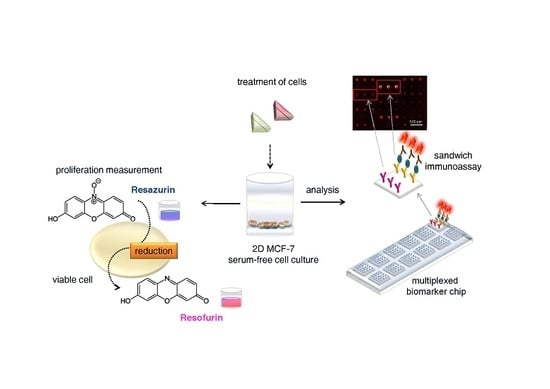
A Chip for Estrogen Receptor Action: Detection of Biomarkers Released by MCF-7 Cells through Estrogenic and Anti-Estrogenic Effects
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Line and Cultivation
2.2. Cell Cultivation in Serum-/Phenol Red-Free Medium
2.3. Cell Treatment for Biomarker and Proliferation Assays
2.4. Resazurin Proliferation Assay
2.5. Materials and Reagents for the Protein Microarray
2.6. Protein Microarray Fabrication, Processing, and Scanning
2.7. Statistical Data Analysis
3. Results
3.1. Compilation of the Biomarker Panel
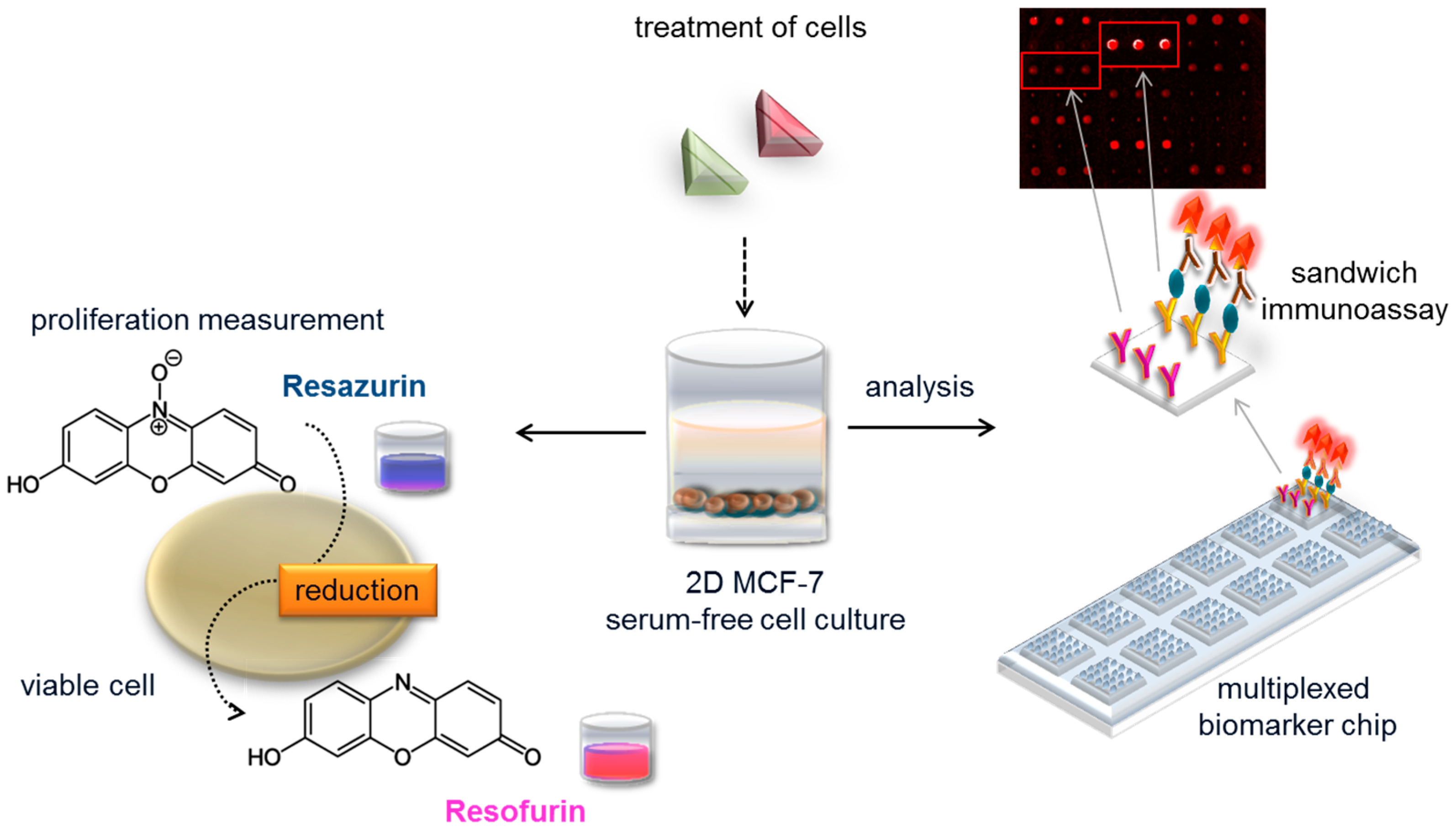
3.2. Assay Performance in Serum-Free Cell Culture Medium
3.3. Quantification of Biomarker Secretion in Cell Culture Supernatant
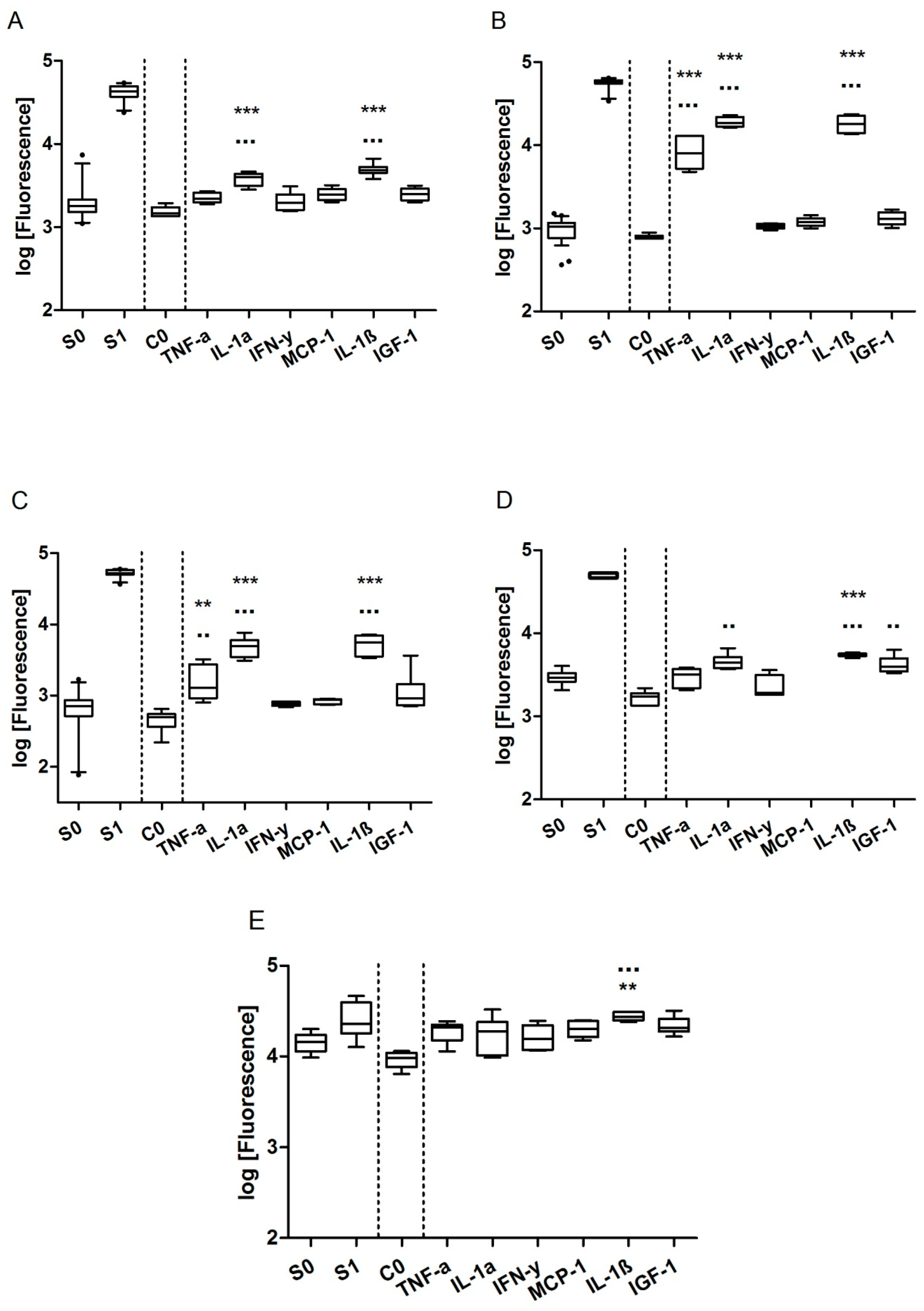
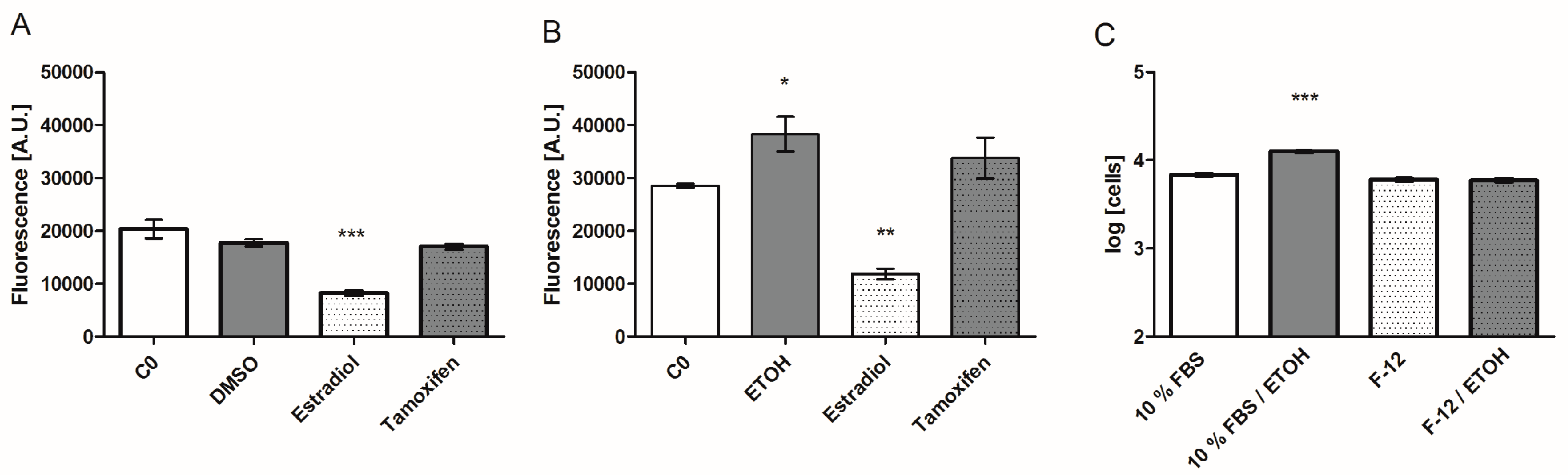
3.4. Effect of Solvents on Biomarker Secretion and Cell Proliferation
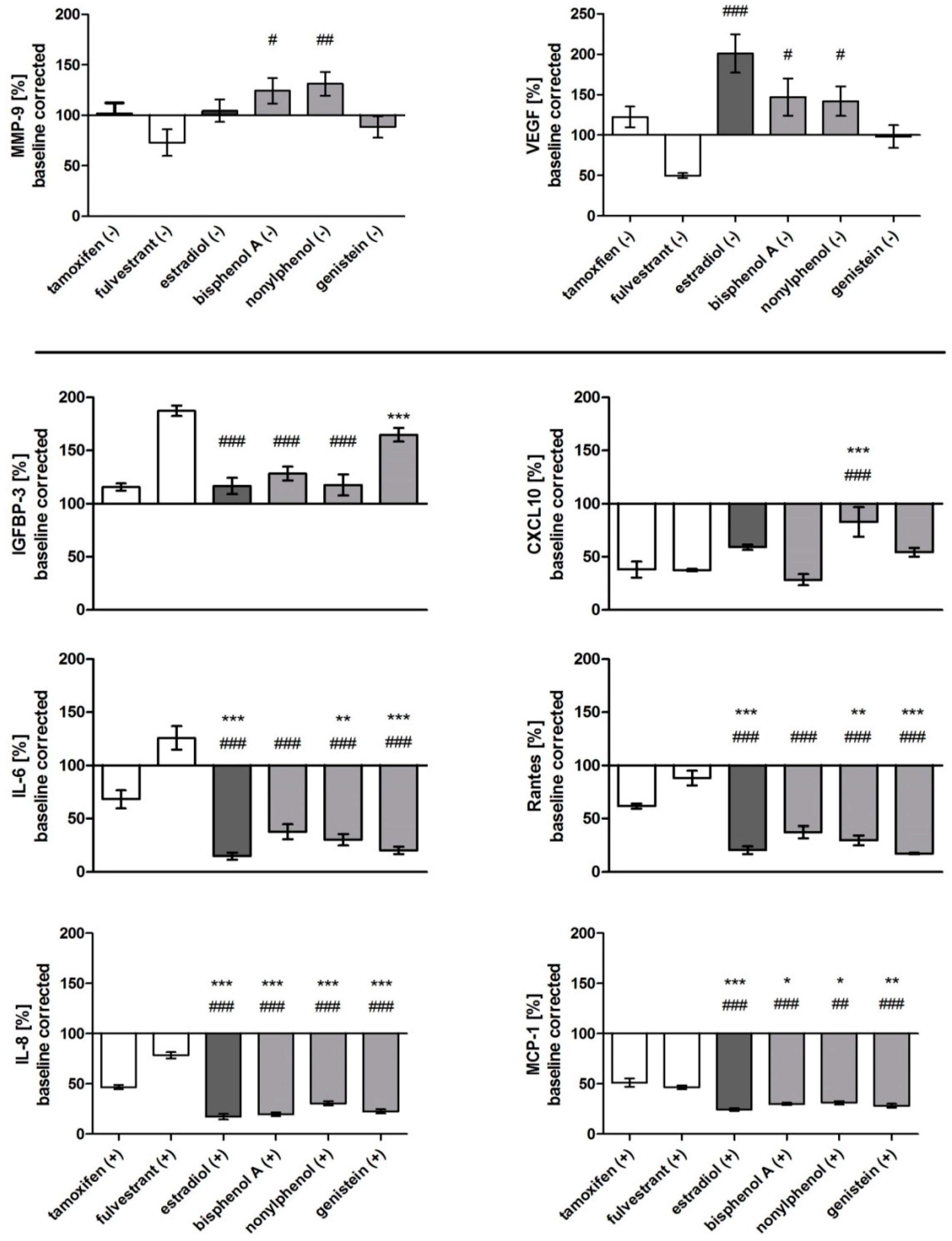
3.5. Specific Biomarker Secretion Patterns and Proliferative Effect of ER Agonists and Antagonists
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Damstra, T.; Barlow, S.; Bergman, A.; Kavlock, R.; Van Der Kraak, G. Global Assessment of the State-Of-The-Science of Endocrine Disruptors; no. WHO/PCS/EDC/02.2 180; WHO Publication: Geneva, Switzerland, 2002. [Google Scholar]
- Rochester, J.R. Bisphenol A and human health: A review of the literature. Reprod. Toxicol. 2013, 42, 132–155. [Google Scholar] [CrossRef] [PubMed]
- Casals-Casas, C.; Desvergne, B. Endocrine Disruptors: From Endocrine to Metabolic Disruption. Annu. Rev. Physiol. 2011, 73, 135–162. [Google Scholar] [CrossRef] [PubMed]
- Mnif, W.; Hassine, A.I.H.; Bouaziz, A.; Bartegi, A.; Thomas, O.; Roig, B. Effect of endocrine disruptor pesticides: A review. Int. J. Environ. Res. Public Health 2011, 8, 2265–2303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patisaul, H.B.; Adewale, H.B. Long-term effects of environmental endocrine disruptors on reproductive physiology and behavior. Front. Behav. Neurosci. 2009, 3, 10. [Google Scholar] [CrossRef] [PubMed]
- Yang, O.; Kim, H.L.; Weon, J.-I.; Seo, Y.R. Endocrine-disrupting Chemicals: Review of Toxicological Mechanisms Using Molecular Pathway Analysis. J. Cancer Prev. 2015, 20, 12–24. [Google Scholar] [CrossRef] [PubMed]
