Identification and Quantification of Celery Allergens Using Fiber Optic Surface Plasmon Resonance PCR
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. qPCR/HRM
2.3. Surface Functionalization of FO-SPR Sensors and Au NPs with DNA
2.4. FO-PCR-MA in Solution
2.5. Biological Sample Preparation
2.6. Data Processing
3. Results
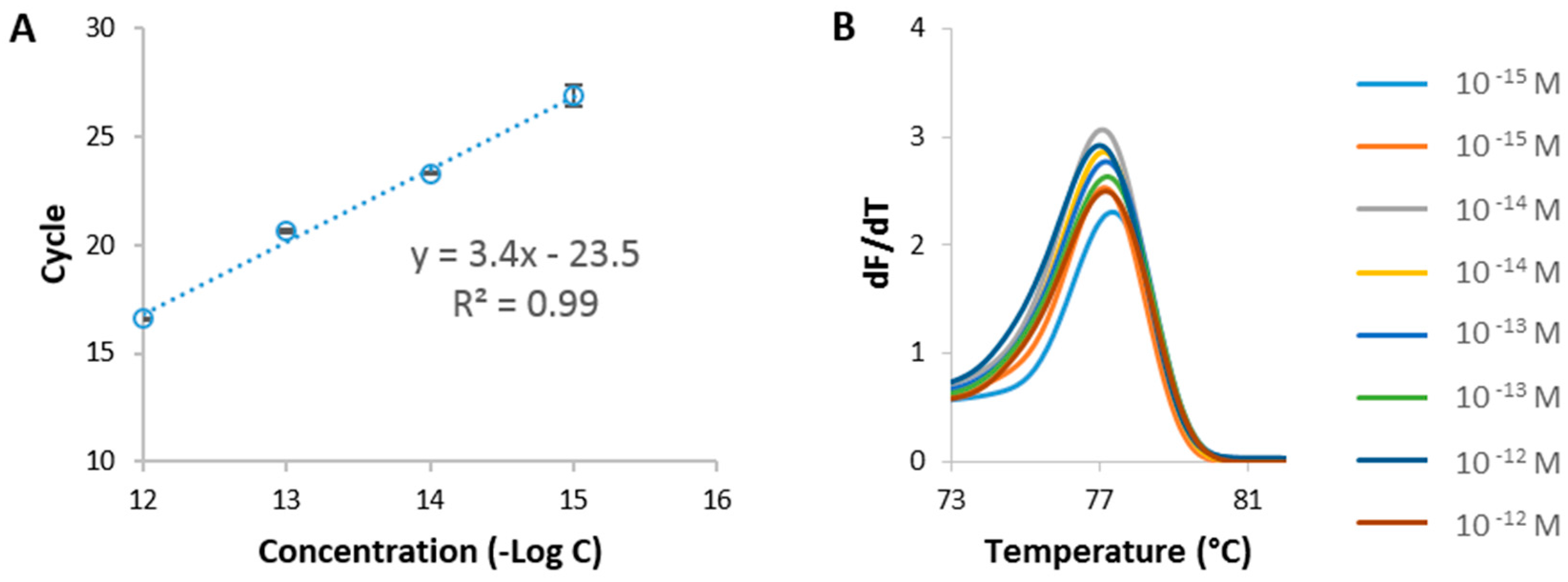
3.1. Selection of Celery Target Sequence Using qPCR/HRM Analysis
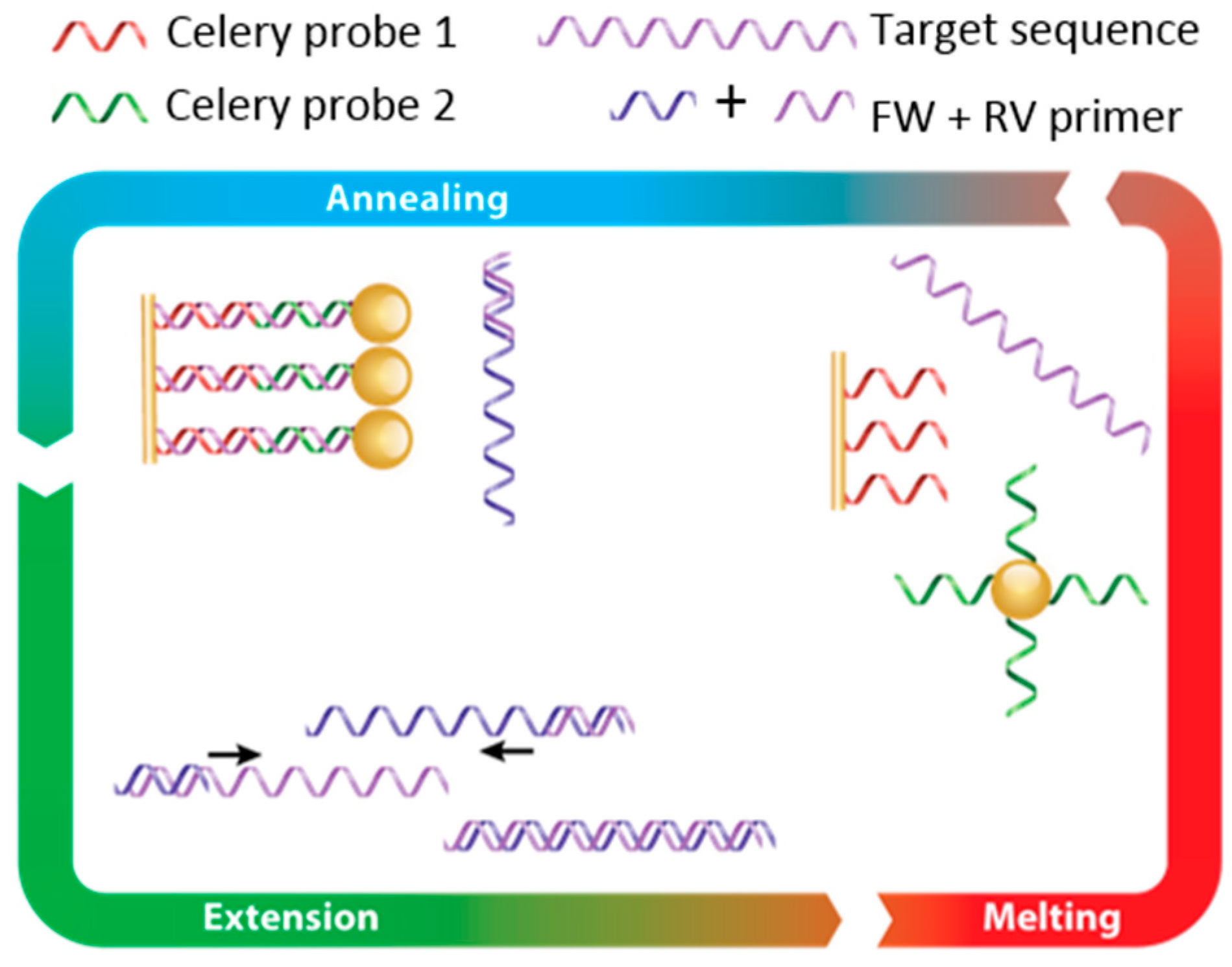
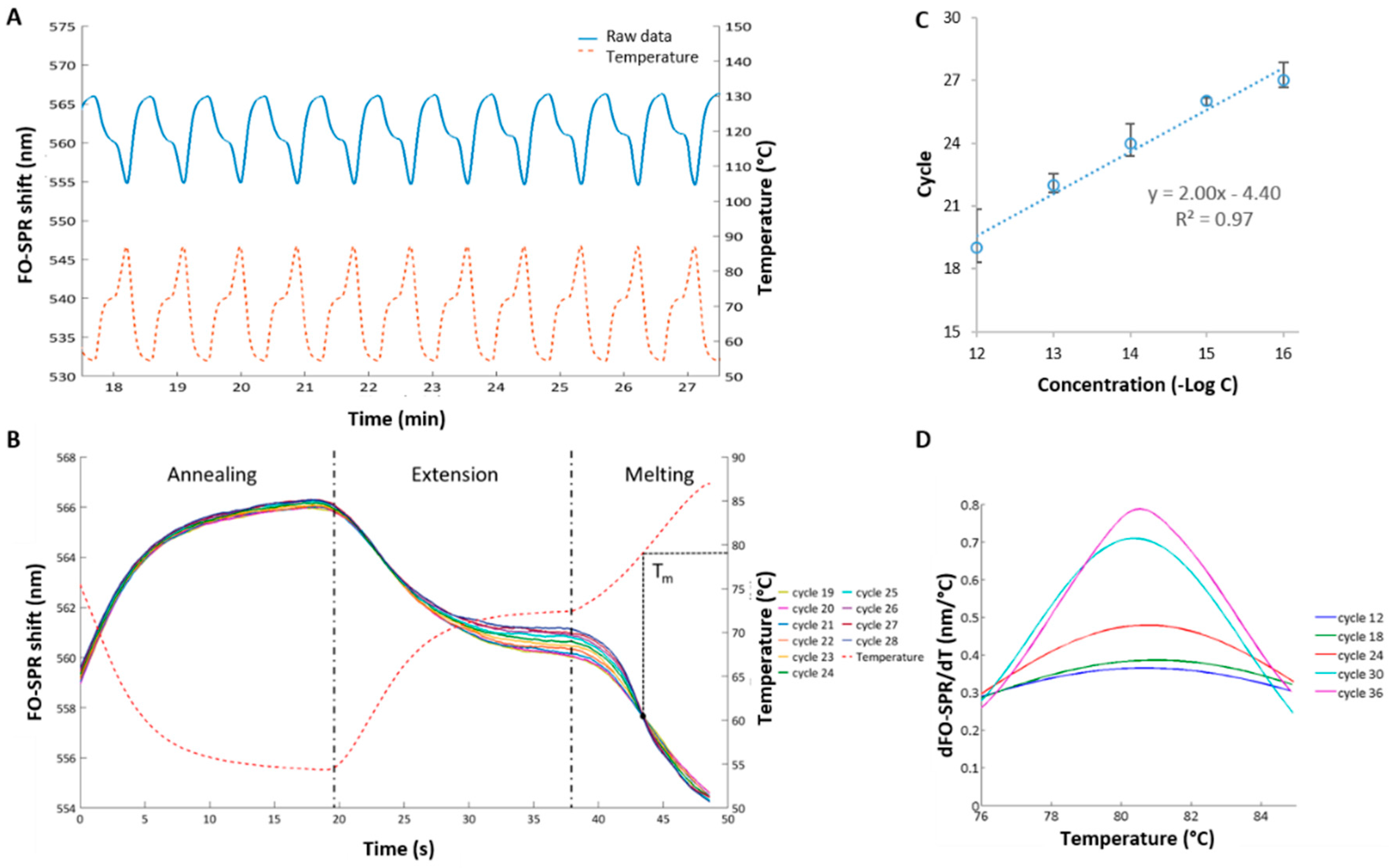
3.2. Establishing FO-PCR-MA for Celery DNA Detection
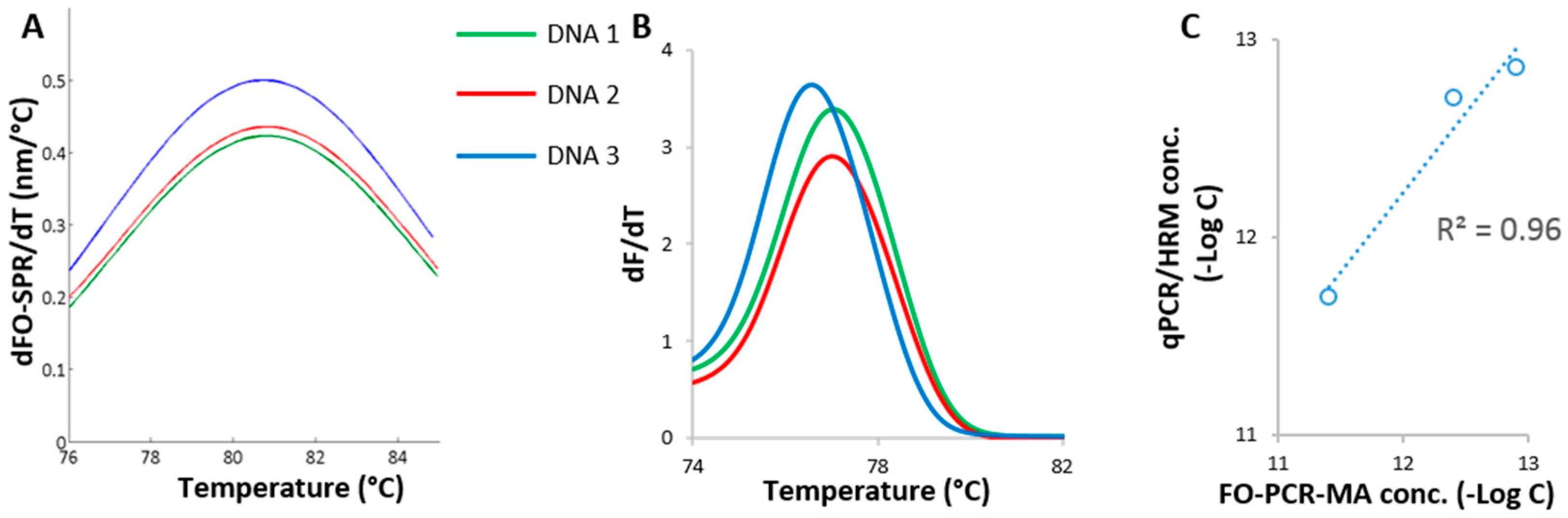
3.3. Validation of Established FO-PCR-MA with Biological Samples
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Wang, H. Detection of the allergenic celery protein component (Api g 1.01) in foods by immunoassay. Eur. Food Res. Technol. 2011, 223, 1023–1028. [Google Scholar] [CrossRef]
- Fuchs, M.; Cichna-Markl, M.; Hochegger, R. Development and validation of a duplex real-time PCR method for the simultaneous detection of celery and white mustard in food. Food Chem. 2013, 141, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Palle-Reisch, M.; Hochegger, R.; Cichna-Markl, M. Development and validation of a triplex real-time PCR assay for the simultaneous detection of three mustard species and three celery varieties in food. Food Chem. 2015, 184, 46–56. [Google Scholar] [CrossRef] [PubMed]
- European Commission. Directive 2003/89/EC of 10 November 2003 amending Directive 2000/13/EC as regards indications of the ingredients present in foodstuffs. Off. J. Eur. Union 2003, L308, 15–18. [Google Scholar]
