High Sensitivity Determination of TNF-α for Early Diagnosis of Neonatal Infections with a Novel and Reusable Electrochemical Sensor
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents and Materials
2.2. Instrumentation
2.3. Preparation of Gold Electrode
2.4. Preparation of the AuNPs
2.5. Preparation of the Signal Molecule
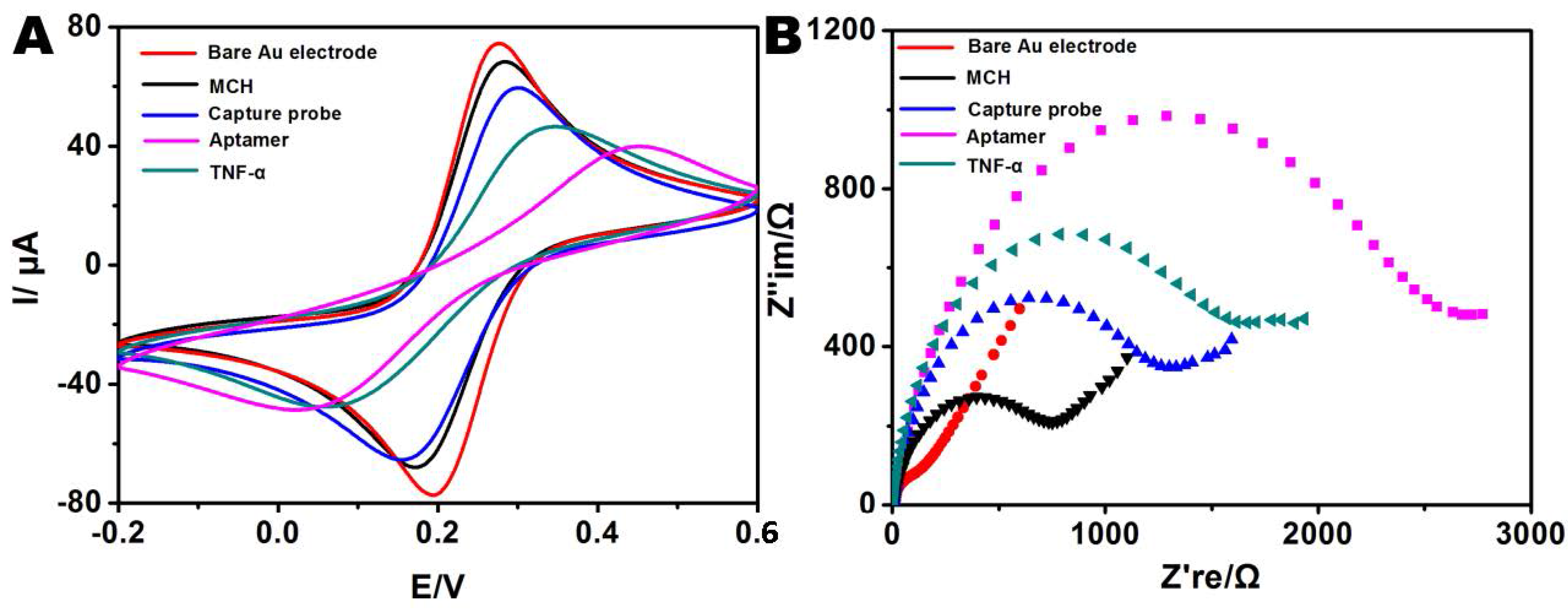
2.6. Fabrication of the TNF-α Sensor
2.7. Electrochemical Detection and Recycling of TNF-α Sensor
2.8. The Preparation of the Serum
2.9. Experimental Protocol
3. Results and Discussion
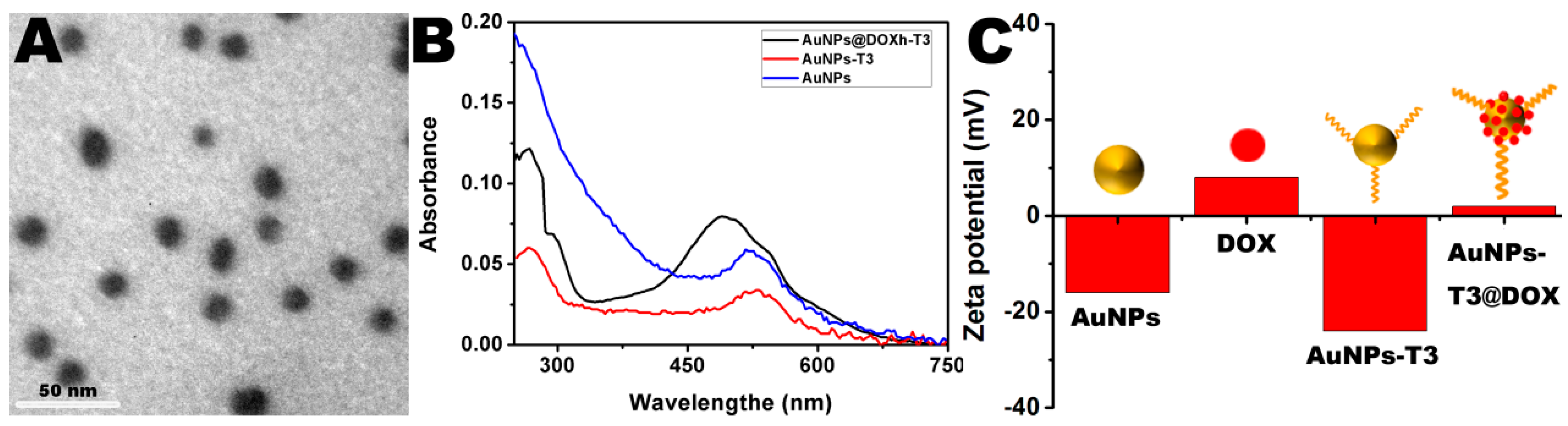
3.1. Characterization of AuNPs and Signal Molecule
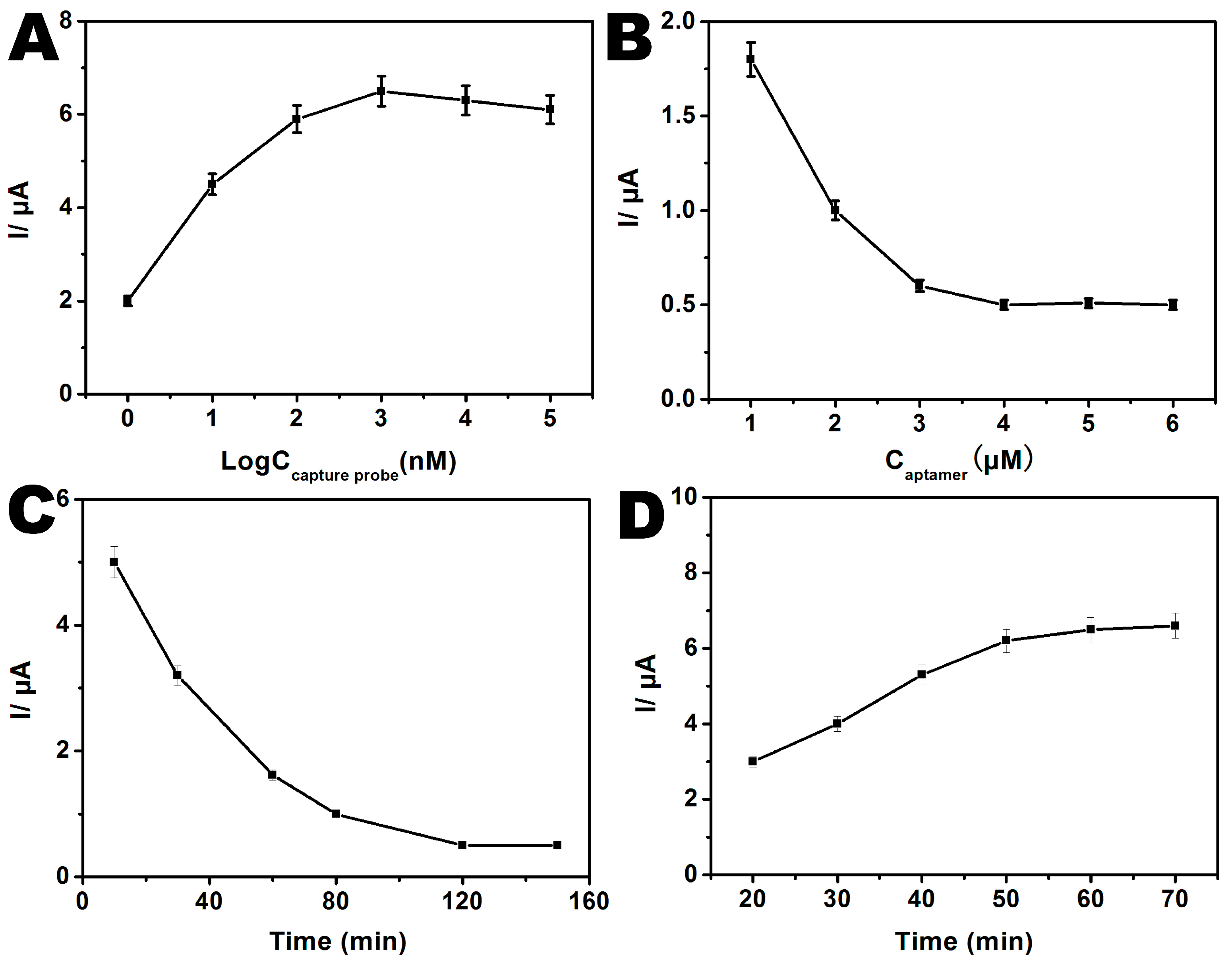
3.2. Optimization of Experimental Parameters
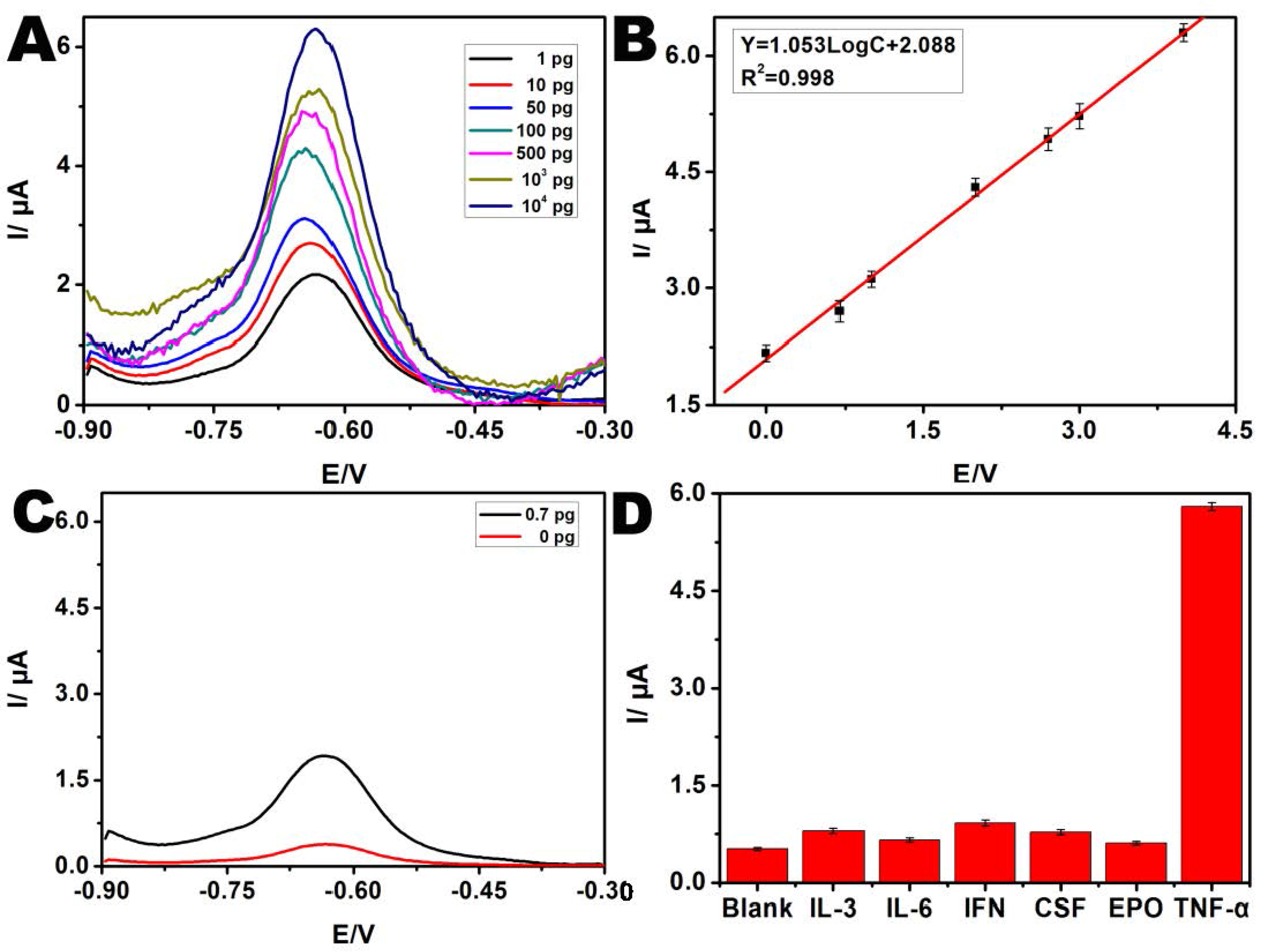
3.3. Detection Sensitivity and Selectivity
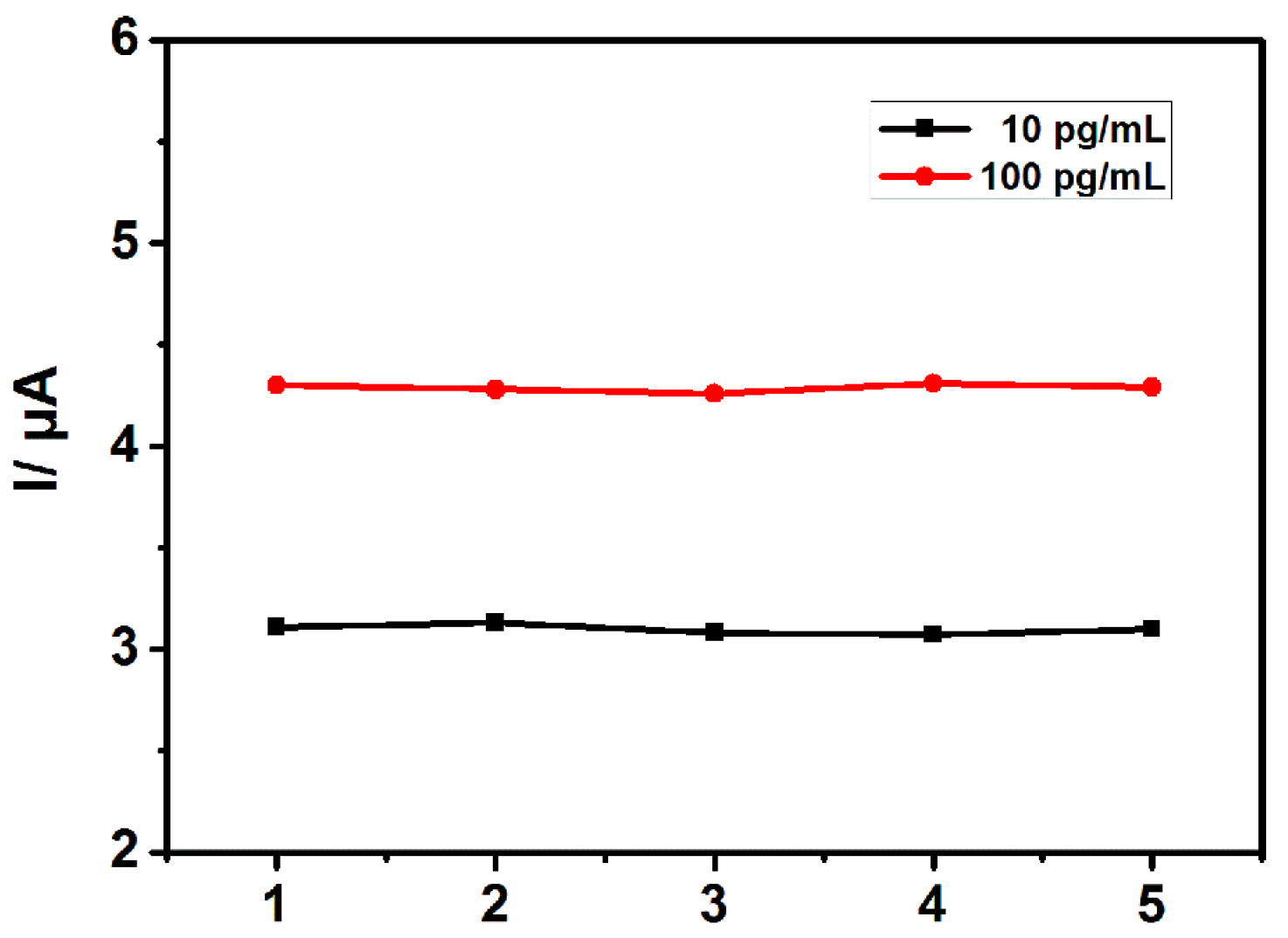
3.4. Reproducibility and Stability of the Sensor
3.5. Reusing of the Sensor
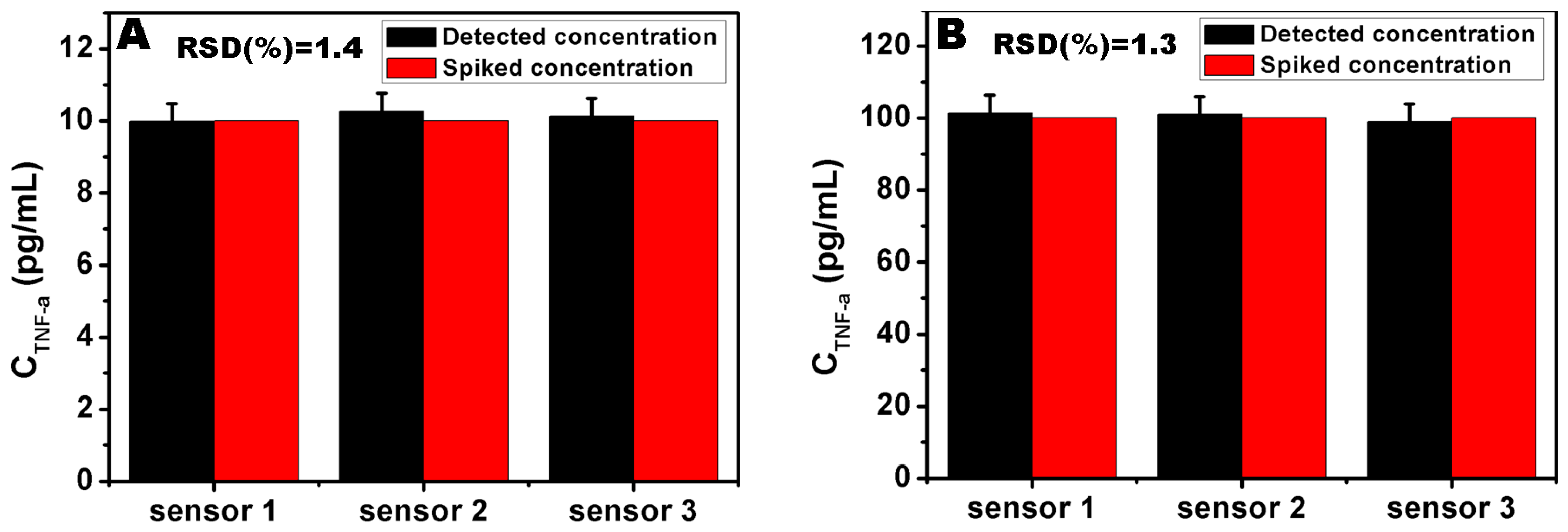
3.6. Detection of the Serum Samples
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Romen, Y.; Artal, R. C-reactive protein in pregnancy and in the postpartum period. Am. J. Obstet. Gynecol. 1985, 151, 380–383. [Google Scholar] [CrossRef]
