phenotypes.
To the best of our knowledge, this is the first documentation of Rh. mucilaginosa to have exhibited QQ activities. Thus, we believe that the both isolated strains of Rh. mucilaginosa have potential as biocontrol agents which would delay food spoilage while novel AHL inactivating enzymes may have utility as therapeutic agents [36,37].
4. Conclusions/Outlook
This study confirms the degradation of AHLs by both isolated Rh. mucilaginosa strains B2 and B3 via lactonase activity. Our data demonstrated that the isolated yeast strains were able to degrade C6-HSL, 3-oxo-C6-HSL and 3-hydroxy-C6-HSL. Further work includes whole genome sequencing of Rh. mucilaginosa in order to identify the QQ gene of these strains.
Acknowledgments
This research was supported by the University of Malaya HIR Grant (UM.C/625/1/HIR/MOHE/CHAN/01, Grant No. A-000001-50001) to Kok-Gan Chan which is gratefully acknowledged.
Author Contributions
X.Y. Chan did the sampling and isolation of strains, N.A. Ghani, J. Sulaiman and Z. Ismail performed the experiments. W.F. Yin, N.A. Ghani and K.G. Chan did the data interpretation and analysis. K.G. Chan obtained the funding and supervised the entire project.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Smith, R.S.; Iglewski, B.H. P. aeruginosa quorum-sensing systems and virulence. Curr. Opin. Microbiol. 2003, 6, 56–60. [Google Scholar]
- Miller, M.B.; Bassler, B.L. Quorum sensing in bacteria. Ann. Rev. Microbiol. 2001, 55, 165–199. [Google Scholar]
- Pearson, J.P.; van Delden, C.; Iglewski, B.H. Active efflux and diffusion are involved in transport of Pseudomonas aeruginosa cell-to-cell signals. J. Bacteriol. 1999, 181, 1203–1210. [Google Scholar]
- Swift, S.; Karlyshev, A.V.; Fish, L.; Durant, E.L.; Winson, M.K.; Chhabra, S.R.; Williams, P.; Macintyre, S.; Stewart, G. Quorum sensing in Aeromonas hydrophila and Aeromonas salmonicida: Identification of the LuxRI homologs AhyRI and AsaRI and their cognate N-acylhomoserine lactone signal molecules. J. Bacteriol. 1997, 179, 5271–5281. [Google Scholar]
- Byers, J.T.; Lucas, C.; Salmond, G.P.; Welch, M. Nonenzymatic turnover of an Erwinia carotovora quorum-sensing signaling molecule. J. Bacteriol. 2002, 184, 1163–1171. [Google Scholar]
- Yates, E.A.; Philipp, B.; Buckley, C.; Atkinson, S.; Chhabra, S.R.; Sockett, R.E.; Goldner, M.; Dessaux, Y.; Cámara, M.; Smith, H. N-acylhomoserine lactones undergo lactonolysis in a pH-, temperature-, and acyl chain length-dependent manner during growth of Yersinia pseudotuberculosis and Pseudomonas aeruginosa. Infect. Immun. 2002, 70, 5635–5646. [Google Scholar]
- Williams, P. Quorum sensing: An emerging target for antibacterial chemotherapy? Expert Opin. Ther. Targets 2002, 6, 257–274. [Google Scholar]
- Bjarnsholt, T.; Givskov, M. Quorum sensing inhibitory drugs as next generation antimicrobials: Worth the effort? Curr. Infect. Dis. Rep. 2008, 10, 22–28. [Google Scholar]
- Lowery, C.A.; Salzameda, N.T.; Sawada, D.; Kaufmann, G.F.; Janda, K.D. Medicinal chemistry as a conduit for the modulation of quorum sensing. J. Med. Chem. 2010, 53, 7467–7489. [Google Scholar]
- Lin, Y.H.; Xu, J.L.; Hu, J.; Wang, L.H.; Ong, S.L.; Leadbetter, J.R.; Zhang, L.H. Acyl-homoserine lactone acylase from Ralstonia strain XJ12B represents a novel and potent class of quorum-quenching enzymes. Mol. Microbiol. 2003, 47, 849–860. [Google Scholar]