- Melnick, R.; Lucier, G.; Wolfe, M.; Hall, R.; Stancel, G.; Prins, G.; Gallo, M.; Reuhl, K.; Ho, S.M.; Brown, T.; et al. Summary of the National Toxicology Program’s report of the endocrine disruptors low-dose peer review. Environ. Health Perspect. 2002, 110, 427–431. [Google Scholar] [CrossRef] [PubMed]
- Vandenberg, L.N.; Colborn, T.; Hayes, T.B.; Heindel, J.J.; Jacobs, D.R.; Lee, D.H.; Soto, A.M.; Saal, F.S.; Welshons, W.V.; Zoeller, R.T.; et al. Hormones and Endocrine—Disrupting Chemicals: Low—Dose Effects and Nonmonotonic Dose Responses. Endocr. Rev. 2014, 33, 378–455. [Google Scholar] [CrossRef] [PubMed]
- Payne, J.; Rajapakse, N.; Wilkins, M.; Kortenkamp, A. Prediction and assessment of the effects of mixtures of four xenoestrogens. Environ. Health Perspect. 2000, 108, 983–987. [Google Scholar] [CrossRef] [PubMed]
- Rajapakse, N.; Silva, E.; Kortenkamp, A. Combining xenoestrogens at levels below individual no-observed-effect concentrations dramatically enhances steroid hormone action. Environ. Health Perspect. 2002, 110, 917–921. [Google Scholar] [CrossRef] [PubMed]
- Gore, A.C.; Chappell, V.A.; Fenton, S.E.; Flaws, J.A.; Nadal, A.; Prins, G.S.; Toppari, J.; Zoeller, R.T. Executive Summary to EDC-2: The Endocrine Society’s second Scientific Statement on endocrine-disrupting chemicals. Endocr. Rev. 2015, 36, E1–E150. [Google Scholar] [CrossRef] [PubMed]
- Routledge, E.J.; Sumpter, J.P. Estrogenic activity of surfactants and some of their degradation products assessed using a recombinant yeast screen. Environ. Toxicol. Chem. 1996, 15, 241–248. [Google Scholar] [CrossRef]
- Seifert, M.; Haindl, S.; Hock, B. Development of an enzyme linked receptor assay (ELRA) for estrogens and xenoestrogens. Anal. Chim. Acta 1999, 386, 191–199. [Google Scholar] [CrossRef]
- Van der Burg, B.; Winter, R.; Weimer, M.; Berckmans, P.; Suzuki, G.; Gijsbers, L.; Jonas, A.; van der Linden, S.; Witters, H.; Aarts, J.; et al. Optimization and prevalidation of the in vitro ERα CALUX method to test estrogenic and antiestrogenic activity of compounds. Reprod. Toxicol. 2010, 30, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Soto, A.M.; Sonnenschein, C.; Chung, K.L.; Fernandez, M.F.; Olea, N.; Olea Serrano, F. The E-SCREEN assay as a tool to identify estrogens: An update on estrogenic environmental pollutants. Environ. Health Perspect. 1995, 103, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Francois, E.; Wang, D.Y.; Fulthorpe, R.; Liss, S.N.; Edwards, E.A. DNA microarrays for detecting endocrine-disrupting compounds. Biotechnol. Adv. 2003, 22, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Sumpter, J.P.; Jobling, S. Vitellogenesis as a biomarker for estrogenic contamination of the aquatic environment. Environ. Health Perspect. 1995, 103, 173–178. [Google Scholar] [CrossRef] [PubMed]
