Adenosine Triphosphate-Encapsulated Liposomes with Plasmonic Nanoparticles for Surface Enhanced Raman Scattering-Based Immunoassays
Abstract
:1. Introduction
2. Experimental
2.1. Materials
2.2. Preparation of SiO2@Au@Ag NPs
2.3. Incorporation of ATP into SiO2@Au@Ag NPs
2.4. Preparation of ATP-Encapsulated Liposomes
2.5. Lysis of ATP-Encapsulated Liposomes and Immobilization of ATP on the SiO2@Au@Ag NPs
2.6. SERS Measurement of the SiO2@Au@Ag NPs
3. Result and Discussion
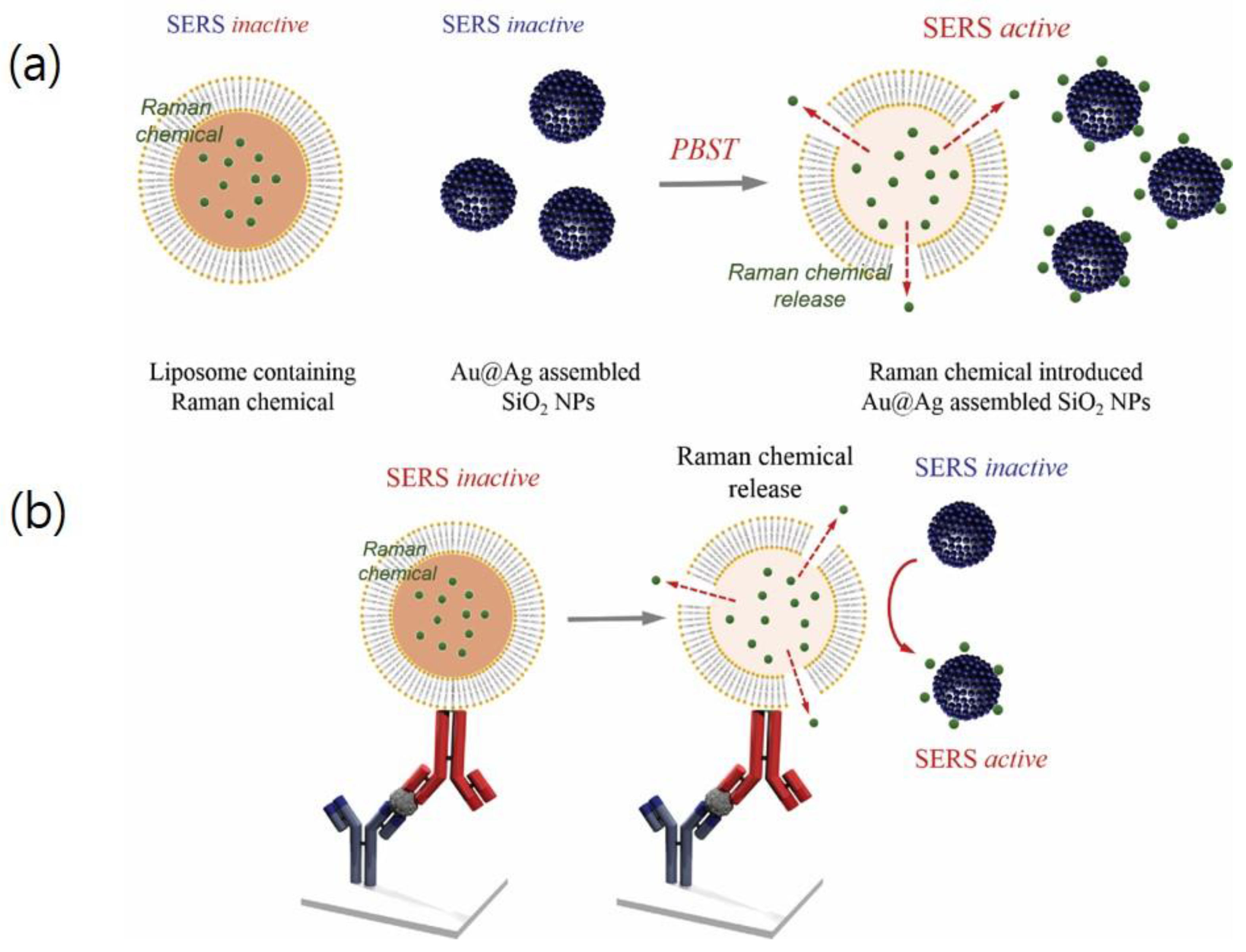
3.1. Preparation of ATP-Encapsulated Liposomes and SiO2@Au@Ag NPs
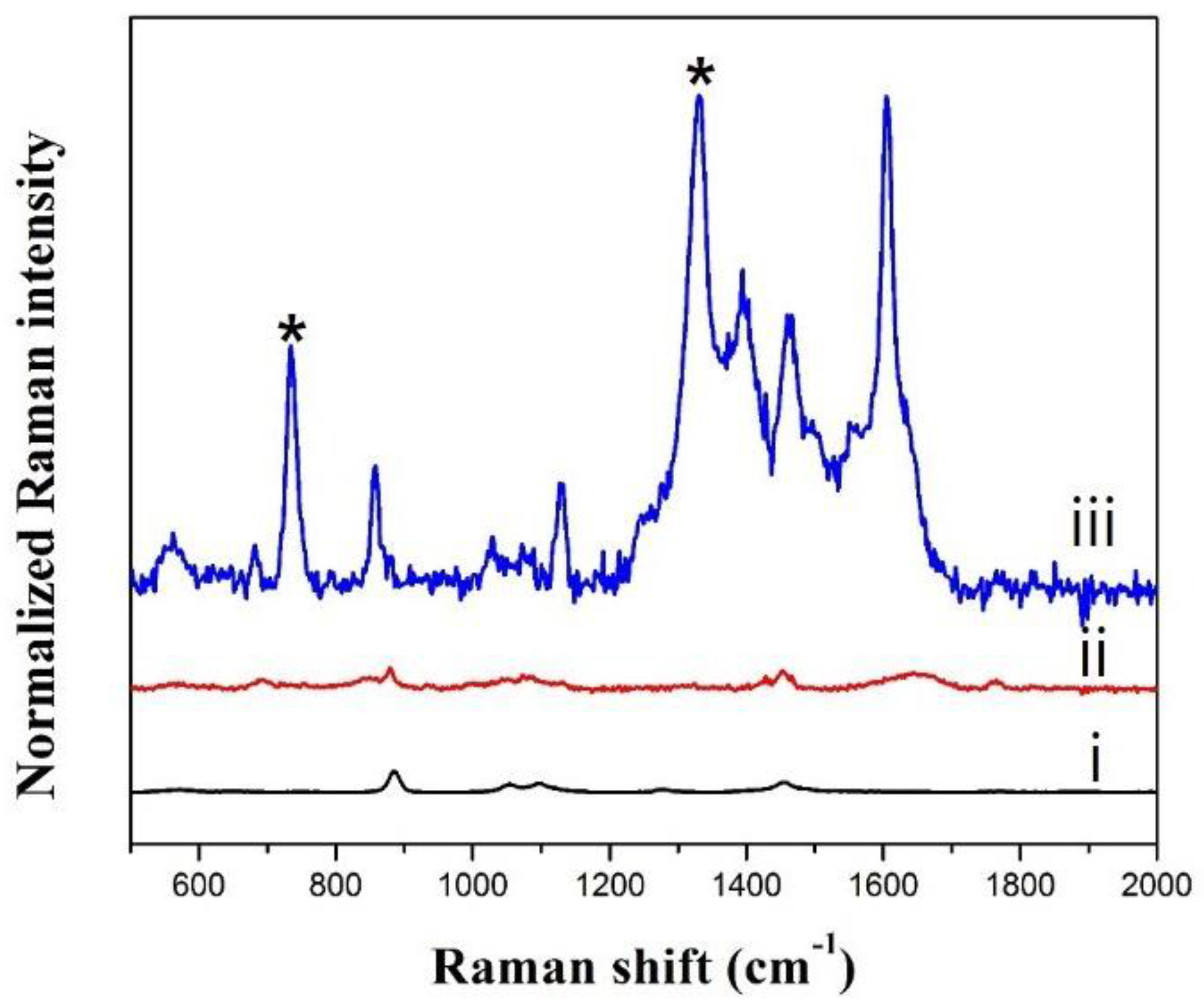
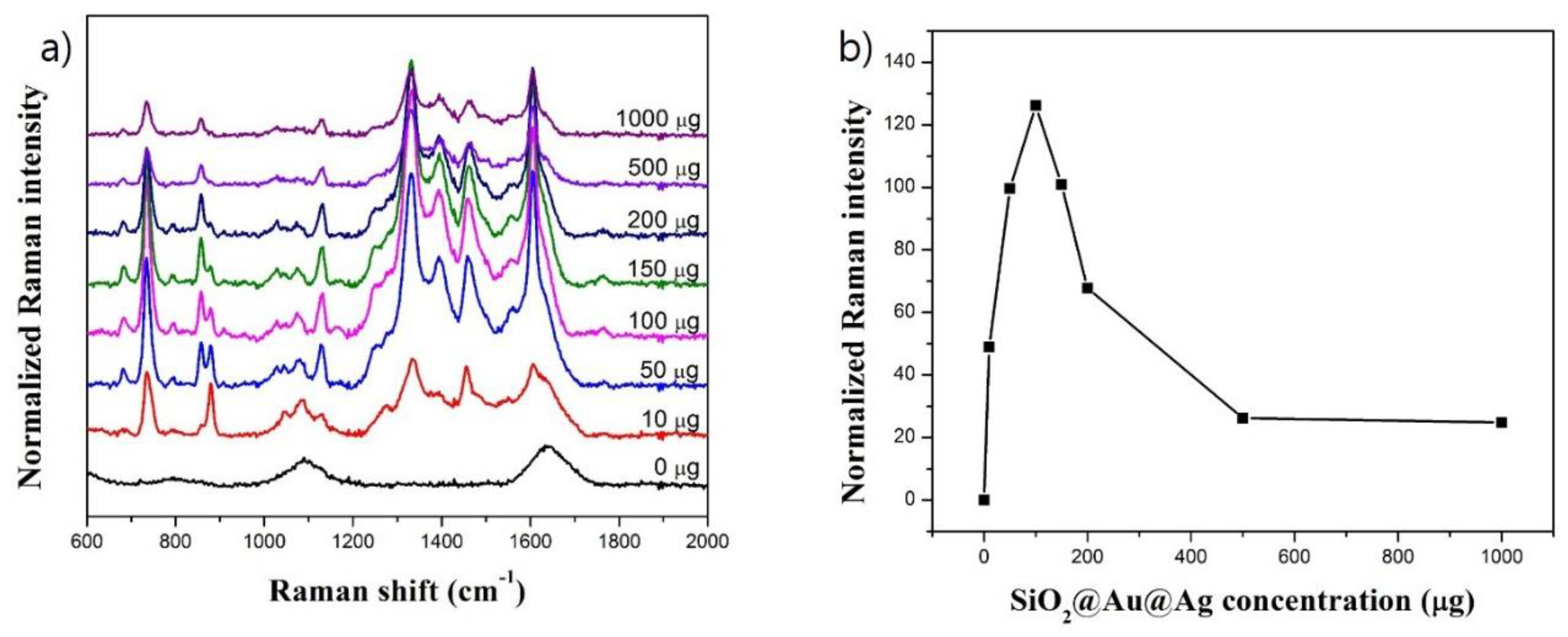
3.2. Optimization of SERS Measurement of ATP in the Presence of SiO2@Au@Ag NPs
3.3. Preparation of ATP-Encapsulated Liposomes
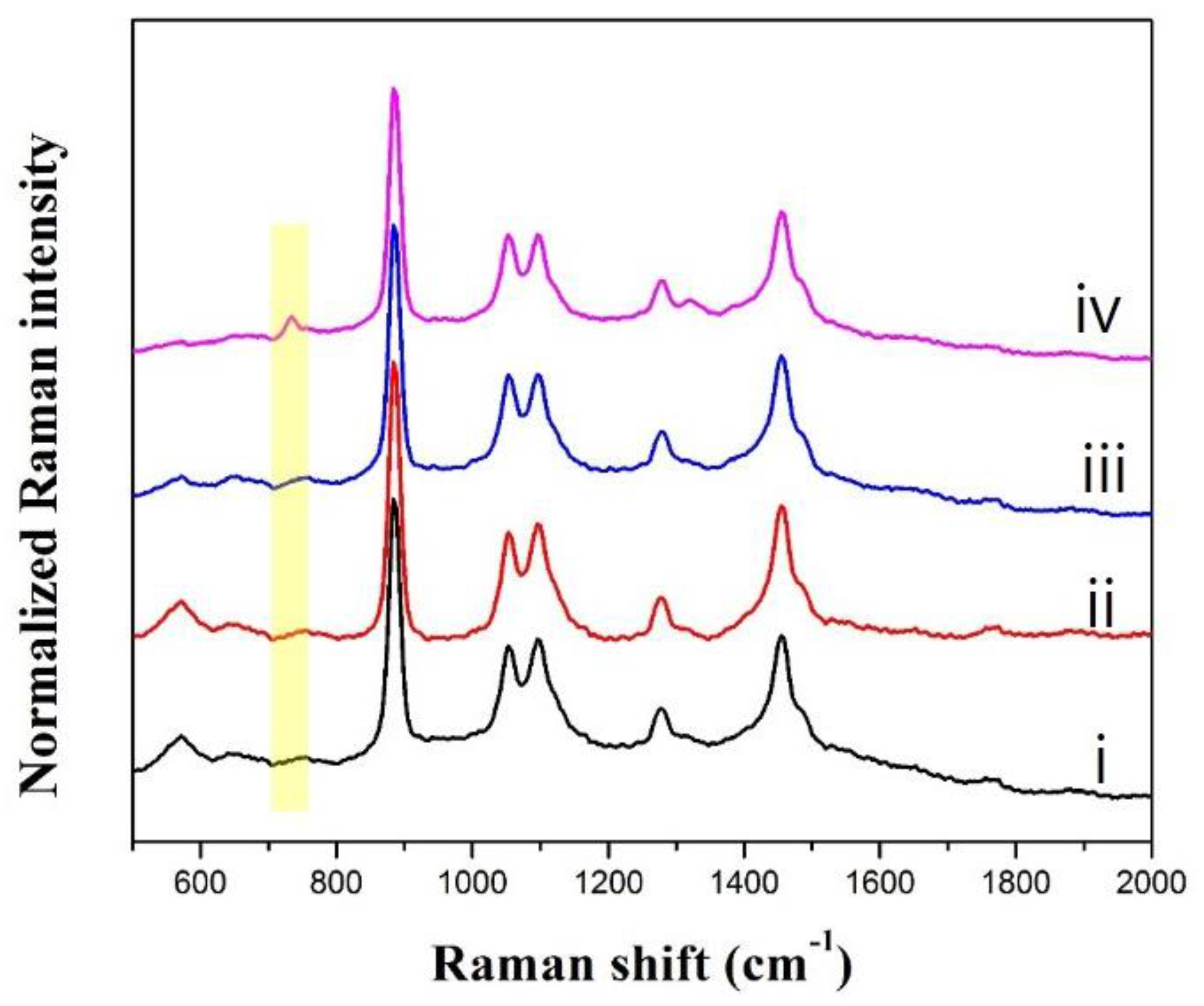
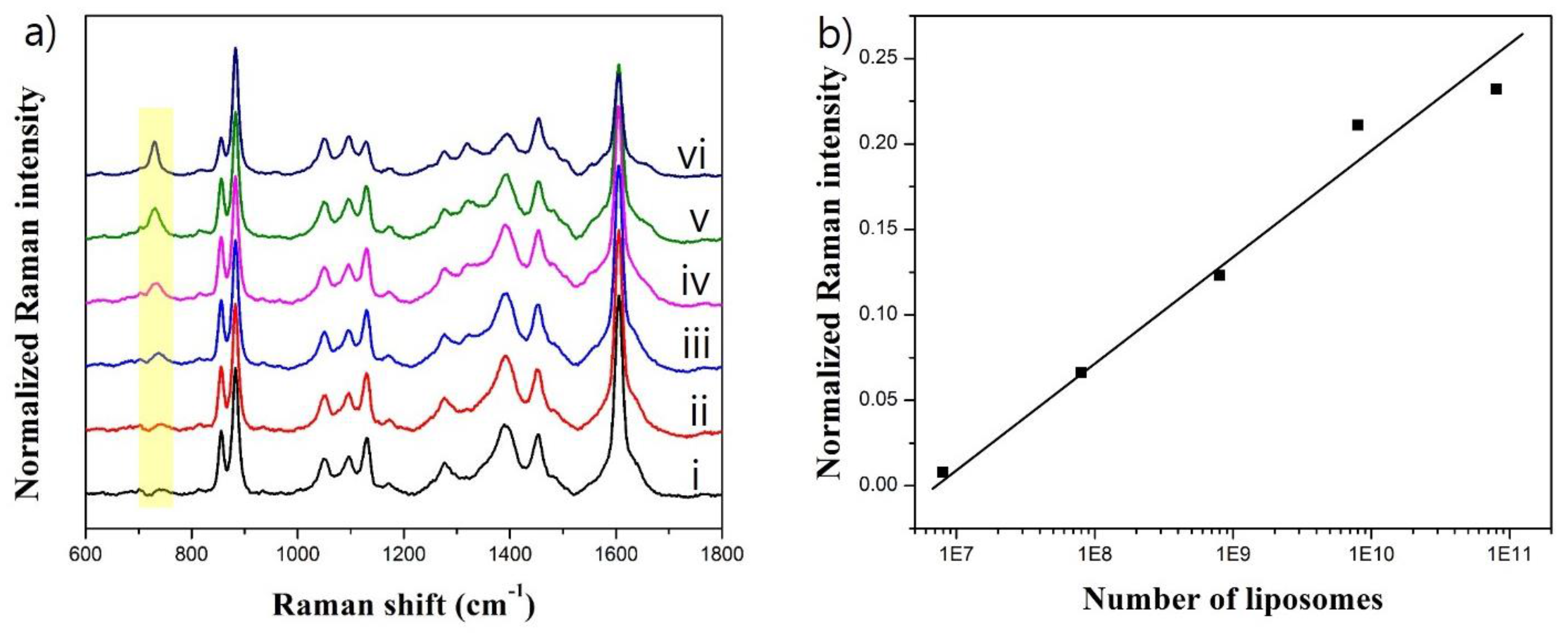
3.4. Effect of ATP-Encapsulated Liposomes on SERS Signal
4. Conclusions
Supplementary Materials
Acknowledgements
Author Contributions
Conflicts of Interest
References
- Smolsky, J.; Kaur, S.; Hayashi, C.; Batra, S.; Krasnoslobodtsev, A. Surface-Enhanced Raman Scattering-Based Immunoassay Technologies for Detection of Disease Biomarkers. Biosensors 2017, 7, 7. [Google Scholar] [CrossRef] [PubMed]
- Klee, G.G.; Post, G. Effect of counting errors on immunoassay precision. Clin. Chem. 1989, 35, 1362–1366. [Google Scholar] [PubMed]
- Hicks, J.M. Fluorescence immunoassay. Hum. Pathol. 1984, 15, 112–116. [Google Scholar] [CrossRef]
- Brown, C.R.; Higgins, K.W.; Frazer, K.; Schoelz, L.K.; Dyminski, J.W.; Marinkovich, V.A.; Miller, S.P.; Burd, J.F. Simultaneous determination of total IgE and allergen-specific IgE in serum by the MAST chemiluminescent assay system. Clin. Chem. 1985, 31, 1500–1505. [Google Scholar] [PubMed]
- Zhang, J.-J.; Liu, Y.; Hu, L.-H.; Jiang, L.-P.; Zhu, J.-J. “Proof-of-principle” concept for ultrasensitive detection of cytokines based on the electrically heated carbon paste electrode. Chem. Commun. 2011, 47, 6551–6553. [Google Scholar] [CrossRef] [PubMed]
