Mobile-Based Analysis of Malaria-Infected Thin Blood Smears: Automated Species and Life Cycle Stage Determination
Abstract
:1. Introduction
2. Malaria Disease Characterization
3. Related Work
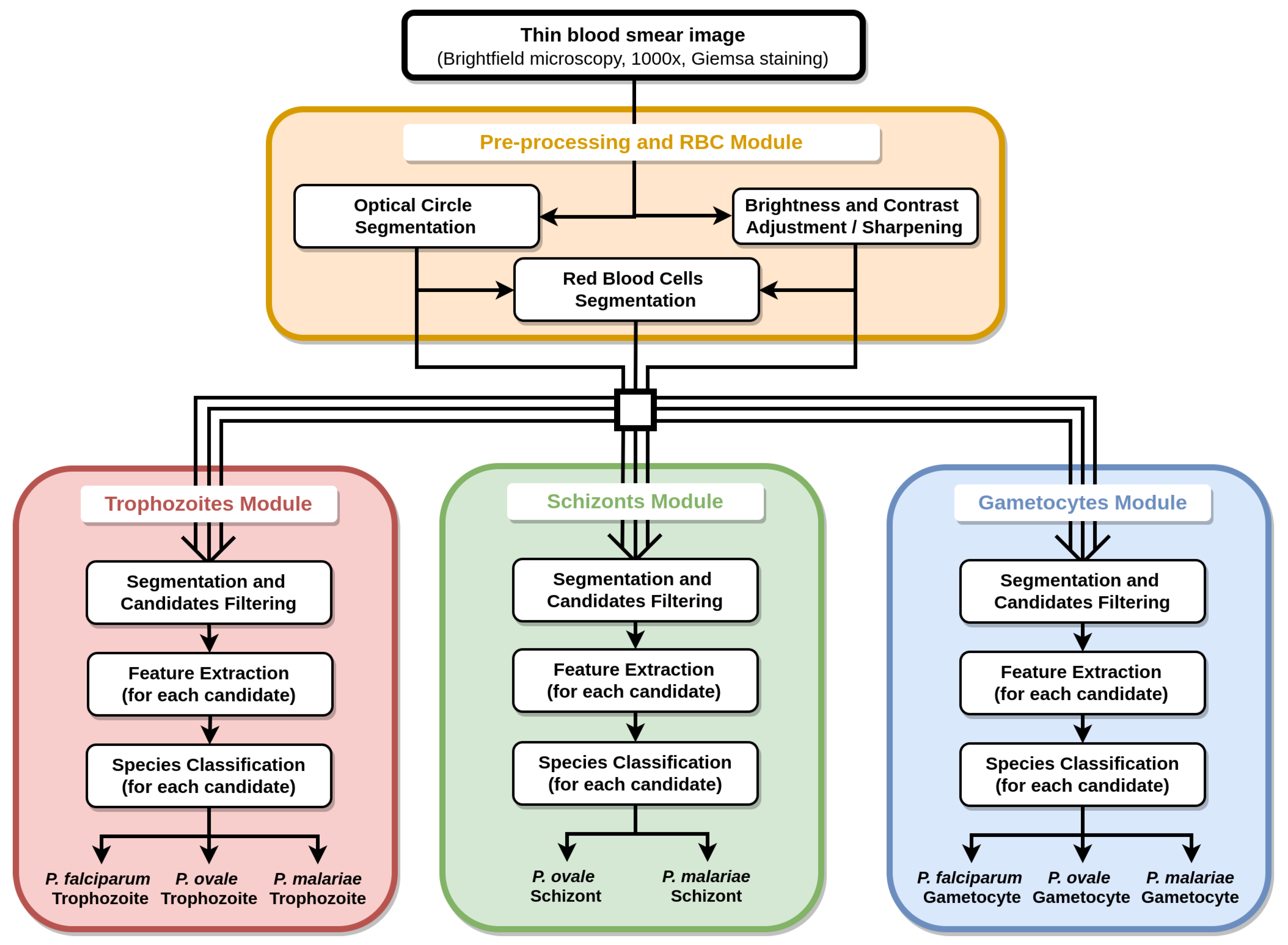
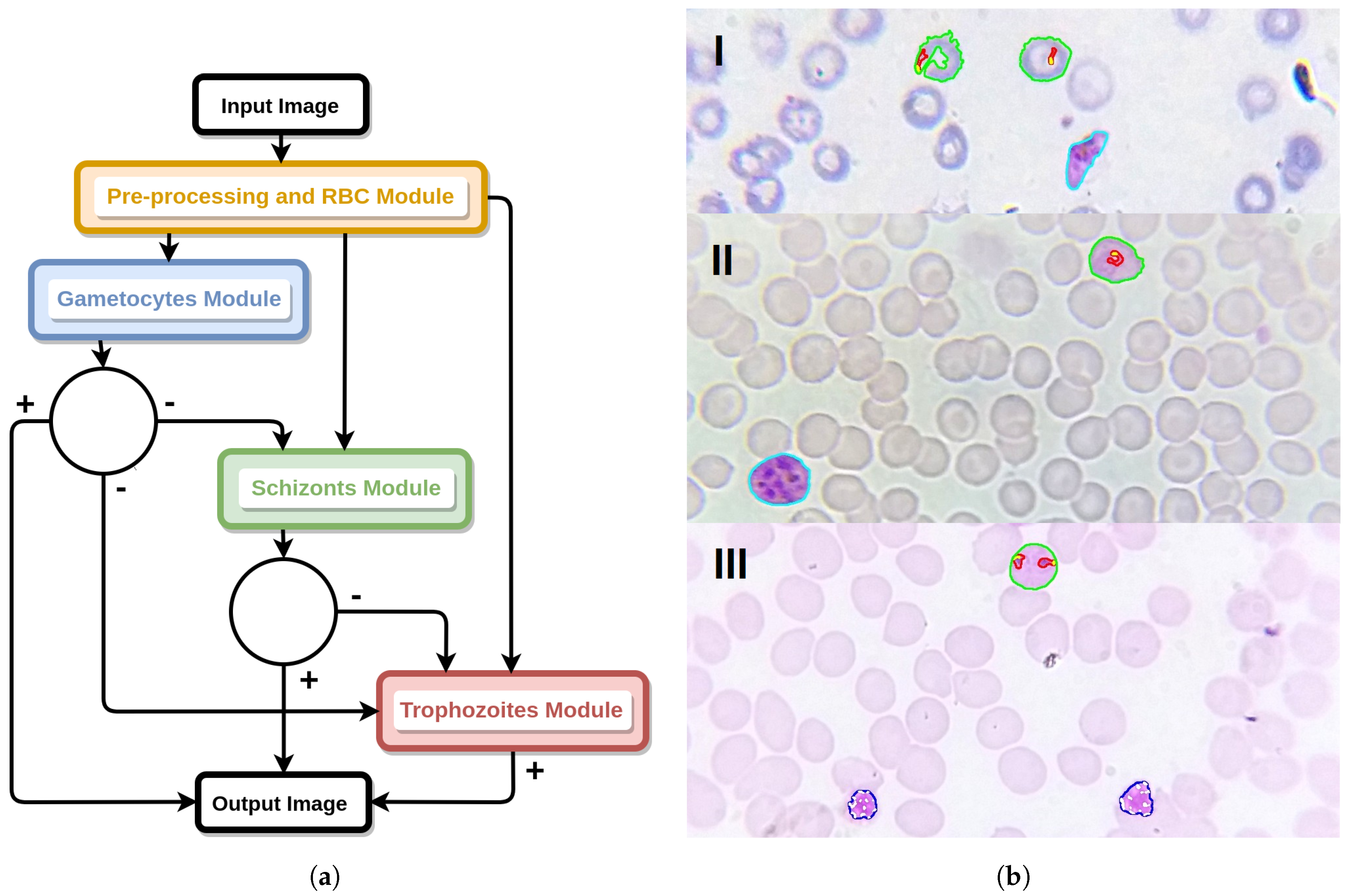
4. Mobile-Based Framework for Malaria Parasites Detection: An Overview
- I
- SmartScope: an inexpensive alternative to the current microscopes that can easily be adapted to a smartphone and used in the field. This gadget guarantees the required 1000× magnification, and the smartphone camera is used to capture images. Moreover, it uses a self-powered motorized automated stage system, in order to move the blood smear and allow the automatic capture of several snapshots of the sample [15];
- II
- Image processing and analysis: consisting of the automated detection of MPs via computer vision and machine learning approaches, on microscopic blood smear images acquired using I. This component consists of two main modules, as detailed in Section 2:
- (a)
- Thick smear module to detect the presence of MPs on thick blood smears [16].
- (b)
- Thin smear module for the determination of MPs species and life cycle stage (main focus of this article).
- III
- Smartphone application: envisioned to be used by technical personnel without specialized knowledge in malaria diagnosis. The user collects and prepares a blood sample of the patient, introducing it in a slot of I. Using the companion mobile application, installed in the smartphone that is coupled to I, the user can take pictures of the blood smear using the smartphone’s camera, being subsequently analyzed by II, so the correct procedures and medication can be administered.