- Hecker, M.; Giesy, J.P. Novel trends in endocrine disruptor testing: The H295R Steroidogenesis Assay for identification of inducers and inhibitors of hormone production. Anal. Bioanal Chem. 2008, 390, 287–291. [Google Scholar] [CrossRef] [PubMed]
- Kiyama, R.; Wada-Kiyama, Y. Estrogenic endocrine disruptors: Molecular mechanisms of action. Environ. Int. 2015, 83, 11–40. [Google Scholar] [CrossRef] [PubMed]
- Bovee, T.F.H.; Hoogenboom, L.A.P.; Thomson, B.M. Bioassays for the detection of hormonal activities. In Endocrine-Disrupting Chemicals in Food; Shaw, I., Ed.; Woodhead Publishing Limited: Sawston, UK, 2009; pp. 259–280. ISBN 9781845692186. [Google Scholar]
- De Paoli, M.; Gogalic, S.; Sauer, U.; Preininger, C.; Pandha, H.; Simpson, G.; Horvath, A.; Marquette, C. Multiplatform Biomarker Discovery for Bladder Cancer Recurrence Diagnosis. Dis. Markers 2016, 2016, 4591910. [Google Scholar] [CrossRef] [PubMed]
- Buchegger, P.; Sauer, U.; Toth-Székély, H.; Preininger, C. Miniaturized protein microarray with internal calibration as point-of-care device for diagnosis of neonatal sepsis. Sensors 2012, 12, 1494–1508. [Google Scholar] [CrossRef] [PubMed]
- Sanjay, S.T.; Fu, G.; Dou, M.; Xu, F.; Liu, R.; Qi, H.; Li, X.J. Biomarker detection for disease diagnosis using cost-effective microfluidic platforms. Analyst 2015, 140, 7062–7081. [Google Scholar] [CrossRef] [PubMed]
- Piraino, F.; Volpetti, F.; Watson, C.; Maerkl, S.J. A digital-analog microfluidic platform for patient-centric multiplexed biomarker diagnostics of ultralow volume samples. ACS Nano 2016, 10, 1699–1710. [Google Scholar] [CrossRef] [PubMed]
- Gogalic, S.; Sauer, U.; Doppler, S.; Heinzel, A.; Perco, P.; Lukas, A.; Simpson, G.; Pandha, H.; Horvath, A.; Preininger, C. Validation of a protein panel for the non-invasive detection of recurrent non-muscle invasive bladder cancer. Biomarkers 2017, 19, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Preininger, C.; Bodrossy, L.; Sauer, U.; Pichler, R.; Weilharter, A. ARChip Epoxy and ARChip UV for covalent on-chip immobilization of pmoA gene-specific oligonucleotides. Anal. Biochem. 2004, 330, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Domnanich, P.; Sauer, U.; Pultar, J.; Preininger, C. Protein microarray for the analysis of human melanoma biomarkers. Sens. Actuator B Chem. 2009, 139, 2–8. [Google Scholar] [CrossRef]
