Optimal Subset Selection of Time-Series MODIS Images and Sample Data Transfer with Random Forests for Supervised Classification Modelling
Abstract
:1. Introduction
2. Study Region and Datasets
2.1. Study Region
2.2. Earth Observation Data
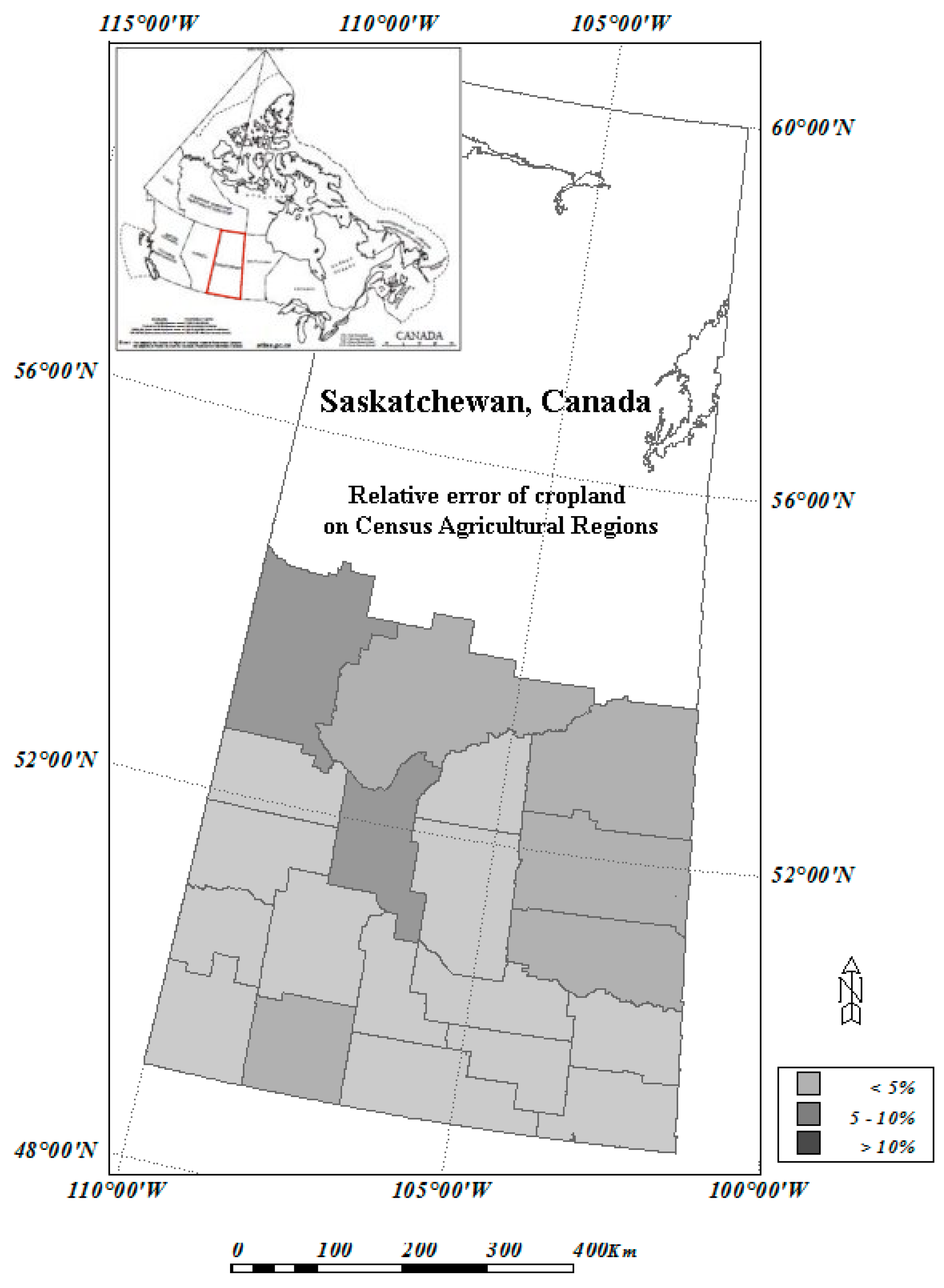
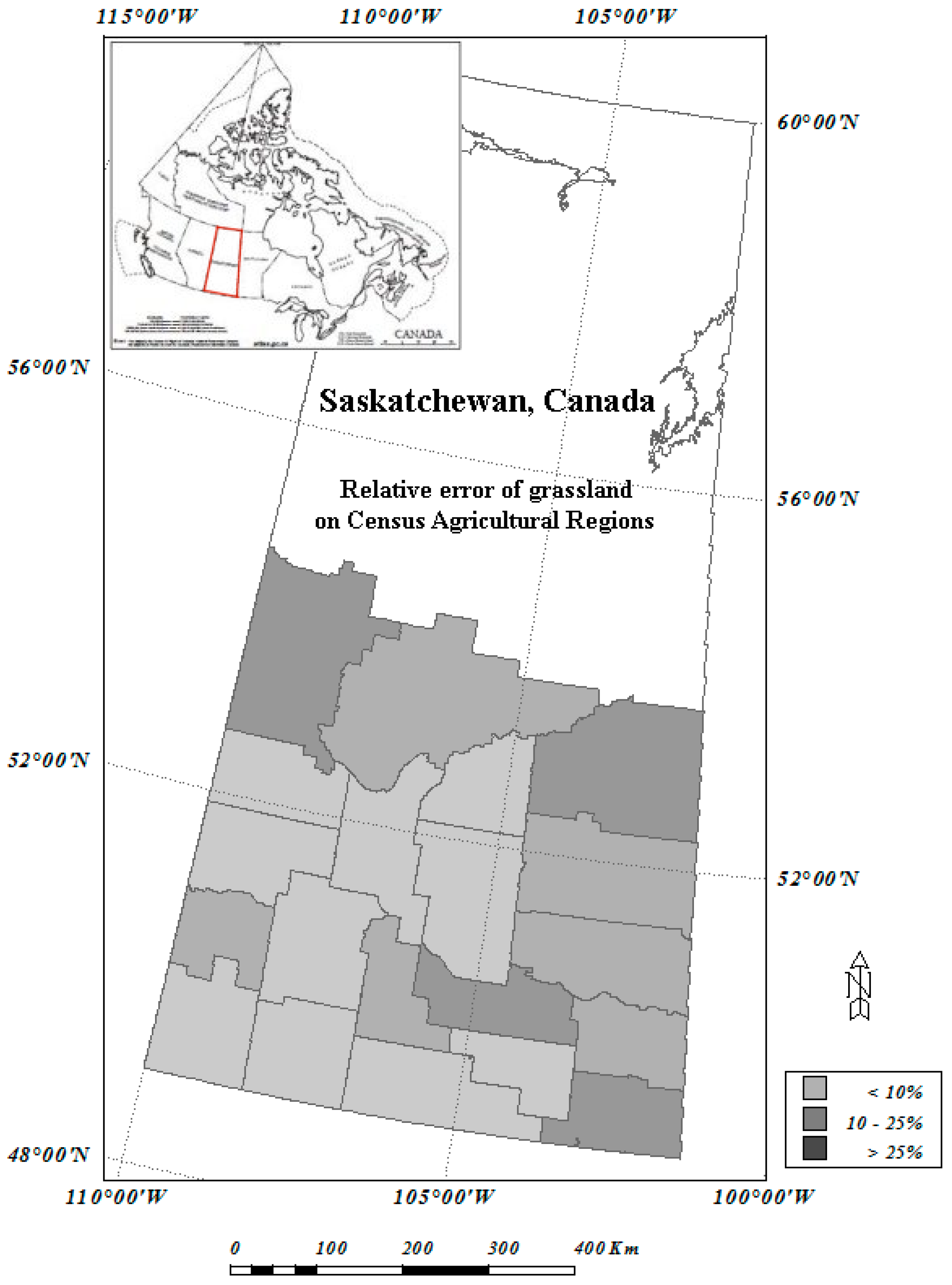
2.3. Land Cover for Agriculture Regions of Canada, Circa 2000
2.4. 2001 Census of Agriculture and Census Agricultural Region
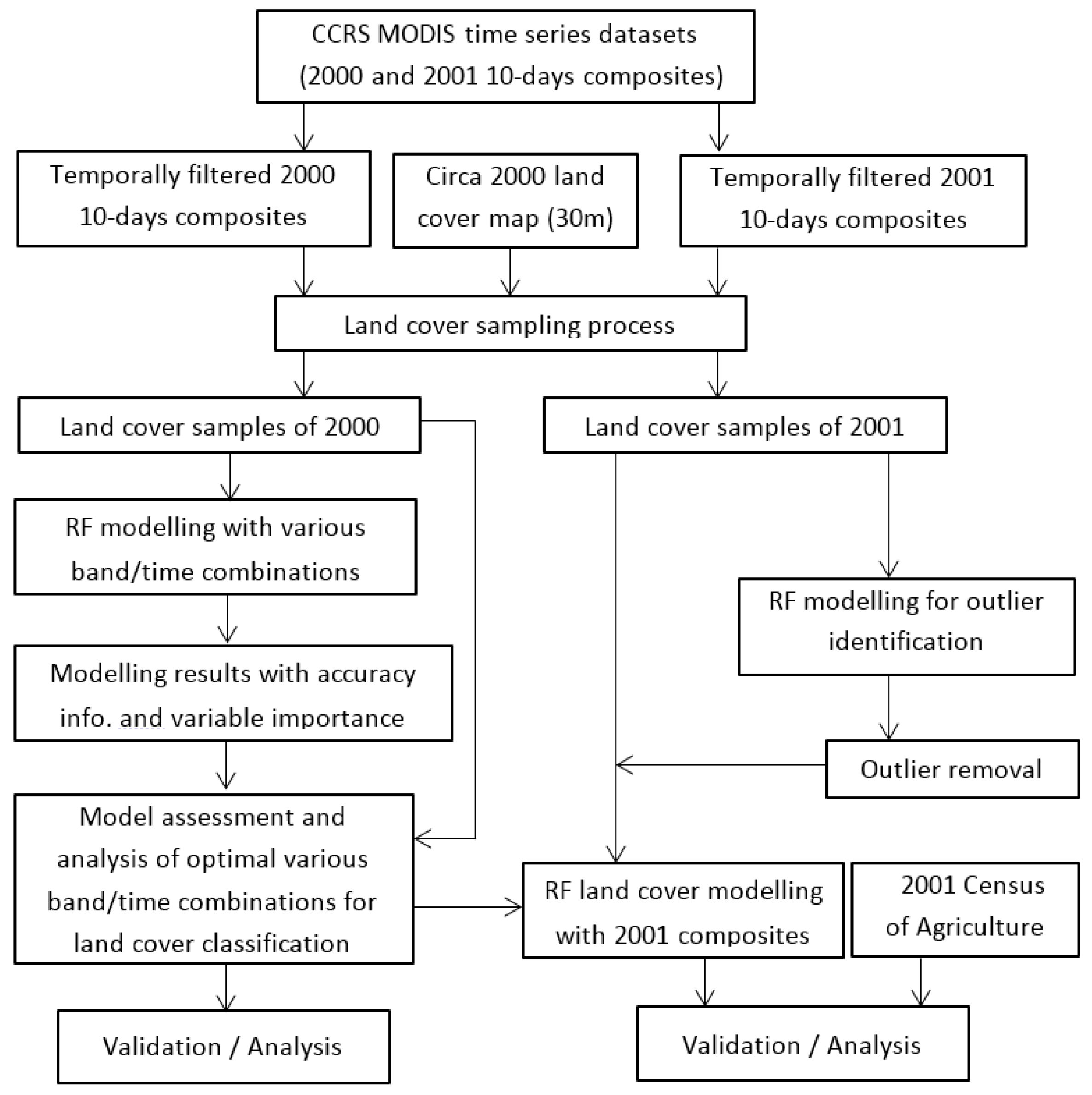
3. Methodology
3.1. Classification Model
3.2. Sampling Strategy
3.3. Sample Data Transferring
4. Results and Analysis
4.1. RF Modelling and Its Validation