- Chong, T.-M.; Koh, C.-L.; Sam, C.-K.; Choo, Y.-M.; Yin, W.-F.; Chan, K.-G. Characterization of quorum sensing and quorum quenching soil bacteria isolated from Malaysian tropical montane forest. Sensors 2012, 12, 4846–4859. [Google Scholar]
- Dong, Y.-H.; Zhang, L.-H. Quorum sensing and quorum-quenching enzymes. J. Microbiol. 2005, 43, 101–109. [Google Scholar]
- Sio, C.F.; Otten, L.G.; Cool, R.H.; Diggle, S.P.; Braun, P.G.; Bos, R.; Daykin, M.; Cámara, M.; Williams, P.; Quax, W.J. Quorum quenching by an N-acyl-homoserine lactone acylase from Pseudomonas aeruginosa PAO1. Infect. Immun. 2006, 74, 1673–1682. [Google Scholar]
- Dong, Y.-H.; Xu, J.-L.; Li, X.-Z.; Zhang, L.-H. AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proc. Natl. Acad. Sci. USA 2000, 97, 3526–3531. [Google Scholar]
- Carlier, A.; Uroz, S.; Smadja, B.; Fray, R.; Latour, X.; Dessaux, Y.; Faure, D. The Ti plasmid of Agrobacterium tumefaciens harbors an attM-paralogous gene, aiiB, also encoding N-acyl homoserine lactonase activity. Appl. Environ. Microbiol. 2003, 69, 4989–4993. [Google Scholar]
- Medina-Martínez, M.S.; Uyttendaele, M.; Rajkovic, A.; Nadal, P.; Debevere, J. Degradation of N-acyl-L-homoserine lactones by Bacillus cereus in culture media and pork extract. Appl. Environ. Microbiol. 2007, 73, 2329–2332. [Google Scholar]
- Hosni, T.; Moretti, C.; Devescovi, G.; Suarez-Moreno, Z.R.; Fatmi, M.B.; Guarnaccia, C.; Pongor, S.; Onofri, A.; Buonaurio, R.; Venturi, V. Sharing of quorum-sensing signals and role of interspecies communities in a bacterial plant disease. ISME 2011, 5, 1857–1870. [Google Scholar]
- Tournas, V.; Katsoudas, E.; Miracco, E. Moulds, yeasts and aerobic plate counts in ginseng supplements. Int. J. Food Microbiol. 2006, 108, 178–181. [Google Scholar]
- Akhtyamova, N.; Sattarova, R. Endophytic yeast Rhodotorula rubra strain TG-1: Antagonistic and plant protection activities. Biochem. Physiol. 2013, 2. [Google Scholar] [CrossRef]
- Seng, P.; Drancourt, M.; Gouriet, F.; La Scola, B.; Fournier, P.-E.; Rolain, J.M.; Raoult, D. Ongoing revolution in bacteriology: Routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Infect. Dis. 2009, 49, 543–551. [Google Scholar]
- Nunes, J.M.; Bizerra, F.C.; Ferreira, R.C.; Colombo, A.L. Molecular identification, antifungal susceptibility profile, and biofilm formation of clinical and environmental Rhodotorula species isolates. Antimicrob. Agents Chemother. 2013, 57, 382–389. [Google Scholar]
- Esteve-Zarzoso, B.; Belloch, C.; Uruburu, F.; Querol, A. Identification of yeasts by RFLP analysis of the 5.8S rRNA gene and the two ribosomal internal transcribed spacers. Int. J. Syst. Bacteriol. 1999, 49, 329–337. [Google Scholar]
- Chan, K.-G.; Wong, C.-S.; Yin, W.-F.; Sam, C.-K.; Koh, C.-L. Rapid degradation of N-3-oxo-acylhomoserine lactones by a Bacillus cereus isolate from Malaysian rainforest soil. Antonie van Leeuwenhoek 2010, 98, 299–305. [Google Scholar]
- Yin, W.-F.; Purmal, K.; Chin, S.; Chan, X.-Y.; Koh, C.-L.; Sam, C.-K.; Chan, K.-G. N-acyl homoserine lactone production by Klebsiella pneumoniae isolated from human tongue surface. Sensors 2012, 12, 3472–3483. [Google Scholar]
- Yin, W.-F.; Tung, H.-J.; Sam, C.-K.; Koh, C.-L.; Chan, K.-G. Quorum quenching Bacillus sonorensis isolated from soya sauce fermentation brine. Sensors 2012, 12, 4065–4073. [Google Scholar]