- Kristóf, K.; Kocsis, E.; Nagy, K. Clinical microbiology of early-onset and late-onset neonatal sepsis, particularly among preterm babies. Acta Microbiol. Immunol. Hung. 2009, 56, 21–51. [Google Scholar] [CrossRef] [PubMed]
- Maniaci, V.; Dauber, A.; Weiss, S.; Nylen, E.; Becker, K.L.; Bachur, R. Procalcitonin in young febrile infants for the detection of serious bacterial infections. Paediatrics 2008, 122, 701–710. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Johnson, H.L.; Cousens, S.; Perin, J.; Scott, S.; Lawn, J.E.; Rudan, I.; Campbell, H.; Cibulskis, R.; Li, M.; et al. Global, regional, and national causes of child mortality: An updated systematic analysis for 2010 with time trends since 2000. Lancet 2012, 379, 2151–2161. [Google Scholar] [CrossRef]
- Verstraete, E.H.; Blot, K.; Mahieu, L.; Vogelaers, D.; Blot, S. Prediction models for neonatal health care-associated sepsis: A meta-analysis. Pediatrics 2015, 135, 1002–1014. [Google Scholar] [CrossRef] [PubMed]
- Kunz, M.; Ibrahim, S.M. Cytokines and Cytokine Profiles in Human Autoimmune Diseases and Animal Models of Autoimmunity. Mediat. Inflamm. 2009, 2009, 979258. [Google Scholar] [CrossRef] [PubMed]