5. Methodology
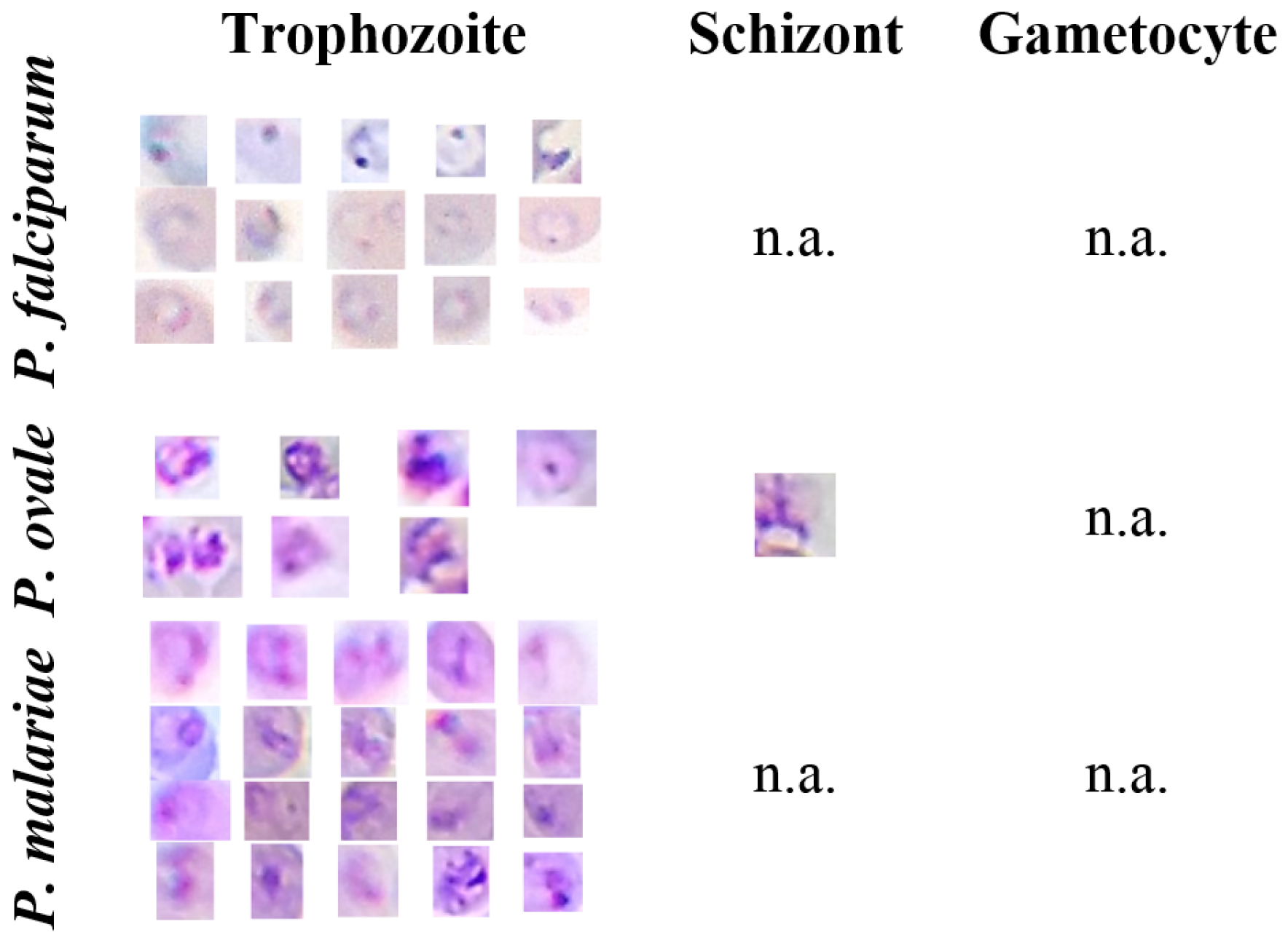
5.1. mThinMPs Database
5.2. Pre-Processing
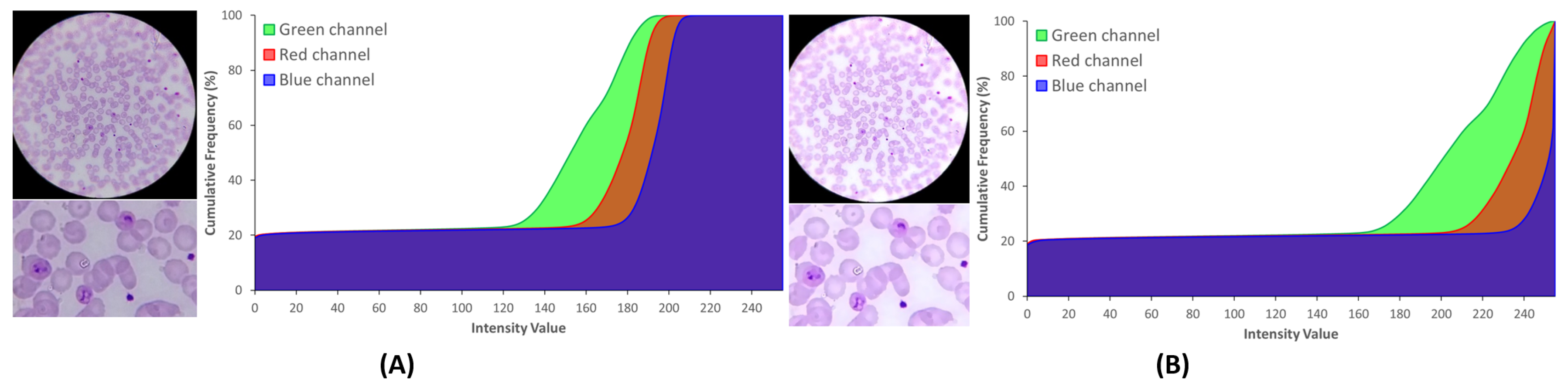
5.2.1. Brightness and Contrast Adjustment
5.2.2. Sharpening
5.3. Segmentation and Filtering
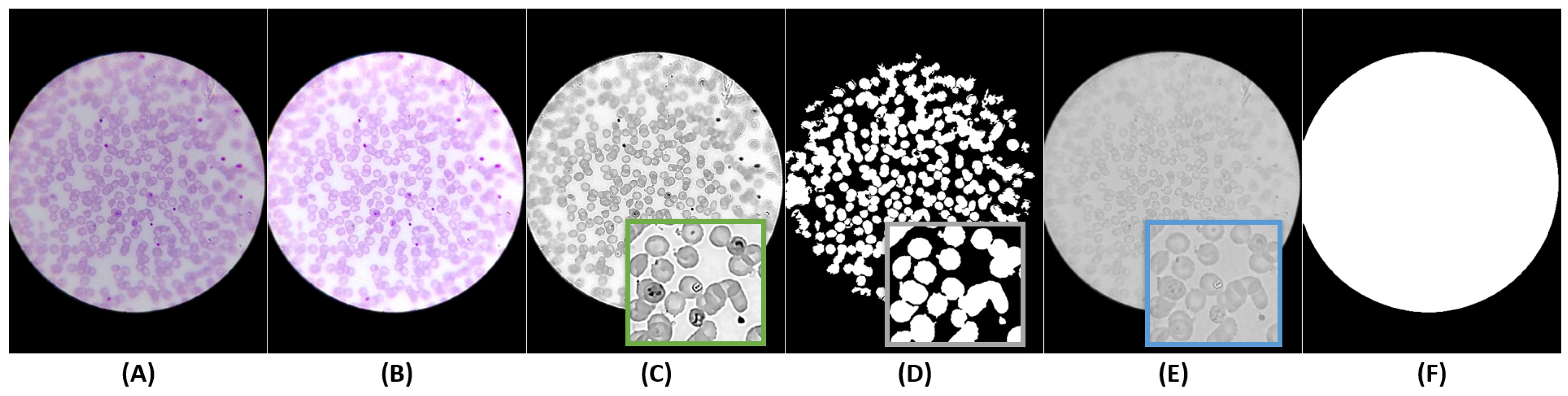
5.3.1. Optical Circle Segmentation
5.3.2. RBCs Segmentation
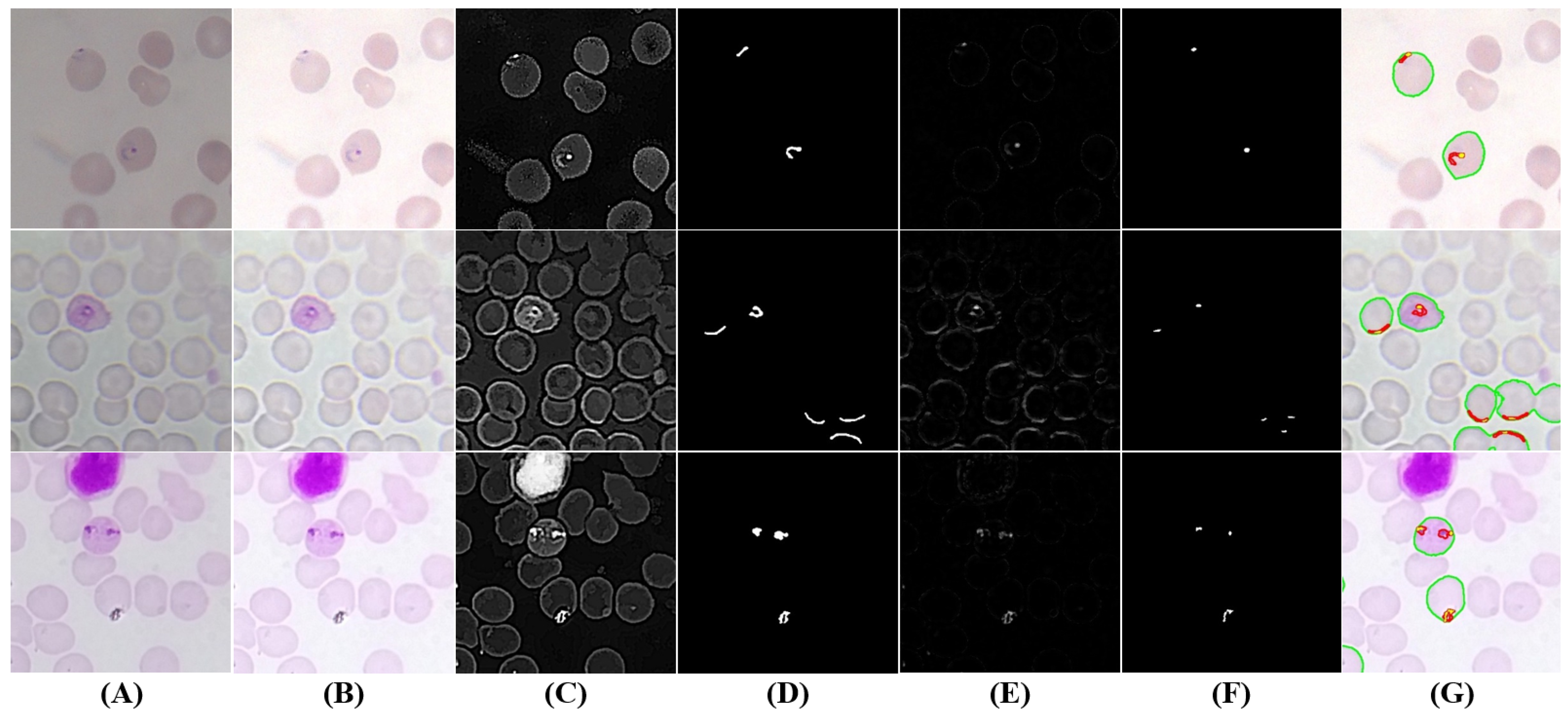
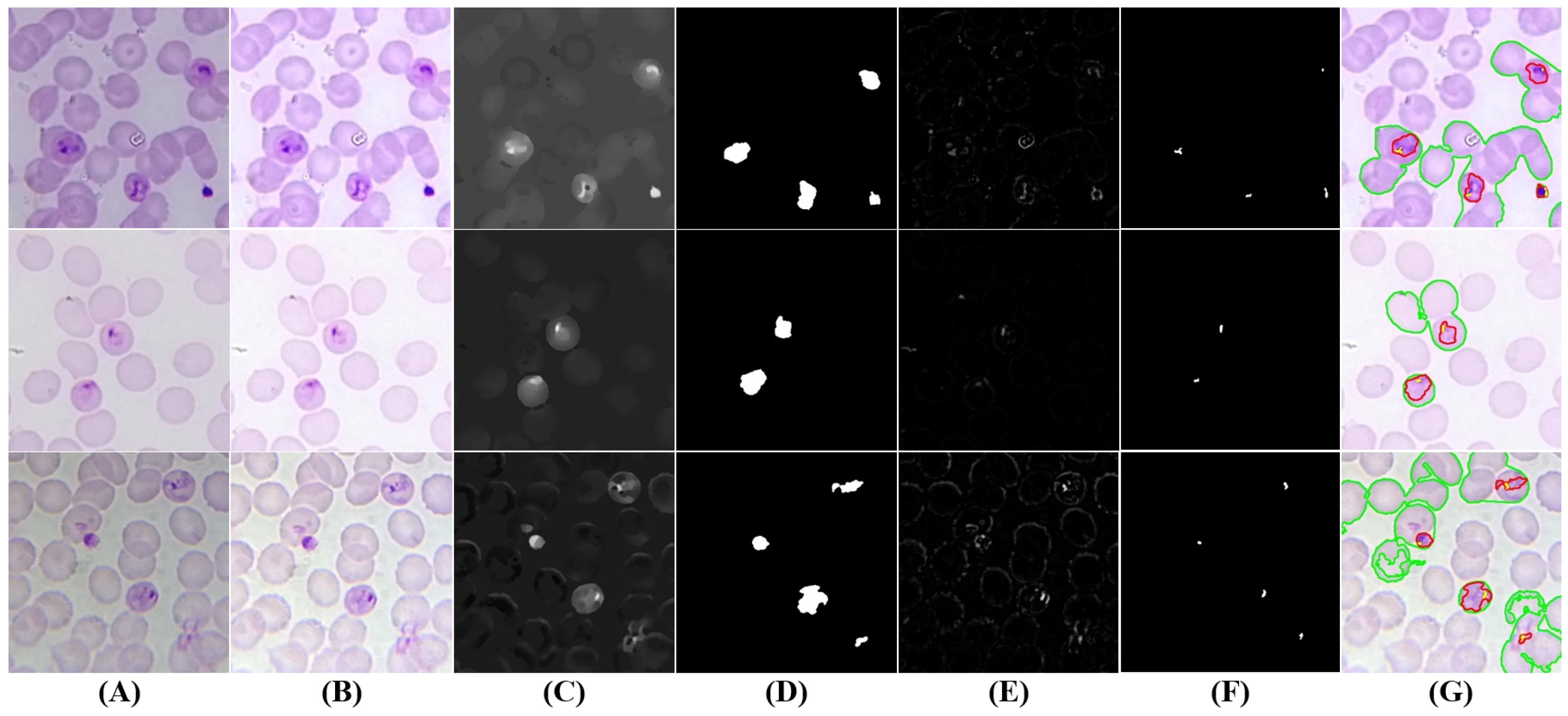
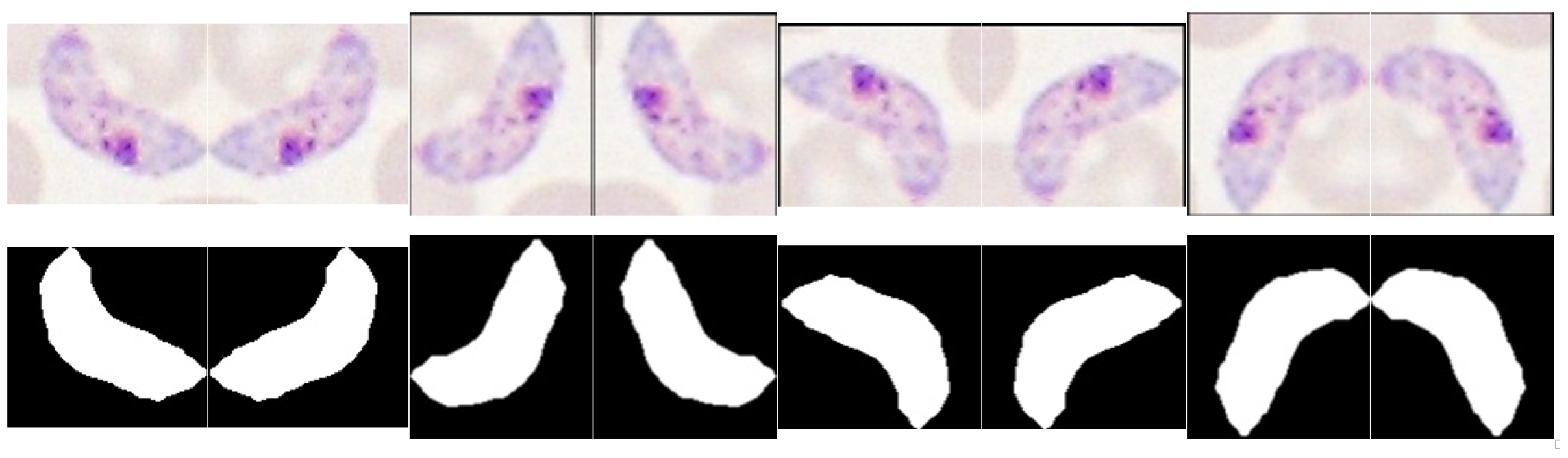
5.3.3. Trophozoites’ Segmentation
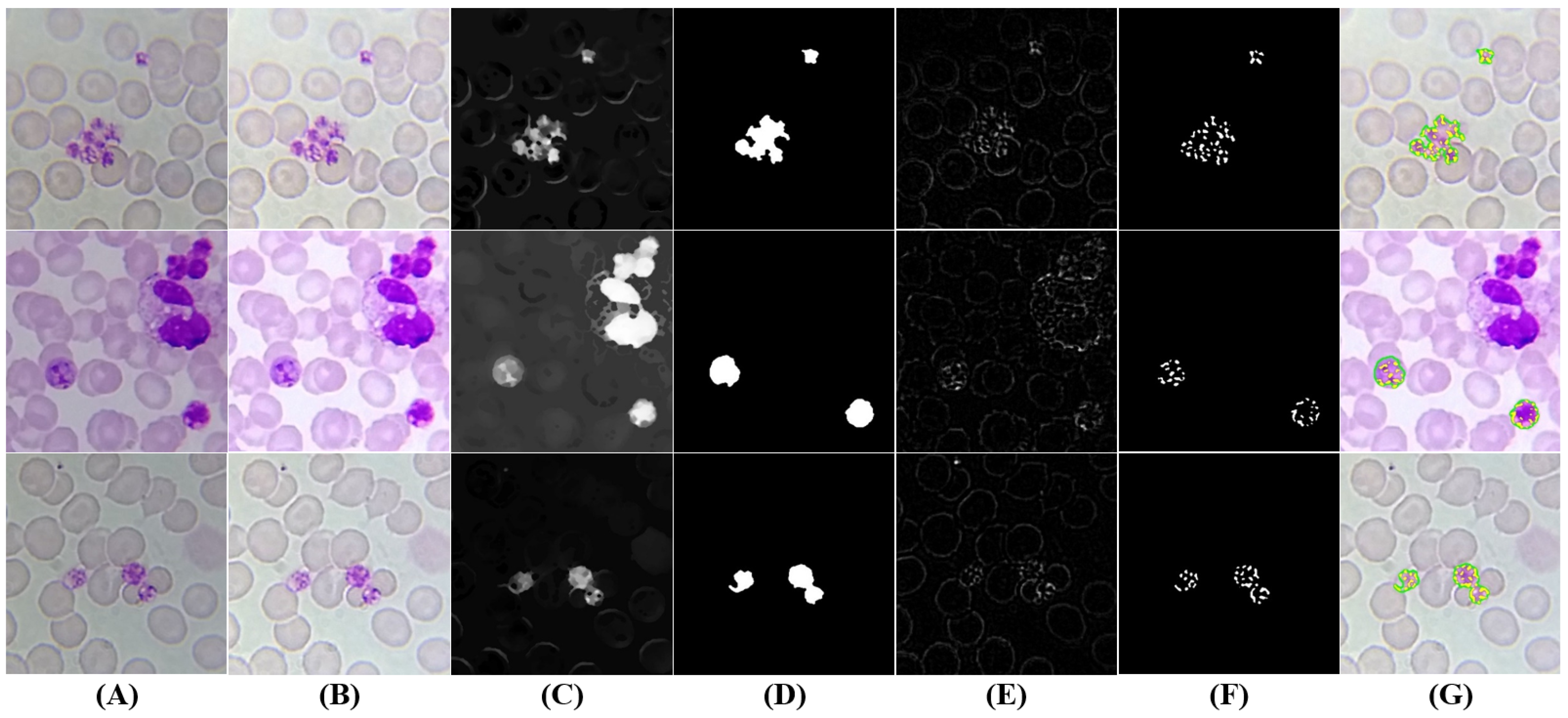
5.3.4. Schizonts’ Segmentation
5.3.5. Gametocytes Segmentation
5.4. Feature Extraction
5.4.1. Trophozoites’ Features
5.4.2. Schizonts’ Features
5.4.3. Gametocytes Features
5.5. Classification
5.5.1. Data Augmentation
5.5.2. SVM Hyperparameters Selection
- All of the features were normalized between .
- Defining the search interval for each hyperparameter, namely and .
- Defining the maximum number of rounds for the individual optimization of each hyperparameter (), namely . In each , the target hyperparameter is optimized, while the other parameter remains with a fixed value. The process starts with the optimization and fixed .
- Defining the maximum number of rounds for the overall optimization of both hyperparameters (), namely . It should be noted that each corresponds to the individual optimization of each , as described in the previous step.
- For each , a hold out strategy is used by randomly splitting the dataset into training and test sets. A total of 20 hold outs was applied for each tested combination, with 90% of the observations of both classes on the training set and the remaining used for testing purposes.