- European Commission. Directive 1169/2011 of the European Parliament and of the Council on the provision of food information to consumers, amending Regulations (EC) No 1924/2006 and (EC) No 1925/2006 of the European Parliament and of the Council, and repealing Commission Directive 87/250/EEC, Council Directive 90/496/EEC, Commission Directive 1999/10/EC, Directive 2000/13/EC of the European Parliament and of the Council, Commission Directives 2002/67/EC and 2008/5/EC and Commission Regulation (EC) No 608/2004. Off. J. Eur. Union 2011, L304, 18. [Google Scholar]
- Faeste, C.; Jonscher, K. Differentiating Cross-Reacting Allergens in the Immunological Analysis of Celery (Apium graveolens) by mass spectrometry. J. AOAC Int. 2010, 93, 451–462. [Google Scholar] [PubMed]
- Sun, M.; Gao, H.; Xiao, X.; Chen, J.; Liu, C.; Feng, L. A novel loop-mediated isothermal amplification method for detection of the carrot materials in food. Eur. Food Res Technol. 2015, 214, 295–302. [Google Scholar] [CrossRef]
- Demmel, A.; Hahn, A. Molekularbiologische Methoden in der Lebensmittelanalytik, 1st ed.; Busch, U., Ed.; Springer: Berlin, Germany, 2010. [Google Scholar]
- Dovicovicova, L.; Olexová, L.; Pangallo, D.; Siekel, P.; Kuchta, T. Polymerase chain reaction (PCR) for the detection of celery (Apium graveolens) in food. Eur. Food Res. Technol. 2004, 218, 493–495. [Google Scholar] [CrossRef]
- Hupfer, C.; Waiblinger, H.; Busch, U. Development and validation of a real-time PCR detection method for celery in food. Eur. Food Res. Technol. 2007, 225, 329–335. [Google Scholar] [CrossRef]
- Mustorp, S.; Engdahl-Axelsson, C.; Svensson, U.; Holck, A. Detection of celery (Apium graveolens), mustard (Sinapis alba, Brassica juncea, Brassica nigra) and sesame (Sesamum indicum) in food by real-time PCR. Eur. Food Res. Technol. 2008, 226, 771–778. [Google Scholar] [CrossRef]