- Somasuntharam, I.; Yehl, K.; Carroll, S.L.; Maxwell, J.T.; Martinez, M.D.; Che, P.L.; Browna, M.E.; Salaita, K.; Davisa, M.E. Knockdown of TNF-α by DNAzyme gold nanoparticles as an anti-inflammatory therapy for myocardial infarction. Biomaterials 2016, 83, 12–22. [Google Scholar] [CrossRef] [PubMed]
- Santana, C.; Guindeo, M.C.; Gonzalez, G.; García-Muñoz, F.; Saavedra, P.; Doménech, E. Cord blood levels of cytokines as predictors of early neonatal sepsis. Acta Paediatr. 2001, 90, 1176–1181. [Google Scholar] [CrossRef] [PubMed]
- Ahrén-Moonga, J.; Lekander, M.; von Blixen, N.; Rönnelid, J.; Holmgren, S.; Af Klinteberg, B. Levels of tumour necrosis factor-alpha and interleukin-6 in severely ill patients with eating disorders. Neuropsychobiology 2011, 63, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Kenison, D.C.; Elsasser, T.H.; Fayer, R.T. Radioimmunoassay for bovine tumor necrosis factor: Concentrations and circulating molecular forms in bovine plasma. Immunoass 1990, 11, 177–198. [Google Scholar] [CrossRef] [PubMed]
- Leister, K.P.; Huang, R.; Goodwin, B.L.; Chen, A.; Austin, C.P.; Xia, M. Two High Throughput Screen Assays for Measurement of TNF-α in THP-1 Cells. Curr. Chem. Genom. 2011, 5, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Vercoutere, W.; Akeson, M. Biosensors for DNA sequence detection. Cur. Opin. Chem. Biol. 2002, 6, 816–822. [Google Scholar] [CrossRef]