- The best hyperparameters combination is selected according to the performance metrics criteria described in Algorithm 1. This new proposed criteria aims to ensure that we select a classification model that has a balanced performance between the detection of both classes, even in data imbalance contexts. Particularly, this criteria allow the selection of a that implies a decrease in one of the two considered metrics (when compared with the found so far), but only if there is a performance improvement of the second metric that is twice greater than the performance loss suffered by the first.
- At the end of each and while is not achieved, the search interval is updated. Particularly, the search interval is updated to , being the main goal of this step the fine tuning of .
- A total number of 50 values of for each search interval was tested. Due to the large value range and possible different orders of magnitude, the logarithmic scale was used to select equally-spaced values on the search interval. Considering the search interval , the value on the i-th position is given by:where is the maximum number of considered values for under the considered search interval, with .
| Algorithm 1: Performance metrics criteria. |
 |
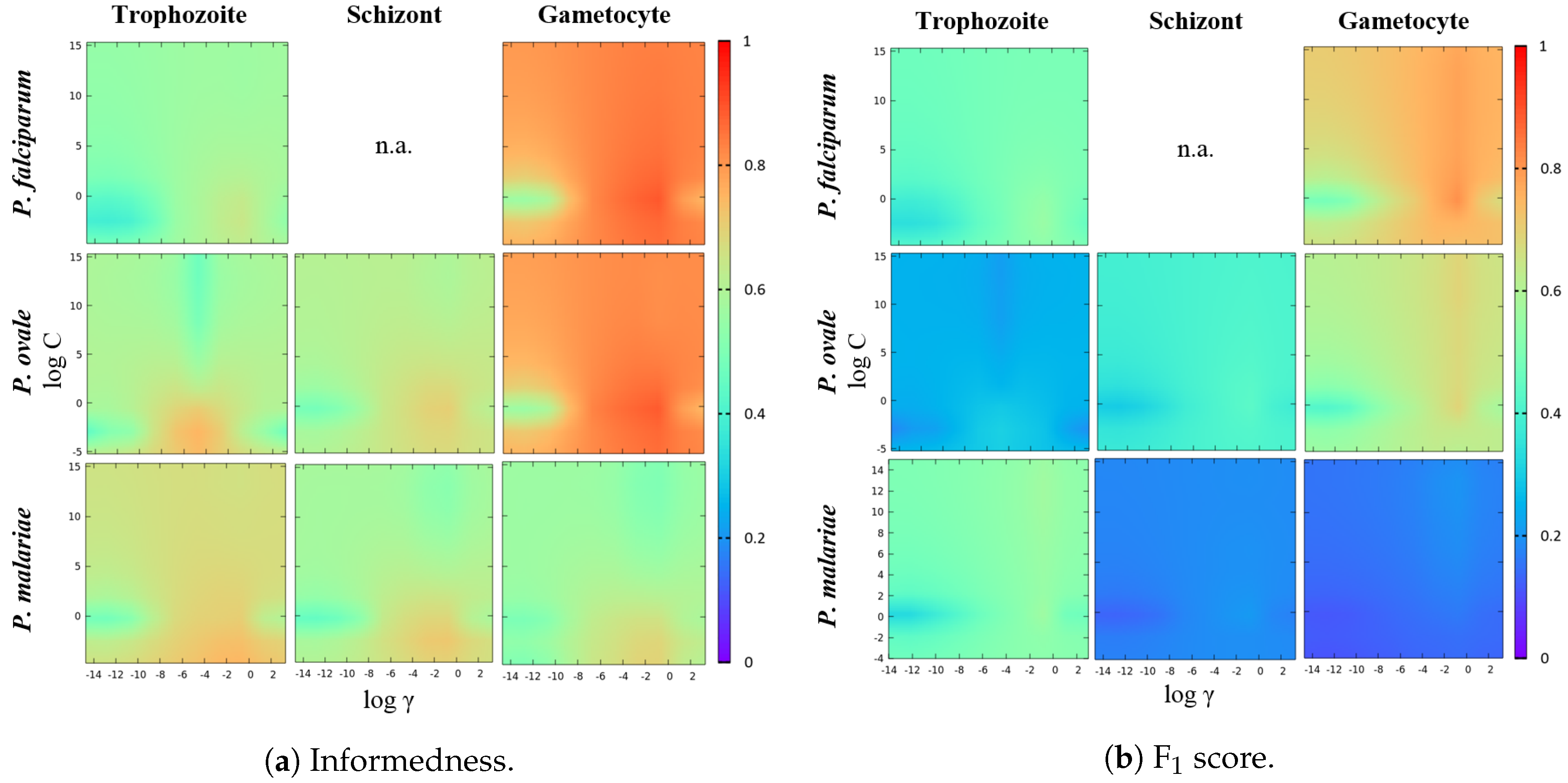
6. Results and Discussion
Classification Models’ Workflow
7. Conclusions and Future Work
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| MPs | Malaria parasites |
| RBCs | Red blood cells |
| RDTs | Rapid diagnostic tests |