- Luber, F.; Demmel, A.; Pankofer, K.; Busch, A.; Engel, K.H. Simultaneous quantification of the food allergens soy bean, celery, white mustard and brown mustard via combination of teraplex real-time PCR and standard addition. Food Control 2015, 47, 246–253. [Google Scholar] [CrossRef]
- Stephan, O.; Weisz, N.; Vieths, S. Protein quantification, sandwich Elisa, and real time PCR used to monitor Industrial cleaning procedures for contamination with peanut and celery allergens. J. AOAC Int. 2004, 87, 1449–1457. [Google Scholar]
- Daems, D.; Knez, K.; Delport, F.; Spasic, D.; Lammertyn, J. Real-time PCR melting analysis with fiber optic SPR enables multiplex DNA identification of bacteria. Analyst 2016, 141, 1906–1911. [Google Scholar] [CrossRef] [PubMed]
- Schasfoort, R.B.M.; Tudos, A.J. Handbook of Surface Plasmon Resonance; Royal Society of Chemistry: Cambridge, UK, 2008. [Google Scholar]
- Homola, J. Surface plasmon resonance sensors for detection of chemical and biological species. Chem. Rev. 2008, 108, 462–493. [Google Scholar] [CrossRef] [PubMed]
- Pollet, J.; Janssen, K.P.F.; Knez, K.; Lammertyn, J. Real-time monitoring of solid-phase PCR using fiber-optic SPR. Small 2011, 7, 1003–1006. [Google Scholar] [CrossRef] [PubMed]
- Aray, A.; Chiavaioli, F.; Arjmand, M.; Trono, C.; Tombelli, S.; Giannetti, A.; Cennamo, N.; Soltanolkotabi, M.; Zeni, L.; Baldini, F. SPR-based plastic optical fibre biosensor for the detection of C-reactive protein in serum. J. Biophotonics 2016, 9, 1077–1084. [Google Scholar] [CrossRef] [PubMed]