- Hayes, F.J.; Halsall, H.B.; Heineman, W.R. Simultaneous Immunoassay Using Electrochemical Detection of Metal Ion Labels. Anal. Chem. 1994, 66, 1860–1865. [Google Scholar] [CrossRef] [PubMed]
- Butler, J.E. Solid Supports in Enzyme-Linked Immunosorbent Assay and Other Solid-Phase Immunoassays. Methods 2000, 22, 4–23. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.-J.; Zheng, T.-T.; Cheng, F.-F.; Zhu, J.-J. Electrochemical sensing for caspase 3 activity and inhibition using quantum dot functionalized carbon nanotube labels. Chem. Commun. 2011, 47, 1178–1180. [Google Scholar] [CrossRef] [PubMed]
- Haynes, C.L.; McFarland, A.D.; Duyne, R.P.V. Surface-Enhanced Raman Spectroscopy. Anal. Chem. 2005, 77, 338A–346A. [Google Scholar] [CrossRef]
- Schlücker, S. Surface-Enhanced Raman Spectroscopy: Concepts and Chemical Applications. Angew. Chem. Int. Ed. 2014, 53, 4756–4795. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yan, B.; Chen, L. SERS Tags: Novel Optical Nanoprobes for Bioanalysis. Chem. Rev. 2013, 113, 1391–1428. [Google Scholar] [CrossRef] [PubMed]
- Culha, M.; Cullum, B.; Lavrik, N.; Klutse, C.K. Surface-Enhanced Raman Scattering as an Emerging Characterization and Detection Technique. J. Nanotechnol. 2012, 2012, 15. [Google Scholar] [CrossRef]
- Jun, B.-H.; Kim, G.; Jeong, S.; Noh, M.S.; Pham, X.-H.; Kang, H.; Cho, M.-H.; Kim, J.-H.; Lee, Y.-S.; Jeong, D.H. Silica Core-Based Surface-Enhanced Raman Scattering (SERS) Tag: Advances in Multifunctional SERS Nanoprobes for Bioimaging and Targeting of Biomarkers. Bull. Korean Chem. Soc. 2015, 36, 963–978. [Google Scholar]
- Nie, S.; Emory, S.R. Probing Single Molecules and Single Nanoparticles by Surface-Enhanced Raman Scattering. Science 1997, 275, 1102–1106. [Google Scholar] [CrossRef] [PubMed]
- Sivashanmugan, K.; Liao, J.-D.; Liu, B.H.; Yao, C.-K.; Luo, S.-C. Ag nanoclusters on ZnO nanodome array as hybrid SERS-active substrate for trace detection of malachite green. Sens. Actuators B Chem. 2015, 207, 430–436. [Google Scholar] [CrossRef]