| SE | Sensitivity |
| SP | Specificity |
| AC | Accuracy |
| INF | Informedness |
| F1 | F1 score |
| ROI | Region of interest |
| SVM | Support vector machines |
| kNN | K-nearest neighbors |
| RGB | Red, green, blue |
| CIE | Commission International de l’Élairage |
| L*a*b* | Lightness, green/red coordinate, blue/yellow coordinate |
| L*C*h | Lightness, chroma, hue |
| HSI | Hue, saturation, intensity |
References
- World Health Organization. World Malaria Report 2016; WHO: Geneva, Switzerland, 2016. [Google Scholar]
- World Health Organization. World Malaria Report 2015; WHO: Geneva, Switzerland, 2015. [Google Scholar]
- Blycroft Limited. Africa & Middle East Mobile Factbook 2Q 2014; Blycroft Publishing: Aylesbury, UK, 2014. [Google Scholar]
- Dolgin, E. Portable pathology for Africa. IEEE Spectr. 2015, 52, 37–39. [Google Scholar] [CrossRef]
- Rosado, L.; Correia da Costa, J.M.; Elias, D.; Cardoso, J.S. A Review of Automatic Malaria Parasites Detection and Segmentation in Microscopic Images. Anti-Infect. Agents 2016, 14, 11–22. [Google Scholar] [CrossRef]
- World Health Organization. Basic Malaria Microscopy: Part II. Tutor’s guide, 2nd ed.; WHO: Geneva, Switzerland, 2010. [Google Scholar]
- Centers for Disease Control and Prevention. Malaria Diagnosis (U.S.)—Rapid Diagnostic Test; CDC: Atlanta, GA, USA, 2017.
- Zou, L.H.; Chen, J.; Zhang, J.; Garcia, N. Malaria Cell Counting Diagnosis within Large Field of View. In Proceedings of the 2010 International Conference on Digital Image Computing: Techniques and Applications (DICTA), Sydney, Australia, 2–3 December 2010; pp. 172–177. [Google Scholar]