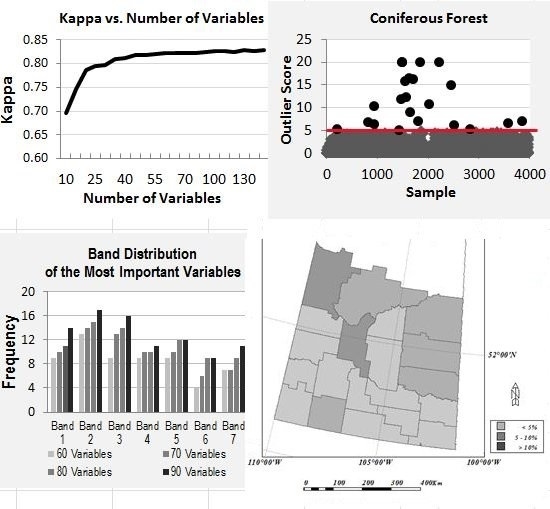
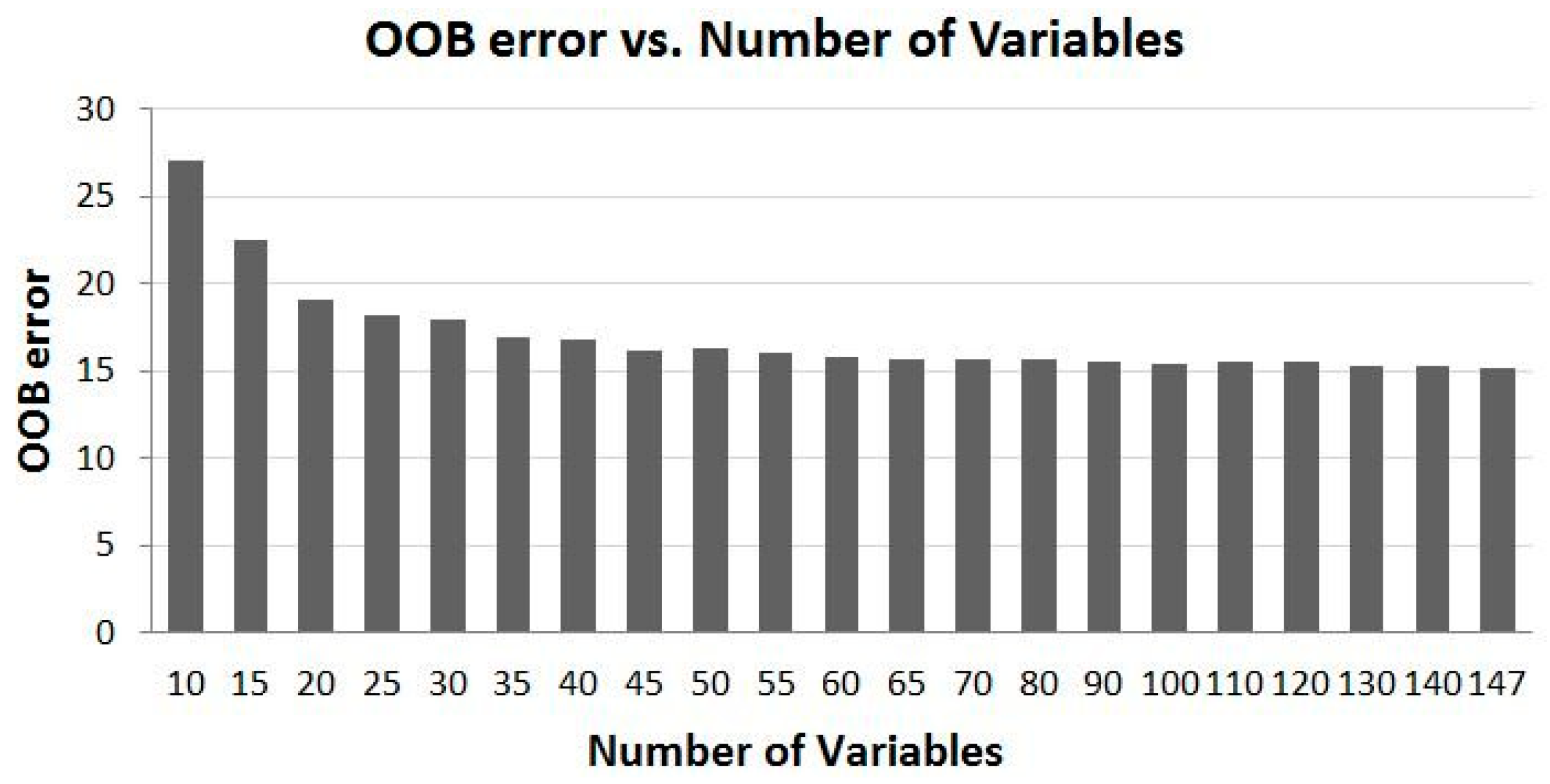
4.2. Overall Trend of Accuracy vs. Number of Variables
4.3. Temporal and Band Frequency Analysis
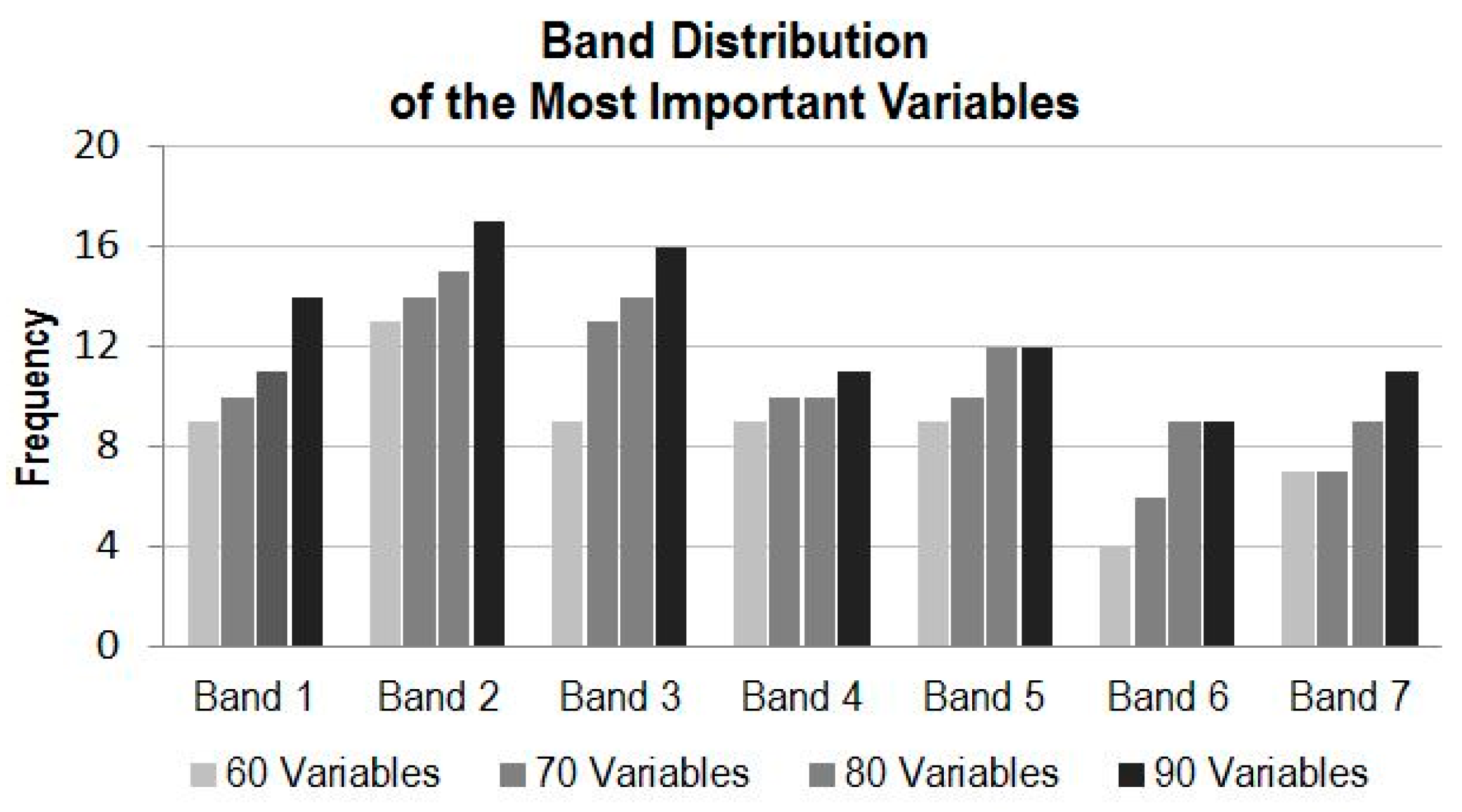
4.3.1. Variable Importance by Band
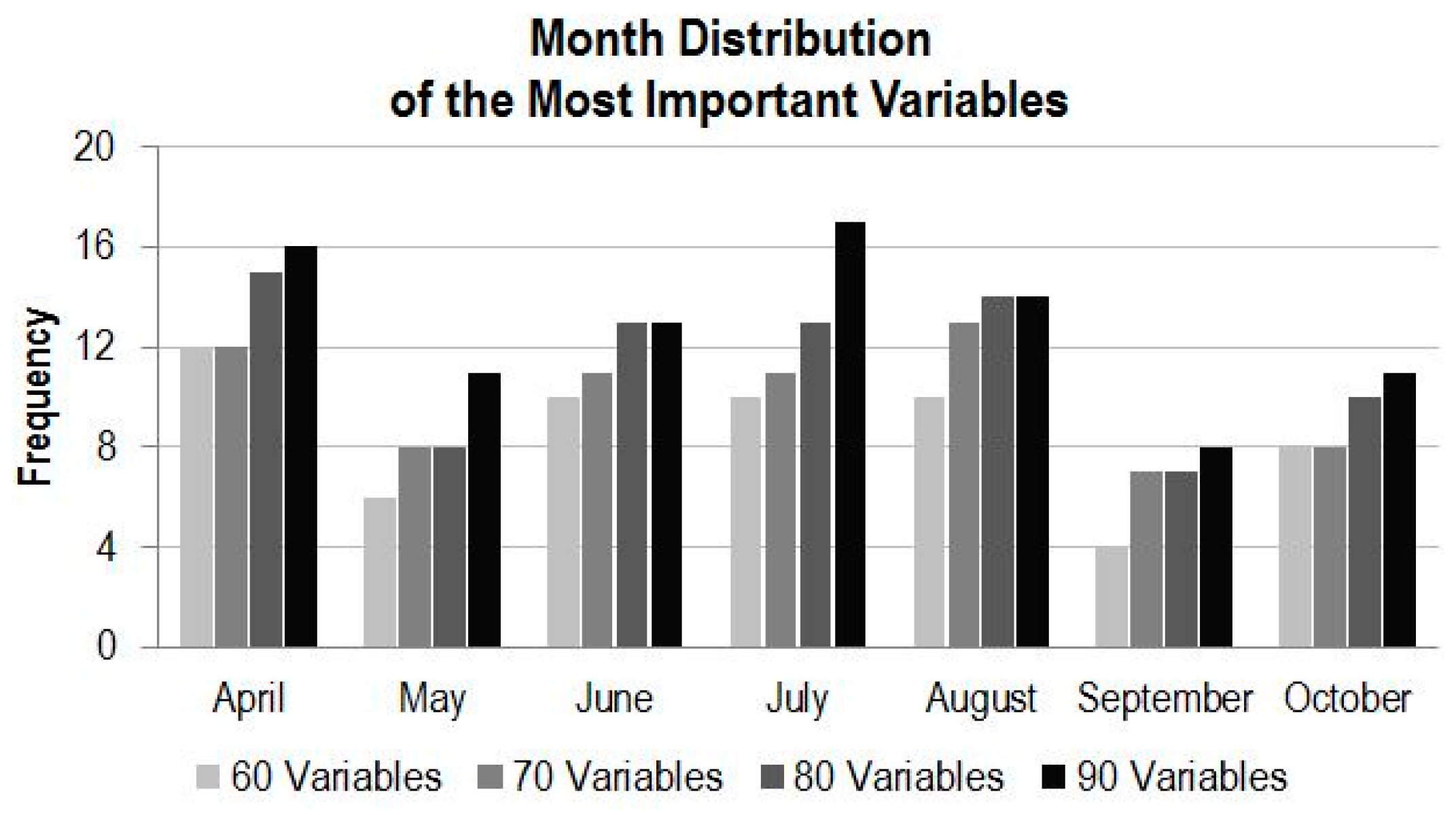
4.3.2. Variable Importance by Time
4.4. Experimental Runs of the Selected Subsets of CCRS MODIS Dataset
4.5. RF Modelling for 2001 MODIS Data
5. Discussion
5.1. Optimal Subset Data Selection
5.2. Knowledge Transfer for Supervised Modelling of Classification
6. Conclusions
- (a)