- Hu, Y.; Liao, J.; Wang, D.; Li, G. Fabrication of Gold Nanoparticle-Embedded Metal–Organic Framework for Highly Sensitive Surface-Enhanced Raman Scattering Detection. Anal. Chem. 2014, 86, 3955–3963. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Nanda, J.; Wang, B.; Chen, G.; Hallinan, D.T. Self-Assembly of Large Gold Nanoparticles for Surface-Enhanced Raman Spectroscopy. ACS Appl. Mater. Interfaces 2017, 9, 13457–13470. [Google Scholar] [CrossRef] [PubMed]
- Bekana, D.; Liu, R.; Amde, M.; Liu, J.-F. Use of Polycrystalline Ice for Assembly of Large Area Au Nanoparticle Superstructures as SERS Substrates. ACS Appl. Mater. Interfaces 2017, 9, 513–520. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Zhao, J.; Xu, H.; Li, Y.; Ji, J.; Liu, B. Multifunctional Paper Strip Based on Self-Assembled Interfacial Plasmonic Nanoparticle Arrays for Sensitive SERS Detection. ACS Appl. Mater. Interfaces 2015, 7, 16767–16774. [Google Scholar] [CrossRef] [PubMed]
- Sugawa, K.; Akiyama, T.; Tanoue, Y.; Harumoto, T.; Yanagida, S.; Yasumori, A.; Tomita, S.; Otsuki, J. Particle size dependence of the surface-enhanced Raman scattering properties of densely arranged two-dimensional assemblies of Au(core)-Ag(shell) nanospheres. Phys. Chem. Chem. Phys. 2015, 17, 21182–21189. [Google Scholar] [CrossRef] [PubMed]
- Tanoue, Y.; Sugawa, K.; Yamamuro, T.; Akiyama, T. Densely arranged two-dimensional silver nanoparticle assemblies with optical uniformity over vast areas as excellent surface-enhanced Raman scattering substrates. Phys. Chem. Chem. Phys. 2013, 15, 15802–15805. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.; Lipert, R.J.; Dawson, G.B.; Porter, M.D. Immunoassay Readout Method Using Extrinsic Raman Labels Adsorbed on Immunogold Colloids. Anal. Chem. 1999, 71, 4903–4908. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Huan, S.; Bu, Y.; Shen, G.; Yu, R. Liposome-mediated enhancement of the sensitivity in immunoassay based on surface-enhanced Raman scattering at gold nanosphere array substrate. Talanta 2008, 75, 797–803. [Google Scholar] [CrossRef] [PubMed]