- Caucheteur, C.; Guo, T.; Albert, J. Review of plasmonic fiber optic biochemical sensors: Improving the limit of detection. Anal. Bioanal. Chem. 2015, 407, 3883–3897. [Google Scholar] [CrossRef] [PubMed]
- Ribaut, C.; Loyez, M.; Larrieu, J.; Chevineau, S.; Lambert, P.; Remmelink, M.; Wattiez, R.; Caucheteur, C. Cancer biomarker sensing using packaged plasmonic optical fiber gratings: Towards in vivo diagnosis. Biosen. Bioelectron. 2017, 92, 449–456. [Google Scholar] [CrossRef] [PubMed]
- Knez, K.; Janssen, K.P.F.; Pollet, J.; Spasic, D.; Lammertyn, J. Fiber-optic high-resolution genetic screening using gold-labeled gene probes. Small 2012, 8, 868–872. [Google Scholar] [CrossRef] [PubMed]
- Knez, K.; Janssen, K.P.F.; Spasic, D.; Declerck, P.; Vanysacker, L.; Denis, C.; Tran, D.T.; Lammertyn, J. Spherical nucleic acid enhanced FO-SPR DNA melting for detection of mutations in Legionella pneumophila. Anal. Chem. 2013, 85, 1734–1742. [Google Scholar] [CrossRef] [PubMed]
- Tran, D.T.; Knez, K.; Janssen, K.P.F.; Pollet, J.; Spasic, D.; Lammertyn, J. Selection of aptamers against Ara h 1 protein for FO-SPR biosensing of peanut allergens in food matrices. Biosens. Bioelectron. 2013, 43, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; van Stappen, T.; Spasic, D.; Delport, F.; Vermeire, S.; Gils, A.; Lammertyn, J. Fiber optic-SPR platform for fast and sensitive infliximab detection in serum of inflammatory bowel disease patients. Biosens. Bioelectron. 2016, 79, 173–179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pollet, J.; Delport, F.; Janssen, K.P.F.; Tran, D.T.; Wouters, J.; Verbiest, T.; Lammertyn, J. Fast and accurate peanut allergen detection with nanobead enhanced optical fiber SPR biosensor. Talanta 2011, 83, 1436–2441. [Google Scholar] [CrossRef] [PubMed]