- For utilization of time-series MODIS data or other time-series EO datasets for applications such as land cover mapping, there are possible hundreds of variables and their combinations available. In general, more variables from a longer time period would produce more accurate results; however, there is a plateau in the accuracy of land cover classification after a certain number of variables are used. In this study, there are 147 variables in the dataset and using about half of the variables reaches the plateau (Kappa of ~0.83). Information produced from the study suggests that in the study region, MODIS composite images from the months of April, July and August, and bands of 1, 2 and 3 contribute the most to the accuracy of land cover classification. In addition, based on the results of the experiment runs, it can be expected that using the early season (from April to August) of the time-series MODIS composites could generate a land cover map similar in accuracy as using the full dataset.
- (b)
- Random Forests modelling produces a variable importance score to measure the variable’s contribution to its modelling. The case study of land cover classification with time-series MODIS dataset demonstrates that the Random Forests’ variable importance could be utilized to efficiently select an optimal subset from the time-series EO data for improving computation efficiency while reaching the highest possible accuracy. This method could be beneficial to other time-series EO dataset analysis, for example, the coming time-series Sentinel 2–3 dataset.
- (c)
- Ground truth or samples are essential for supervised classification modelling. One common problem of supervised modelling of time-series EO data analysis is the lack of sample data for model training and validation. Knowledge or sample data transferring from one timeframe to another provides a way to tackle this critical issue. The proposed simple but effective method used in this study could be practical for applications where ground truth is lacking but is available at an adjacent timeframe with similar environmental conditions, for example in the case of historical dataset. The proposed method also has large economic benefit as field work for data sampling is usually costly. Random Forests’ outlier identification feature is found useful for achieving this goal. Studies focused on improving the understanding of the outlier characteristics of ground features for sample transfer for supervised land cover classification modelling should get more attention in the future.