- Porter, M.D.; Lipert, R.J.; Siperko, L.M.; Wang, G.; Narayanan, R. SERS as a bioassay platform: Fundamentals, design, and applications. Chem. Soc. Rev. 2008, 37, 1001–1011. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Hu, Y.; Gao, J.; Zhong, L. Highly sensitive detection of clenbuterol using competitive surface-enhanced Raman scattering immunoassay. Anal. Chim. Acta 2011, 697, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Lee, K.; Kim, K.H.; Oh, K.W.; Choo, J. SERS-based immunoassay using a gold array-embedded gradient microfluidic chip. Lab Chip 2012, 12, 3720–3727. [Google Scholar] [CrossRef] [PubMed]
- Pham, X.-H.; Lee, M.; Shim, S.; Jeong, S.; Kim, H.-M.; Hahm, E.; Lee, S.H.; Lee, Y.-S.; Jeong, D.H.; Jun, B.-H. Highly sensitive and reliable SERS probes based on nanogap control of a Au-Ag alloy on silica nanoparticles. RSC Adv. 2017, 7, 7015–7021. [Google Scholar] [CrossRef]
- Shim, S.; Pham, X.-H.; Cha, M.G.; Lee, Y.-S.; Jeong, D.H.; Jun, B.-H. Size effect of gold on Ag-coated Au nanoparticle-embedded silica nanospheres. RSC Adv. 2016, 6, 48644–48650. [Google Scholar] [CrossRef]
- Deng, Z.J.; Morton, S.W.; Ben-Akiva, E.; Dreaden, E.C.; Shopsowitz, K.E.; Hammond, P.T. Layer-by-layer nanoparticles for systemic codelivery of an anticancer drug and siRNA for potential triple-negative breast cancer treatment. ACS Nano 2013, 7, 9571–9584. [Google Scholar] [CrossRef] [PubMed]
- Bozzuto, G.; Molinari, A. Liposomes as nanomedical devices. Int. J. Nanomed. 2015, 10, 975–999. [Google Scholar] [CrossRef] [PubMed]
- Wicki, A.; Witzigmann, D.; Balasubramanian, V.; Huwyler, J. Nanomedicine in cancer therapy: Challenges, opportunities, and clinical applications. J. Controll. Release 2015, 200, 138–157. [Google Scholar] [CrossRef] [PubMed]