- Anggraini, D.; Nugroho, A.; Pratama, C.; Rozi, I.; Pragesjvara, V.; Gunawan, M. Automated status identification of microscopic images obtained from malaria thin blood smears using bayes decision: A study case in plasmodium falciparum. In Proceedings of the 2011 International Conference on Advanced Computer Science and Information System (ICACSIS), Jakarta, Indonesia, 17–18 December 2011; pp. 347–352. [Google Scholar]
- Malihi, L.; Ansari-Asl, K.; Behbahani, A. Malaria parasite detection in giemsa-stained blood cell images. In Proceedings of the 2013 8th Iranian Conference on Machine Vision and Image Processing (MVIP), Zanjan, Iran, 10–12 September 2013; pp. 360–365. [Google Scholar]
- Khan, N.; Pervaz, H.; Latif, A.; Musharraf, A.; Saniya. Unsupervised identification of malaria parasites using computer vision. In Proceedings of the 11th International Joint Conference on Computer Science and Software Engineering (JCSSE), Chon Buri, Thailand, 14–16 May 2014; pp. 263–267. [Google Scholar]
- Nugroho, H.A.; Akbar, S.A.; Murhandarwati, E.E.H. Feature extraction and classification for detection malaria parasites in thin blood smear. In Proceedings of the 2015 2nd International Conference on Information Technology, Computer, and Electrical Engineering (ICITACEE), Semarang, Indonesia, 16–18 October 2015; pp. 197–201. [Google Scholar]
- Nanoti, A.; Jain, S.; Gupta, C.; Vyas, G. Detection of malaria parasite species and life cycle stages using microscopic images of thin blood smear. In Proceedings of the 2016 International Conference on Inventive Computation Technologies (ICICT), Coimbatore, India, 26–27 August 2016; Volume 1, pp. 1–6. [Google Scholar]