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Cihlar, J. Land cover mapping of large areas from satellites: Status and research priorities. Int. J. Remote Sens. 2000, 21, 1093–1114. [Google Scholar] [CrossRef]
- Friedl, M.A.; McIver, D.K.; Hodges, J.C.F.; Zhang, X.Y.; Muchoney, D.; Strahler, A.H.; Woodcock, C.E.; Gopal, S.; Schneider, A.; Cooper, A.; et al. Global land cover mapping from MODIS: Algorithms and early results. Remote Sens. Environ. 2002, 83, 287–302. [Google Scholar] [CrossRef]
- Knight, J.F.; Lunetta, R.S.; Ediriwickrema, J.; Khorram, S. Regional scale land cover characterization using MODIS-NDVI 250 m multi-temporal imagery: A phenology-based approach. GISci. Remote Sens. 2006, 43, 1–23. [Google Scholar] [CrossRef]
- Wardlow, B.D.; Egbert, S.L. Large-area crop mapping using time-series MODIS 250 m NDVI data: An assessment for the U.S. Central Great Plains. Remote Sens. Environ. 2008, 112, 1096–1116. [Google Scholar] [CrossRef]
- Alcantara, C.; Kuemmerle, T.; Prishchepov, A.V.; Radeloff, V.C. Mapping abandoned agriculture with multi-temporal MODIS satellite data. Remote Sens. Environ. 2012, 124, 334–347. [Google Scholar] [CrossRef]
- Guindin-Garcia, N.; Gitelson, A.A.; Arkebauer, T.J.; Shanahan, J.; Weiss, A. An evaluation of MODIS 8- and 16-day composite products for monitoring maize green leaf area index. Agric. Forest Meteorol. 2012, 161, 15–25. [Google Scholar] [CrossRef]
- Xiao, Z.; Liang, S.; Wang, J.; Xie, D.; Song, J.; Fensholt, R. A framework for consistent estimation of leaf area index, fraction of absorbed photosynthetically active radiation, and surface albedo from MODIS time-series data. IEEE Trans. Geosci. Remote Sens. 2015, 53, 3178–3197. [Google Scholar] [CrossRef]
- Vermote, E.; Salleous, N.E.; Justice, C.O. Atmospheric correction of MODIS data in the visible to near infrared: First results. Remote Sens. Environ. 2002, 83, 97–111. [Google Scholar] [CrossRef]
- Didan, K.; Huete, A. MODIS Vegetation Index Product Series Collection 5 Changes Document. Available online: http://landweb.nascom.nasa.gov/QA WWW/forPage/MOD13 (accessed on 20 July 2015).
- Luo, Y.; Trishchenko, A.P.; Khlopenkov, K.V. Developing clear-sky, cloud and cloud shadow mask for producing clear-sky composites at 250-meter spatial resolution for the seven MODIS land bands over Canada and North America. Remote Sens. Environ. 2008, 112, 4167–4185. [Google Scholar] [CrossRef]
- Zhou, F.; Zhang, A.; Townley-Smith, L. A data mining approach for evaluation of optimal time-series of MODIS data for land cover mapping at a regional level. ISPRS J. Photogramm. Remote Sens. 2003, 84, 114–129. [Google Scholar] [CrossRef]
- Nitze, I.; Barrett, B.; Cawkwell, F. Temporal optimisation of image acquisition for land coverclassification with Random Forest and MODIS time-series. Int. J. Appl. Earth Obs. Geoinform. 2015, 34, 136–146. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Pal, M. Random forest classifier for remote sensing classification. Int. J. Remote Sens. 2005, 26, 217–222. [Google Scholar] [CrossRef]
- Gislason, P.O.; Benediktsson, J.A.; Sveinsson, J.R. Random Forests for land cover classification. Pattern Recognit. Lett. 2006, 27, 294–300. [Google Scholar] [CrossRef]
- Rodriguez-Galiano, V.F.; Chica-Olmo, M.; Abarca-Hernandez, F.; Atkinson, P.M.; Jeganathan, C. Random Forest classification of Mediterranean land cover using multi-seasonal imagery and multi-seasonal texture. Remote Sens. Environ. 2012, 121, 93–107. [Google Scholar] [CrossRef]
- Vintrou, E.; Sourmare, M.; Bernard, S.; Begue, A.; Baron, C.; Lo Seen, D. Mapping fragmented agricultural systems in the Sudano-Sahelian environements of Africa using Random Forest and ensemble metrics of coarse resolution MODIS imagery. Photogramm. Eng. Remote Sens. 2012, 78, 839–848. [Google Scholar] [CrossRef]
- Zhu, X. Land cover classification using moderate resolution satellite imagery and random forests with post-hoc smoothing. J. Spat. Sci. 2013, 58, 323–337. [Google Scholar] [CrossRef]
- Breiman, L.; Cutler, A. Random Forests Description and Manual. Available online: https://www.stat.berkeley.edu/~breiman/RandomForests/cc_home.htm (accessed on 10 June 2016).