- MacDougall, D.; Crummett, W.B. Guidelines for data acquisition and data quality evaluation in environmental chemistry. Anal. Chem. 1980, 52, 2242–2249. [Google Scholar] [CrossRef]
- Freund, A.; Jolivel, V.; Durand, S.; Kersual, N.; Chalbos, D.; Chavey, C.; Vignon, F.; Lazennec, G. Mechanisms underlying differential expression of interleukin-8 in breast cancer cells. Oncogene 2004, 23, 6105–6114. [Google Scholar] [CrossRef] [PubMed]
- Hollingshead, B.D.; Beischlag, T.V.; DiNatale, B.C.; Ramadoss, P.; Perdew, G.H. Inflammatory signaling and aryl hydrocarbon receptor mediate synergistic induction of interleukin 6 in MCF-7 cells. Cancer Res. 2008, 68, 3609–3617. [Google Scholar] [CrossRef] [PubMed]
- Bronger, H.; Kraeft, S.; Schwarz-Boeger, U.; Cerny, C.; Stöckel, A.; Avril, S.; Kiechle, M.; Schmitt, M. Modulation of CXCR3 ligand secretion by prostaglandin E2 and cyclooxygenase inhibitors in human breast cancer. Breast Cancer Res. 2012, 14, R30. [Google Scholar] [CrossRef] [PubMed]
- Mira, E.; Lacalle, R.A.; González, M.A.; Gómez-Moutón, C.; Abad, J.L.; Bernad, A.; Martínez, C.; Santos, M. A role for chemokine receptor transactivation in growth factor signaling. EMBO Rep. 2001, 2, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Jamalzadeh, L.; Ghafoori, H.; Sariri, R.; Rabuti, H.; Nasirzade, J.; Hasani, H.; Aghamaali, M.R. Cytotoxic Effects of Some Common Organic Solvents on MCF-7, RAW-264.7 and Human Umbilical Vein Endothelial Cells. Avicenna J. Med. Biochem. 2016, 4, 1–6. [Google Scholar] [CrossRef]
- Wetherill, Y.B.; Akingbemi, B.T.; Kanno, J.; McLachlan, J.A.; Nadal, A.; Sonnenschein, C.; Watson, C.S.; Zoeller, R.T.; Belcher, S.M. In vitro molecular mechanisms of bisphenol A action. Reprod. Toxicol. 2007, 24, 178–198. [Google Scholar] [CrossRef] [PubMed]
- Payne, J.; Jones, C.; Lakhani, S.; Kortenkamp, A. Improving the reproducibility of the MCF-7 cell proliferation assay for the detection of xenoestrogens. Sci. Total Environ. 2000, 248, 51–62. [Google Scholar] [CrossRef]
- Van Meeuwen, J.A.; ter Burg, W.; Piersma, A.H.; van den Berg, M.; Sanderson, J.T. Mixture effects of estrogenic compounds on proliferation and pS2 expression of MCF-7 human breast cancer cells. Food Chem. Toxicol. 2007, 45, 2319–2330. [Google Scholar] [CrossRef] [PubMed]