- Oberoi, H.S.; Nukolova, N.V.; Kabanov, A.V.; Bronich, T.K. Nanocarriers for delivery of platinum anticancer drugs. Adv. Drug Deliv. Rev. 2013, 65, 1667–1685. [Google Scholar] [CrossRef] [PubMed]
- Edwards, K.A.; Bolduc, O.R.; Baeumner, A.J. Miniaturized bioanalytical systems: Enhanced performance through liposomes. Curr. Opin. Chem. Biol. 2012, 16, 444–452. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, J.P.; Campbell, R.L.; Fleming, B.M.; Mercolino, T.J.; Johnson, M.D.; McLaurin, D.A. A highly sensitive immunoassay system involving antibody-coated tubes and liposome-entrapped dye. Clin. Chem. 1985, 31, 1424–1426. [Google Scholar] [PubMed]
- Kannuck, R.M.; Bellama, J.M.; Durst, R.A. Measurement of liposome-released ferrocyanide by a dual-function polymer modified electrode. Anal. Chem. 1988, 60, 142–147. [Google Scholar] [CrossRef] [PubMed]
- Monnard, P.-A.; Oberholzer, T.; Luisi, P. Entrapment of nucleic acids in liposomes. Biochim. Biophy. Acta 1997, 1329, 39–50. [Google Scholar] [CrossRef]
- Colletier, J.P.; Chaize, B.; Winterhalter, M.; Fournier, D. Protein encapsulation in liposomes: Efficiency depends on interactions between protein and phospholipid bilayer. BMC Biotechnol. 2002, 2, 9. [Google Scholar] [CrossRef]
- Khreich, N.; Lamourette, P.; Boutal, H.; Devilliers, K.; Créminon, C.; Volland, H. Detection of Staphylococcus enterotoxin B using fluorescent immunoliposomes as label for immunochromatographic testing. Anal. Biochem. 2008, 377, 182–188. [Google Scholar] [CrossRef] [PubMed]
- Locascio-Brown, L.; Plant, A.L.; Horvath, V.; Durst, R.A. Liposome flow injection immunoassay: Implications for sensitivity, dynamic range, and antibody regeneration. Anal. Chem. 1990, 62, 2587–2593. [Google Scholar] [CrossRef] [PubMed]
- Edwards, K.A.; Baeumner, A.J. Optimization of DNA-tagged liposomes for use in microtiter plate analyses. Anal. Bioanal. Chem. 2006, 386, 1613–1623. [Google Scholar] [CrossRef] [PubMed]