- Saskatchewan (Province). Available online: http://www.thecanadianencyclopedia.ca/en/article/saskatchewan (accessed on 6 May 2016).
- Pouliot, D.; Latifovic, R.; Fernandes, R.; Olthof, I. Evaluation of annual forest disturbance monitoring using a static decision tree approach and 250 m MODIS data. Remote Sens. Environ. 2009, 113, 1749–1759. [Google Scholar] [CrossRef]
- Agriculture and Agri-food Canada (AAFC). ISO 19131 Land Cover for Agricultural Regions of Canada, Circa 2000—Data Product Specification; Revision A; AAFC: Ottawa, ON, USA, 2002; p. 13.
- Statistics Canada. Census of Agriculture. Available online: http://www23.statcan.gc.ca/imdb/p2SV.pl?Function=getSurvey&SDDS=3438 (accessed on 9 March 2016).
- Bruzzone, L.; Marconcini, M. Domain adaptation problems: A DASVM classification technique and a circular validation strategy. IEEE Trans. Pattern Anal. Mach. Intell. 2010, 32, 770–787. [Google Scholar] [CrossRef] [PubMed]
- Hwa, R. Supervised grammar induction using training data with limited constituent information. In Proceedings of the 37th Annual Meeting of the Association for Computational Linguistics, College Park, MD, USA, 20–26 June 1999; pp. 73–79.
- Gildea, D. Corpus variation and parser performance. In Proceedings of the 2001 Conference on Empirical Methods in Natural Language Processing (EMNLP), Pittsburgh, PA, USA, 3–4 June 2001; 2001. [Google Scholar]
- Hong, G.; Zhang, A.; Zhou, F.; Townley-Smith, L.; Brisco, B.; Olthof, I. Crop-type identification potential of Radarsat-2 and MODIS images for the Canadian prairies. Can. J. Remote Sens. 2011, 37, 45–54. [Google Scholar] [CrossRef]
- Yin, H.; Pflugmacher, D.; Kennedy, R.E.; Sulla-Menashe, D.; Hostert, P. Mapping annual land use and land cover changes using MODIS time series. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 3421–3427. [Google Scholar] [CrossRef]
- Rulequest Research. Data Mining Tools See5 and C5.0. Available online: https://www.rulequest.com/see5-info.html (accessed on 6 June 2016).


| Subset Data | Number of Months | Number of Bands | Number of Variables | Kappa | OOB Error (%) |
|---|---|---|---|---|---|
| 1 | 4 | 6 | 72 | 0.816 | 16.344 |
| 2 | 4 | 7 | 84 | 0.817 | 16.268 |
| 3 | 5 | 5 | 75 | 0.817 | 16.285 |
| 4 | 5 | 6 | 90 | 0.821 | 15.936 |
| 5 | 5 | 7 | 105 | 0.819 | 16.118 |
| 6 | 6 | 4 | 72 | 0.822 | 15.825 |
| 7 | 6 | 5 | 90 | 0.823 | 15.704 |
| 8 | 6 | 6 | 108 | 0.825 | 15.537 |
| 9 | 4 (April, May, June, July) | 7 | 84 | 0.814 | 16.525 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, F.; Zhang, A. Optimal Subset Selection of Time-Series MODIS Images and Sample Data Transfer with Random Forests for Supervised Classification Modelling. Sensors 2016, 16, 1783. https://doi.org/10.3390/s16111783
Zhou F, Zhang A. Optimal Subset Selection of Time-Series MODIS Images and Sample Data Transfer with Random Forests for Supervised Classification Modelling. Sensors. 2016; 16(11):1783. https://doi.org/10.3390/s16111783
Chicago/Turabian StyleZhou, Fuqun, and Aining Zhang. 2016. "Optimal Subset Selection of Time-Series MODIS Images and Sample Data Transfer with Random Forests for Supervised Classification Modelling" Sensors 16, no. 11: 1783. https://doi.org/10.3390/s16111783