- Thorp, H.H.; March, T. Cutting out the middleman: DNA biosensors based on electrochemical oxidation. Trends Biotechnol. 1998, 16, 117–121. [Google Scholar] [CrossRef]
- Huang, K.J.; Niu, D.J.; Sun, J.Y.; Han, C.H.; Wu, Z.W.; Li, Y.L.; Xiong, X.Q. Novel electrochemical sensor based on functionalized graphene for simultaneous determination of adenine and guanine in DNA. Colloids Surf. B Biointerfaces 2011, 82, 543–549. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Li, S.; Yu, X.; Zhang, G.; Zhang, S.; Yao, J.; Keita, B.; Nadjo, L.; Zhi, L. Facile Synthesis of Au-Nanoparticle/Polyoxometalate/Graphene Tricomponent Nanohybrids: An Enzyme-Free Electrochemical Biosensor for Hydrogen Peroxide. Small 2012, 8, 1398–1406. [Google Scholar] [CrossRef] [PubMed]
- Kongsuphol, P.; Ng, H.H.; Pursey, J.P.; Arya, S.K.; Wong, C.C.; Stulz, E.; Park, M.K. EIS-based biosensor for ultra-sensitive detection of TNF-α from non-diluted human serum. Biosens. Bioelectron. 2014, 61, 274–279. [Google Scholar] [CrossRef] [PubMed]
- Castro-López, V.; Elizalde, J.; Pacek, M.; Hijona, E.; Bujanda, L. A simple and portable device for the quantification of TNF-α in human plasma by means of on-chip magnetic bead-based proximity ligation assay. Biosens. Bioelectron. 2014, 54, 499–505. [Google Scholar] [CrossRef] [PubMed]