- Yang, D.; Subramanian, G.; Duan, J.; Gao, S.; Bai, L.; Chandramohanadas, R.; Ai, Y. A portable image-based cytometer for rapid malaria detection and quantification. PLoS ONE 2017, 12, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Rosado, L.; Oliveira, J.; Vasconcelos, M.J.M.; da Costa, J.M.C.; Elias, D.; Cardoso, J.S. μSmartScope: 3D-printed Smartphone Microscope with Motorized Automated Stage. In Proceedings of the 10th International Joint Conference on Biomedical Engineering Systems and Technologies, Porto, Portugal, 21–23 February 2017; pp. 38–48. [Google Scholar]
- Rosado, L.; Costa, J.M.C.D.; Elias, D.; Cardoso, J.S. Automated Detection of Malaria Parasites on Thick Blood Smears via Mobile Devices. Proc. Comput. Sci. 2016, 90, 138–144. [Google Scholar] [CrossRef]
- Fang, Y.; Xiong, W.; Lin, W.; Chen, Z. Unsupervised malaria parasite detection based on phase spectrum. In Proceedings of the 2011 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Boston, MA, USA, 20 August–3 September 2011; pp. 7997–8000. [Google Scholar]
- Wang, L.; Liu, G.; Dai, Q. Optimization of Segmentation Algorithms Through Mean-Shift Filtering Preprocessing. IEEE Geosci. Remote Sens. Lett. 2014, 11, 622–626. [Google Scholar] [CrossRef]
- Crutcher, J.M.; Hoffman, S.L. Malaria. In Medical Microbiology, 4th ed.; Baron, S., Ed.; University of Texas Medical Branch at Galveston: Galveston, TX, USA, 1996. [Google Scholar]
- Bradski, G. The OpenCV Library. Dr. Dobb’s J. Softw. Tools 2000, 25, 120–123. [Google Scholar]




| Trophozoites | Schizonts | Gametocytes | |