- Edwards, K.A.; Baeumner, A.J. Optimization of DNA-tagged dye-encapsulating liposomes for lateral-flow assays based on sandwich hybridization. Anal. Bioanal. Chem. 2006, 386, 1335–1343. [Google Scholar] [CrossRef] [PubMed]
- Kobatake, E.; Sasakura, H.; Haruyama, T.; Laukkanen, M.-L.; Keinänen, K.; Aizawa, M. A Fluoroimmunoassay Based on Immunoliposomes Containing Genetically Engineered Lipid-Tagged Antibody. Anal. Chem. 1997, 69, 1295–1298. [Google Scholar] [CrossRef] [PubMed]
- Städler, B.; Falconnet, D.; Pfeiffer, I.; Höök, F.; Vörös, J. Micropatterning of DNA-Tagged Vesicles. Langmuir 2004, 20, 11348–11354. [Google Scholar] [CrossRef] [PubMed]
- Rongen, H.A.H.; Bult, A.; Van Bennekom, W.P. Liposomes and immunoassays. J. Immunol. Methods 1997, 204, 105–133. [Google Scholar] [CrossRef]
- Edwards, K.A.; Baeumner, A.J. Liposomes in analyses. Talanta 2006, 68, 1421–1431. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Hens, A.; Manuel Fernández-Romero, J. The role of liposomes in analytical processes. Trends Anal. Chem. 2005, 24, 9–19. [Google Scholar] [CrossRef]
- Zhu, D.; Wang, Z.; Zong, S.; Chen, H.; Wu, X.; Pei, Y.; Chen, P.; Ma, X.; Cui, Y. Ag@4ATP-coated liposomes: SERS traceable delivery vehicles for living cells. Nanoscale 2014, 6, 8155–8161. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Wang, Z.Y.; Zong, S.F.; Chen, H.; Chen, P.; Li, M.Y.; Wu, L.; Cui, Y.P. In Gold nanoparticles decorated liposomes and their SERS performance in tumor cells. Proc. SPIE 2015, 9543, 954316. [Google Scholar] [CrossRef]
- Gomes, J.F.P.d.S.; Sonnen, A.F.P.; Kronenberger, A.; Fritz, J.; Coelho, M.Á.N.; Fournier, D.; Fournier-Nöel, C.; Mauzac, M.; Winterhalter, M. Stable Polymethacrylate Nanocapsules from Ultraviolet Light-Induced Template Radical Polymerization of Unilamellar Liposomes. Langmuir 2006, 22, 7755–7759. [Google Scholar] [CrossRef] [PubMed]
- An, S.Y.; Bui, M.-P.N.; Nam, Y.J.; Han, K.N.; Li, C.A.; Choo, J.; Lee, E.K.; Katoh, S.; Kumada, Y.; Seong, G.H. Preparation of monodisperse and size-controlled poly(ethylene glycol) hydrogel nanoparticles using liposome templates. J. Colloid Interface Sci. 2009, 331, 98–103. [Google Scholar] [CrossRef] [PubMed]