- Mazloum-Ardakani, M.; Hosseinzadeh, L.; Taleat, Z. Synthesis and electrocatalytic effect of Ag@Pt core–shell nanoparticles supported on reduced graphene oxide for sensitive and simple label-free electrochemical aptasensor. Biosens. Bioelectron. 2015, 74, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Spain, E.; Keyes, T.E.; Forster, R.J. DNA sensor based on vapour polymerised pedot films functionalised with gold nanoparticles. Biosens. Bioelectron. 2013, 41, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Hua, E.; Liang, M.; Ma, C.; Liu, Z.; Sheng, S.; Liu, M.; Xie, G.; Feng, W. Graphene sheets, polyaniline and AuNPs based DNA sensor for electrochemical determination of BCR/ABL fusion gene with functional hairpin probe. Biosens. Bioelectron. 2014, 51, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Doroshow, J.H.; Reeves, J. Daunorubicin-stimulated reactive oxygen metabolism in cardiac sarcosomes. Biochem. Pharmacol. 1981, 30, 259–262. [Google Scholar] [CrossRef]
- Zhu, X.; Feng, C.; Ye, Z.; Chen, Y.; Li, G. Fabrication of magneto-controlled moveable architecture to develop reusable electrochemical biosensors. Sci. Rep. 2014, 4, 4196. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Yang, J.; Chen, Z.; Chen, X.; Wang, L.; Hu, J.; Ji, F.; Xie, G.; Feng, W. A novel label-free and reusable electrochemical cytosensor for highly sensitive detection and specific collection of CTCs. Biosens. Bioelectron. 2016, 81, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Hermann, T. Adaptive recognition by nucleic acid aptamers. Science 2000, 287, 820–825. [Google Scholar] [CrossRef] [PubMed]
- Josephs, E.A.; Ye, T. Nanoscale Spatial Distribution of Thiolated DNA on Model Nucleic Acid Sensor Surfaces. ACS Nano 2013, 7, 3653–3660. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Shen, H.W.; Zhang, X.; Tao, Y.Y.; Xiang, H.; Xie, G.M. A novel platform for high sensitivity determination of PbP2a based on gold nanoparticles composited graphitized mesoporous carbon and doxorubicin loaded hollow gold nanospheres. Biosens. Bioelectron. 2016, 15, 1119–1125. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.Y.; Wang, J.Q.; Huang, H.Y.; Chen, H.M.; Guan, R.L.; Chen, Y.; Ji, L.N.; Chao, H. RuNH2@AuNPs as two-photon luminescent probes for thiols in living cells and tissues. Biomaterials 2014, 35, 9003–9011. [Google Scholar] [CrossRef] [PubMed]
- Li, C.M.; Zheng, L.L.; Yang, X.X.; Wan, X.X.; Wu, W.B.; Zhen, S.J.; Li, Y.F.; Luo, L.F.; Huang, C.Z. DNA-AuNP networks on cell membranes as a protective barrier to inhibit viral attachment, entry and budding. Biomaterials 2016, 77, 216–226. [Google Scholar] [CrossRef] [PubMed]