- Chang, C.-Y.; Koh, C.-L.; Sam, C.-K.; Chan, X.-Y.; Yin, W.F.; Chan, K.G. Unusual long-chain N-acyl homoserine lactone production by and presence of quorum quenching activity in bacterial isolates from diseased tilapia fish. PLoS One 2012, 7, e44034. [Google Scholar]
- Wong, C.-S.; Yin, W.-F.; Choo, Y.-M.; Sam, C.-K.; Koh, C.-L.; Chan, K.-G. Coexistence of quorum-quenching and quorum-sensing in tropical marine Pseudomonas aeruginosa strain MW3A. World J. Microbiol. Biotechnol. 2012, 28, 453–461. [Google Scholar]
- Dong, Y.-H.; Wang, L.-H.; Xu, J.-L.; Zhang, H.-B.; Zhang, X.-F.; Zhang, L.-H. Quenching quorum-sensing-dependent bacterial infection by an N-acyl homoserine lactonase. Nature 2001, 411, 813–817. [Google Scholar]
- McClean, K.H.; Winson, M.K.; Fish, L.; Taylor, A.; Chhabra, S.R.; Camara, M.; Daykin, M.; Lamb, J.H.; Swift, S.; Bycroft, B.W. Quorum sensing and Chromobacterium violaceum: Exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology 1997, 143, 3703–3711. [Google Scholar]
- Miceli, M.H.; Díaz, J.A.; Lee, S.A. Emerging opportunistic yeast infections. Lancet Infect. Dis. 2011, 11, 142–151. [Google Scholar]
- Baradkar, V.; Kumar, S. Meningitis caused by Rhodotorula mucilaginosa in human immunodeficiency virus seropositive patient. Ann. Indian Acad. Neurol. 2008, 11, 245–247. [Google Scholar]
- Li, R.; Zhang, H.; Liu, W.; Zheng, X. Biocontrol of postharvest gray and blue mold decay of apples with Rhodotorula mucilaginosa and possible mechanisms of action. Int. J. Food Microbiol. 2011, 146, 151–156. [Google Scholar]
- Saravanakumar, D.; Ciavorella, A.; Spadaro, D.; Garibaldi, A.; Gullino, M.L. Metschnikowia pulcherrima strain MACH1 outcompetes Botrytis cinerea, Alternaria alternata and Penicillium expansum in apples through iron depletion. Postharvest Biol. Technol. 2008, 49, 121–128. [Google Scholar]
- Jonathan, A.C.; Andrew, N.W.B.; Prashanth, B.; Allen, Y.M.; David, J.T.; John, E.H. The biology of habitat dominance; Can microbes behave as weeds? Microb. Biotechnol. 2013, 6, 453–492. [Google Scholar]
- Wong, C.-S.; Koh, C.-L.; Sam, C.-K.; Chen, J.W.; Chong, Y.M.; Yin, W.-F.; Chan, K.-G. Degradation of bacterial quorum sensing signaling molecules by the microscopic yeast Trichosporon loubieri isolated from tropical wetland waters. Sensors 2013, 13, 12943–12957. [Google Scholar]
- Uroz, S.; Dessaux, Y.; Oger, P. Quorum sensing and quorum quenching: The yin and yang of bacterial communication. ChemBioChem 2009, 10, 205–216. [Google Scholar]
- Hong, K.-W.; Koh, C.-L.; Sam, C.-K.; Yin, W.-F.; Chan, K.-G. Quorum quenching revisited—From signal decays to signalling confusion. Sensors 2012, 12, 4661–4696. [Google Scholar]







© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Ghani, N.A.; Sulaiman, J.; Ismail, Z.; Chan, X.-Y.; Yin, W.-F.; Chan, K.-G. Rhodotorula Mucilaginosa, a Quorum Quenching Yeast Exhibiting Lactonase Activity Isolated from a Tropical Shoreline. Sensors 2014, 14, 6463-6473. https://doi.org/10.3390/s140406463
Ghani NA, Sulaiman J, Ismail Z, Chan X-Y, Yin W-F, Chan K-G. Rhodotorula Mucilaginosa, a Quorum Quenching Yeast Exhibiting Lactonase Activity Isolated from a Tropical Shoreline. Sensors. 2014; 14(4):6463-6473. https://doi.org/10.3390/s140406463
Chicago/Turabian StyleGhani, Norshazliza Ab, Joanita Sulaiman, Zahidah Ismail, Xin-Yue Chan, Wai-Fong Yin, and Kok-Gan Chan. 2014. "Rhodotorula Mucilaginosa, a Quorum Quenching Yeast Exhibiting Lactonase Activity Isolated from a Tropical Shoreline" Sensors 14, no. 4: 6463-6473. https://doi.org/10.3390/s140406463