- Roberts, M.A.; Locascio-Brown, L.; MacCrehan, W.A.; Durst, R.A. Liposome Behavior in Capillary Electrophoresis. Anal. Chem. 1996, 68, 3434–3440. [Google Scholar] [CrossRef] [PubMed]
- Bui, M.-P.N.; Ahmed, S.; Abbas, A. Single-Digit Pathogen and Attomolar Detection with the Naked Eye Using Liposome-Amplified Plasmonic Immunoassay. Nano Lett. 2015, 15, 6239–6246. [Google Scholar] [CrossRef] [PubMed]
- Meneghetti, M.; Scarsi, A.; Litti, L.; Marcolongo, G.; Amendola, V.; Gobbo, M.; Di Chio, M.; Boscaini, A.; Fracasso, G.; Colombatti, M. Serrs: Plasmonic Nanostructures for SERRS Multiplexed Identification of Tumor-Associated Antigens. Small 2012, 8, 3860. [Google Scholar] [CrossRef]
- Maley, A.M.; Arbiser, J.L. Gentian Violet: A 19(th) Century Drug Re-Emerges in the 21(st) Century. Exp. Dermatol. 2013, 22, 775–780. [Google Scholar] [CrossRef] [PubMed]
- Alford, R.; Simpson, H.M.; Duberman, J.; Hill, G.C.; Ogawa, M.; Regino, C.; Kobayashi, H.; Choyke, P.L. Toxicity of Organic Fluorophores Used in Molecular Imaging: Literature Review. Mol. Imaging 2009, 8, 341–354. [Google Scholar] [PubMed]
- Duff, D.G.; Baiker, A.; Edwards, P.P. A new hydrosol of gold clusters. 1. Formation and particle size variation. Langmuir 1993, 9, 2301–2309. [Google Scholar] [CrossRef]
- Hammami, K.; Feki, H.E.; Marsan, O.; Drouet, C. Adsorption of nucleotides on biomimetic apatite: The case of adenosine 5′ monophosphate (AMP). Appl. Surf. Sci. 2015, 353, 165–172. [Google Scholar] [CrossRef]
- Chen, T.T.; Kuo, C.S.; Chou, Y.C.; Liang, N.T. Surface-enhanced Raman scattering of adenosine triphosphate molecules. Langmuir 1989, 5, 887–891. [Google Scholar] [CrossRef]
- Zhang, C.; Man, B.Y.; Jiang, S.Z.; Yang, C.; Liu, M.; Chen, C.S.; Xu, S.C.; Qiu, H.W.; Li, Z. SERS detection of low-concentration adenosine by silver nanoparticles on silicon nanoporous pyramid arrays structure. Appl. Surf. Sci. 2015, 347, 668–672. [Google Scholar] [CrossRef]