- Hizir, M.S.; Top, M.; Balcioglu, M.; Rana, M.; Robertson, N.M.; Shen, F.S.; Sheng, J.; Yigit, M.V. Multiplexed Activity of perAuxidase: DNA-Capped AuNPs Act as Adjustable Peroxidase. Anal. Chem. 2016, 88, 600–605. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Lu, Y. Preparation of aptamer-linked gold nanoparticle purple aggregates for colorimetric sensing of analytes. Nat. Protoc. 2006, 1, 246–252. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Xiang, H.; Hua, E.; Wang, L.; Jing, X.; Cao, X.; Sheng, S.; Xie, G. Ultrasensitive Electrochemical Biosensor for the Detection of the mecA Gene Sequence in Methicillin Resistant Strains of Employing Gold Nanoparticles. Anal. Lett. 2014, 47, 579–591. [Google Scholar] [CrossRef]
- Deng, R.; Wang, L.; Yi, G.; Hua, E.; Xie, G. Target-induced aptamer release strategy based on electrochemical detection of staphylococcal enterotoxin B using GNPs-ZrO2-Chits film. Colloids Surf. B Biointerfaces 2014, 120, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhu, X.; Zhang, W.; Xie, G.; Feng, W. Hydrogen Peroxide Biosensor Based on Gold Nanoparticles/Thionine/Gold Nanoparticles/Multi-walled Carbon Nanotubes–chitosans Composite Film-modified Electrode. Appl. Surf. Sci. 2012, 91, 2802–2807. [Google Scholar] [CrossRef]