- Pollet, J.; Delport, F.; Janssen, K.P.F.; Jans, K.; Maes, G.; Pfeiffer, H.; Wevers, M.; Lammertyn, J. Fiber optic SPR biosensing of DNA hybridization and DNA-protein interactions. Biosens. Bioelectron. 2009, 25, 864–869. [Google Scholar] [CrossRef] [PubMed]
- Pafundo, S.; Gullì, M.; Marmiroli, N. Comparison of DNA extraction methods and development of duplex PCR and real-time PCR to detect tomato, carrot, and celery in food. J. Agric. Food Chem. 2011, 59, 10414–10424. [Google Scholar] [CrossRef] [PubMed]
- Malanoski, A.P.; Lin, B.; Wang, Z.; Schnur, J.M.; Stenger, S.A. Automated identification of multiple micro-organisms from resequencing DNA microarrays. Nucleic Acids Res. 2006, 34, 5300–5311. [Google Scholar] [CrossRef] [PubMed]
- Hurst, S.J.; Lytton-Jean, A.K.; Mirkin, C.A. Maximizing DNA loading on a range of gold nanoparticle sizes. Anal. Chem. 2006, 78, 8313–8318. [Google Scholar] [CrossRef] [PubMed]
- Kock, G.G. Intraclass correlation coefficient. In Encyclopedia of Statistical Sciences; Kotz, S., Johnson, N.L., Eds.; John Wiley and Sons: New York, NY, USA, 1982; pp. 213–217. [Google Scholar]
| Targets (5′ → 3′) | Sequence |
|---|---|
| Api g 1 (76-mer) | GGG CTT TGT CAT TGA TGT TGA CAC AGT CCT TCC CAA GGC TGC GCC TGG AGC TTA CAA GAG TGT CGA AAT CAA GGG A |
| Mtd (101-mer) | CGA TGA GCG TGT ACT GAG TCA GTG TTA TGT TTG GAT TAC GGT GTG ATG AGT CAG CGT TAT CTG TTT TTA TAT GTT TGG TAT GAT TAA TGT TAG TTC CTA TT |
| Mtd 2 (101-mer) | CCT TGT TAG CGG AGT CTA AAT CGG AAT CTA AAT CTA AAT CAT TTT AAG CAT GTT AGC CCT TGT ATT TTG GCT TTT GGC TTT TAA CCA TTT TGT TC |