- Pagliai, M.; Caporali, S.; Muniz-Miranda, M.; Pratesi, G.; Schettino, V. SERS, XPS, and DFT Study of Adenine Adsorption on Silver and Gold Surfaces. J. Phys. Chem. Lett. 2012, 3, 242–245. [Google Scholar] [CrossRef] [PubMed]
- Cialla, D.; Pollok, S.; Steinbrücker, C.; Weber, K.; Popp, J. SERS-based detection of biomolecules. In Nanophotonics; Walter de Gruyter GmbH: Berlin, Germany, 2014; Volume 3, p. 383. [Google Scholar]
- Barhoumi, A.; Zhang, D.; Tam, F.; Halas, N.J. Surface-Enhanced Raman Spectroscopy of DNA. J. Am. Chem. Soc. 2008, 130, 5523–5529. [Google Scholar] [CrossRef] [PubMed]
- Ding, G.; Xie, S.; Liu, Y.; Wang, L.; Xu, F. Graphene oxide-silver nanocomposite as SERS substrate for dye detection: Effects of silver loading amount and composite dosage. Appl. Surface Sci. 2015, 345, 310–318. [Google Scholar] [CrossRef]
- Leekumjorn, S.; Sum, A.K. Molecular Simulation Study of Structural and Dynamic Properties of Mixed DPPC/DPPE Bilayers. Biophys. J. 2006, 90, 3951–3965. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pham, X.-H.; Hahm, E.; Kim, T.H.; Kim, H.-M.; Lee, S.H.; Lee, Y.-S.; Jeong, D.H.; Jun, B.-H. Adenosine Triphosphate-Encapsulated Liposomes with Plasmonic Nanoparticles for Surface Enhanced Raman Scattering-Based Immunoassays. Sensors 2017, 17, 1480. https://doi.org/10.3390/s17071480
Pham X-H, Hahm E, Kim TH, Kim H-M, Lee SH, Lee Y-S, Jeong DH, Jun B-H. Adenosine Triphosphate-Encapsulated Liposomes with Plasmonic Nanoparticles for Surface Enhanced Raman Scattering-Based Immunoassays. Sensors. 2017; 17(7):1480. https://doi.org/10.3390/s17071480
Chicago/Turabian StylePham, Xuan-Hung, Eunil Hahm, Tae Han Kim, Hyung-Mo Kim, Sang Hun Lee, Yoon-Sik Lee, Dae Hong Jeong, and Bong-Hyun Jun. 2017. "Adenosine Triphosphate-Encapsulated Liposomes with Plasmonic Nanoparticles for Surface Enhanced Raman Scattering-Based Immunoassays" Sensors 17, no. 7: 1480. https://doi.org/10.3390/s17071480