- Johnstone, C.N.; Chand, A.; Putoczki, T.L.; Ernst, M. Emerging roles for IL-11 signaling in cancer development and progression: Focus on breast cancer. Cytokine Growth Factor Rev. 2015, 26, 489–498. [Google Scholar] [CrossRef] [PubMed]
- Soria, G.; Ben-Baruch, A. The inflammatory chemokines CCL2 and CCL5 in breast cancer. Cancer Lett. 2008, 267, 271–285. [Google Scholar] [CrossRef] [PubMed]
- Chiu, J.J.; Sgagias, M.K.; Cowan, K.H. Interleukin 6 acts as a paracrine growth factor in human mammary carcinoma cell lines. Clin. Cancer Res. 1996, 2, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Schafer, Z.T.; Brugge, J.S. IL-6 involvement in epithelial cancers. J. Clin. Investig. 2007, 117, 3660–3663. [Google Scholar] [CrossRef] [PubMed]
- Bendrik, C.; Dabrosin, C. Estradiol increases IL-8 secretion of normal human breast tissue and breast cancer in vivo. J. Immunol. 2009, 182, 371–378. [Google Scholar] [CrossRef] [PubMed]
- Garvin, S.; Nilsson, U.W.; Dabrosin, C. Effects of oestradiol and tamoxifen on VEGF, soluble VEGFR-1, and VEGFR-2 in breast cancer and endothelial cells. Br. J. Cancer 2005, 93, 1005–1010. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, U.W.; Garvin, S.; Dabrosin, C. MMP-2 and MMP-9 activity is regulated by estradiol and tamoxifen in cultured human breast cancer cells. Breast Cancer Res. Treat. 2007, 102, 253–261. [Google Scholar] [CrossRef] [PubMed]
- Roomi, M.W.; Monterrey, J.C.; Kalinovsky, T.; Rath, M.; Niedzwiecki, A. Patterns of MMP-2 and MMP-9 expression in human cancer cell lines. Oncol. Rep. 2009, 21, 1323–1333. [Google Scholar] [CrossRef] [PubMed]
- Goldberg-Bittman, L.; Neumark, E.; Sagi-Assif, O.; Azenshtein, E.; Meshel, T.; Witz, I.P.; Ben-Baruch, A. The expression of the chemokine receptor CXCR3 and its ligand, CXCL10, in human breast adenocarcinoma cell lines. Immunol. Lett. 2004, 92, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Cardona, S.M.; Garcia, J.A.; Cardona, A.E. The fine balance of chemokines during disease: trafficking, inflammation, and homeostasis. Methods Mol. Biol. 2013, 1013, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Colston, K.W.; Perks, C.M.; Xie, S.P.; Holly, J.M. Growth inhibition of both MCF-7 and Hs578T human breast cancer cell lines by vitamin D analogues is associated with increased expression of insulin-like growth factor binding protein-3. J. Mol. Endocrinol. 1998, 20, 157–162. [Google Scholar] [CrossRef] [PubMed]