| Mtd 3 (109-mer) | CCC GTA CGA GAT ATA TTT TTG TCT GGT TTG AGA TAT ATA TTA CAT GCT GAG TCA CGA TGA GCG TGT ACT GAG TCA GTG TTA TGT TTG GAT TAC GGT GTG ATG AGT CAG C |
| Primers (5′ → 3′) | |
| Api g 1 forward (22-mer) | GGG CTT TGT CAT TGA TGT TGA C |
| Api g 1 reverse (24-mer) | TCC CTT GAT TTC GAC ACT CTT GTA |
| Mtd forward (20-mer) | CGA TGA GCG TGT ACT GAG TC |
| Mtd reverse (29-mer) | AAT AGG AAC TAA CAT TAA TCA TAC CAA AC |
| Mtd 2 forward (24-mer) | CCT TGT TAG CGG AGT CTA AAT CGG |
| Mtd 2 reverse (20-mer) | GAA CAA AAT GGT TAA AAG CC |
| Mtd 3 forward (26-mer) | CCC GTA CGA GAT ATA TTT TTG TCT GG |
| Mtd 3 reverse (23-mer) | GCT GAC TCA TCA CAC CGT AAT CC |
| Hybridization probes (5′ → 3′) | |
| Mtd 3 probe 1 (66-mer) | GTG ACT CAG CAT GTA ATA TAT ATC TCA AAC CAG ACA AAA ATA TAT CTC GTA CGG GTT TTT TTT TT |
| Mtd 3 probe 2 (64-mer) | TTT TTT TTT TGC TGA CTC ATC ACA CCG TAA TCC AAA CAT AAC ACT GAC TCA GTA CAC GCT CAT C |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Daems, D.; Peeters, B.; Delport, F.; Remans, T.; Lammertyn, J.; Spasic, D. Identification and Quantification of Celery Allergens Using Fiber Optic Surface Plasmon Resonance PCR. Sensors 2017, 17, 1754. https://doi.org/10.3390/s17081754
Daems D, Peeters B, Delport F, Remans T, Lammertyn J, Spasic D. Identification and Quantification of Celery Allergens Using Fiber Optic Surface Plasmon Resonance PCR. Sensors. 2017; 17(8):1754. https://doi.org/10.3390/s17081754
Chicago/Turabian StyleDaems, Devin, Bernd Peeters, Filip Delport, Tony Remans, Jeroen Lammertyn, and Dragana Spasic. 2017. "Identification and Quantification of Celery Allergens Using Fiber Optic Surface Plasmon Resonance PCR" Sensors 17, no. 8: 1754. https://doi.org/10.3390/s17081754