|---|---|---|---|
| P. falciparum | 585 | n.a. | 58 |
| P. ovale | 122 | 80 | 62 |
| P. malariae | 164 | 27 | 29 |
| Length (m) | Approximate Ratio | |
|---|---|---|
| 215 | - | |
| RBCs | 7~8 | ~ |
| Trophozoites | 1~7 | ~ |
| Schizonts | 5~10 | ~ |
| Gametocytes | 7~14 | ~ |
| Group | Family | Channels | Features |
|---|---|---|---|
| Geometry | Binary | Maximum Diameter a, Minimum Diameter a, Perimeter a, Eccentricity, Convex Hull Area a, Area a, Elongation Bounding Box Area a, Solidity, Extent, Circularity, Elliptical Symmetry, Principal Axis Ratio, Radial Variance, Asymmetry Indexes/ Ratios, Compactness Index, Irregularity Indexes, Bounding Box Ratio, Lengthening Index, Equivalent Diameter a, Asymmetry Celebi. | |
| Color | C* and h (from L*C*h) | Mean b, Standard Deviation b, L1 Norm b, L2 Norm b Entropy b, Energy b, Skewness b, Kurtosis. b | |
| Discrete Fourier Transform | Grayscale | Mean, Standard Deviation, Maximum, Minimum. | |
| Texture | Gray Level Run Length Matrix | Grayscale | Short run emphasis c, long run emphasis c, run percentage c, long run high grey level emphasis c, low grey level runs emphasis c, high grey level runs emphasis c, short run low grey level emphasis c, short run high grey level emphasis c, grey level non-uniformity c, long run low grey level emphasis c. |
| Gray Level Co-occurrence Matrix | R, G, B (from RGB) | Energy c, Entropy c, Contrast c, Correlation c, Maximum probability c, Dissimilarity c, Homogeneity c. | |
| Laplacian | Grayscale | Mean, Standard deviation, Maximum, Minimum. |
| True Positives | False Positives | False Negatives | |
|---|---|---|---|