- Lorincz, A.M.; Sukumar, S. Molecular links between obesity and breast cancer. Endocr. Relat. Cancer 2006, 13, 279–292. [Google Scholar] [CrossRef] [PubMed]
- Dupont, J.; Le Roith, D. Insulin-like growth factor 1 and oestradiol promote cell proliferation of MCF-7 breast cancer cells: new insights into their synergistic effects. Mol. Pathol. 2001, 54, 149–154. [Google Scholar] [CrossRef] [PubMed]
- Figueroa, J.A.; Jackson, J.G.; McGuire, W.L.; Krywicki, R.F.; Yee, D. Expression of insulin-like growth factor binding proteins in human breast cancer correlates with estrogen receptor status. J. Cell. Biochem. 1993, 52, 196–205. [Google Scholar] [CrossRef] [PubMed]
- Salahifar, H.; Baxter, R.C.; Martin, J.L. Insulin-like growth factor binding protein (IGFBP)-3 protease activity secreted by MCF-7 breast cancer cells: inhibition by IGFs does not require IGF-IGFBP interaction. Endocrinology 1997, 138, 1683–1690. [Google Scholar] [CrossRef] [PubMed]
- Kawashima, I.; Ohsumi, J.; Mita-Honjo, K.; Shimoda-Takano, K.; Ishikawa, H.; Sakakibara, S.; Miyadai, K.; Takiguchi, Y. Molecular cloning of cDNS encoding adipogenisis inhibitory factor and identity with interleukin-11. FEBS Lett. 1991, 283, 199–202. [Google Scholar] [CrossRef]
- Dubois, V.; Couissi, D.; Schonne, E.; Remacle, C.; Trouet, A. Intracellular levels and secretion of insulin-like-growth-factor-binding proteins in MCF-7/6, MCF-7/AZ and MDA-MB-231 breast cancer cells. Differential modulation by estrogens in serum-free medium. Eur. J. Biochem. 1995, 232, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Inadera, H.; Sekiya, T.; Yoshimura, T.; Matsushima, K. Molecular analysis of the inhibition of monocyte chemoattractant protein-1 gene expression by estrogen and xenoestrogens in MCF-7 cells. Endocrinology 2000, 141, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Kanda, N.; Watanabe, S. 17β-estradiol inhibits the production of RANTES in human keratinocytes. J. Investig. Dermatol. 2003, 120, 420–427. [Google Scholar] [CrossRef] [PubMed]