- Armbruster, D.A.; Tillman, M.D.; Hubbs, L.M. Limit of detection (LQD)/limit of quantitation (LOQ): Comparison of the empirical and the statistical methods exemplified with GC-MS assays of abused drugs. Clin. Chem. 1994, 40, 1233–1238. [Google Scholar] [PubMed]


© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, L.; Li, M.; Wang, W.; Zhang, Q.; Liu, D.; Li, X.; Jiang, H. High Sensitivity Determination of TNF-α for Early Diagnosis of Neonatal Infections with a Novel and Reusable Electrochemical Sensor. Sensors 2017, 17, 992. https://doi.org/10.3390/s17050992
Li L, Li M, Wang W, Zhang Q, Liu D, Li X, Jiang H. High Sensitivity Determination of TNF-α for Early Diagnosis of Neonatal Infections with a Novel and Reusable Electrochemical Sensor. Sensors. 2017; 17(5):992. https://doi.org/10.3390/s17050992
Chicago/Turabian StyleLi, Liangliang, Miaomiao Li, Wenwen Wang, Qian Zhang, Dongyun Liu, Xianghong Li, and Hong Jiang. 2017. "High Sensitivity Determination of TNF-α for Early Diagnosis of Neonatal Infections with a Novel and Reusable Electrochemical Sensor" Sensors 17, no. 5: 992. https://doi.org/10.3390/s17050992