| Trophozoites | 811 | 13,701 | 60 |
| Schizonts | 106 | 5733 | 1 |
| Gametocytes | 149 | 4190 | 0 |
| SVM Parameters | Sensitivity | Specificity | Informedness | F1 Score | Accuracy | |
|---|---|---|---|---|---|---|
| P. falciparum Trophozoites | = C = | 73.9% | 97.0% | 70.9% | 60.0% | 96.1% |
| P. falciparum Gametocytes | = C = 1 | 94.8% | 99.3% | 94.1% | 87.4% | 99.2% |
| P. ovale Trophozoites | = C = | 84.6% | 97.0% | 81.6% | 34.8% | 96.9% |
| P. ovale Schizonts | = C = 1.55 | 82.7% | 97.9% | 80.6% | 52.6% | 97.7% |
| P. ovale Gametocytes | = C = 1 | 96.2% | 99.0% | 95.2% | 77.1% | 99.0% |
| P. malariae Trophozoites | = C = 1 | 82.0% | 99.1% | 81.1% | 63.5% | 98.9% |
| P. malariae Schizonts | = C = 1 | 87.8% | 96.5% | 84.3% | 25.9% | 96.5% |
| P. malariae Gametocytes | = C = | 94.9% | 92.6% | 87.5% | 18.8% | 92.6% |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rosado, L.; Da Costa, J.M.C.; Elias, D.; Cardoso, J.S. Mobile-Based Analysis of Malaria-Infected Thin Blood Smears: Automated Species and Life Cycle Stage Determination. Sensors 2017, 17, 2167. https://doi.org/10.3390/s17102167
Rosado L, Da Costa JMC, Elias D, Cardoso JS. Mobile-Based Analysis of Malaria-Infected Thin Blood Smears: Automated Species and Life Cycle Stage Determination. Sensors. 2017; 17(10):2167. https://doi.org/10.3390/s17102167
Chicago/Turabian StyleRosado, Luís, José M. Correia Da Costa, Dirk Elias, and Jaime S. Cardoso. 2017. "Mobile-Based Analysis of Malaria-Infected Thin Blood Smears: Automated Species and Life Cycle Stage Determination" Sensors 17, no. 10: 2167. https://doi.org/10.3390/s17102167