- Chrzan, B.G.; Bradford, P.G. Phytoestrogens activate estrogen receptor beta1 and estrogenic responses in human breast and bone cancer cell lines. Mol. Nutr. Food Res. 2007, 51, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Kayisli, U.A.; Mahutte, N.G.; Arici, A. Uterine chemokines in reproductive physiology and pathology. Am. J. Reprod. Immunol. 2002, 47, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Hull, K.L.; Harvey, S. Growth Hormone and Reproduction: A Review of Endocrine and Autocrine/Paracrine Interactions. Int. J. Endocrinol. 2014. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, J. Effective Blocking Procedures; ELISA Tech Bull: Kennebunk, ME, USA, 2001; pp. 1–6. [Google Scholar]
- Kishore, V.; Sangeetha Gowda, K.R.; Krishna, S.; Sharma, K.; Rashmi, M.; Nishita, K.P. Bovine Serum Albumin a Potential Thermostabilizer: a Study on α—Amylase. J. Appl. Environ. Microbiol. 2014, 2, 37–41. [Google Scholar] [CrossRef]
- Seo, H.S.; DeNardo, D.G.; Jacquot, Y.; Laïos, I.; Vidal, D.S.; Zambrana, C.R.; Leclercq, G.; Brown, P.H. Stimulatory effect of genistein and apigenin on the growth of breast cancer cells correlates with their ability to activate ER alpha. Breast Cancer Res. Treat. 2006, 99, 121–134. [Google Scholar] [CrossRef] [PubMed]
- Sonnenschein, C.; Soto, A.M.; Fernandez, M.F.; Olea, N.; Olea-Serrano, M.F.; Ruiz-Lopez, M.D. Development of a marker of estrogenic exposure in human serum. Clin. Chem. 1995, 41, 1888–1895. [Google Scholar] [PubMed]
- Etique, N.; Chardard, D.; Chesnel, A.; Merlin, J.L.; Flament, S.; Grillier-Vuissoz, I. Ethanol stimulates proliferation, ERalpha and aromatase expression in MCF-7 human breast cancer cells. Int. J. Mol. Med. 2004, 13, 149–155. [Google Scholar] [PubMed]
- Etique, N.; Grillier-Vuissoz, I.; Flament, S. Ethanol stimulates the secretion of matrix metalloproteinases 2 and 9 in MCF-7 human breast cancer cells. Oncol. Rep. 2006, 15, 603–608. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Meng, Q.; Gao, B.; Grossman, J.; Yadegari, M.; Goldberg, I.D.; Rosen, E.M. Alcohol stimulates estrogen receptor signaling in human breast cancer cell lines. Cancer Res. 2000, 60, 5635–5639. [Google Scholar] [PubMed]
- Katzenellenbogen, B.S.; Kendra, K.L.; Norman, M.J.; Katzenellenbogen, B.S.; Rendra, K.L.; Norman, M.J.; Berthois, Y. Proliferation, Hormonal Responsiveness, and Estrogen Receptor Content of MCF-7 Human Breast Cancer Cells Grown in the Short-Term and Long-Term Absence of Estrogens Proliferation. Cancer Res. 1987, 47, 4355–4360. [Google Scholar] [PubMed]
- Maggiolini, M.; Bonofiglio, D.; Marsico, S.; Panno, M.L.; Cenni, B.; Picard, D.; Andò, S. Estrogen receptor alpha mediates the proliferative but not the cytotoxic dose-dependent effects of two major phytoestrogens on human breast cancer cells. Mol. Pharmacol. 2001, 60, 595–602. [Google Scholar] [PubMed]
- Scott, S.M.; Brown, M.; Come, S.E. Emerging data on the efficacy and safety of fulvestrant, a unique antiestrogen therapy for advanced breast cancer. Expert Opin. Drug Saf. 2011, 10, 819–826. [Google Scholar] [CrossRef] [PubMed]

| Biomarker | LOD pg/mL | LOQ pg/mL | EC50 pg/mL | Recovery Rate % | CV % |
|---|---|---|---|---|---|
| MCP-1 (CCL2) | 4 ± 2 | 16 ± 7 | 2431± 1668 | 83 | 14 |
| IL-6 | 5 ± 2 | 16 ± 6 | 3916 ± 1155 | 132 | 13 |
| Rantes (CCL5) | 16 ± 7 | 46 ± 21 | 4400 ± 1841 | 144 | 11 |
| VEGF | 20 ± 12 | 61 ± 25 | 3923 ± 938 | 131 | 15 |
| IL-8 | 53 ± 17 | 196 ± 97 | 3216 ± 678 | 80 | 22 |
| CXCL10 (IP-10) | 91 ± 24 | 321 ± 119 | 7852 ± 2275 | 116 | 19 |
| IL-11 | 157 ± 26 | 530 ± 199 | 26,330 ± 13,022 | 105 | 13 |
| IGFBP-3 | 581 ± 254 | 1630 ± 477 | 30,644 ± 3011 | 104 | 17 |
| MMP-9 | 669 ± 197 | 2035 ± 447 | 49,071 ± 7070 | 118 | 19 |
| IGF-1 | 803 ± 122 | 2553 ± 453 | 39,523 ± 10,874 | 87 | 15 |
| Tamoxifen | Fulvestrant | Estradiol | Genistein | BPA | Nonylphenol | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MMP-9 - | 102 | 73 | 105 | 89 | 124 | 131 | |||||
| VEGF - | 122 | 50 | 201 | 98 | 147 | 142 | |||||
| IGFBP-3 + | 133 | 213 | 130 | 190 | 145 | 136 | |||||
| IL-6 + | 68 | 126 | 15 | 20 | 38 | 30 | |||||
| IL-8 + | 47 | 78 | 17 | 23 | 20 | 31 | |||||
| MCP-1 + | 51 | 47 | 24 | 28 | 30 | 31 | |||||
| Rantes + | 62 | 88 | 21 | 17 | 37 | 30 | |||||
| CXCL-10 + | 38 | 38 | 59 | 54 | 28 | 83 | |||||
| IL-11 + | 139 | 328 | 192 | 268 | 295 | 198 | |||||
| IGF-1 + | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. |
 .
.© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gier, K.; Preininger, C.; Sauer, U. A Chip for Estrogen Receptor Action: Detection of Biomarkers Released by MCF-7 Cells through Estrogenic and Anti-Estrogenic Effects. Sensors 2017, 17, 1760. https://doi.org/10.3390/s17081760
Gier K, Preininger C, Sauer U. A Chip for Estrogen Receptor Action: Detection of Biomarkers Released by MCF-7 Cells through Estrogenic and Anti-Estrogenic Effects. Sensors. 2017; 17(8):1760. https://doi.org/10.3390/s17081760
Chicago/Turabian StyleGier, Konstanze, Claudia Preininger, and Ursula Sauer. 2017. "A Chip for Estrogen Receptor Action: Detection of Biomarkers Released by MCF-7 Cells through Estrogenic and Anti-Estrogenic Effects" Sensors 17, no. 8: 1760. https://doi.org/10.3390/s17081760