Online Removal of Baseline Shift with a Polynomial Function for Hemodynamic Monitoring Using Near-Infrared Spectroscopy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Detrending Method
2.2. Solid Phantom
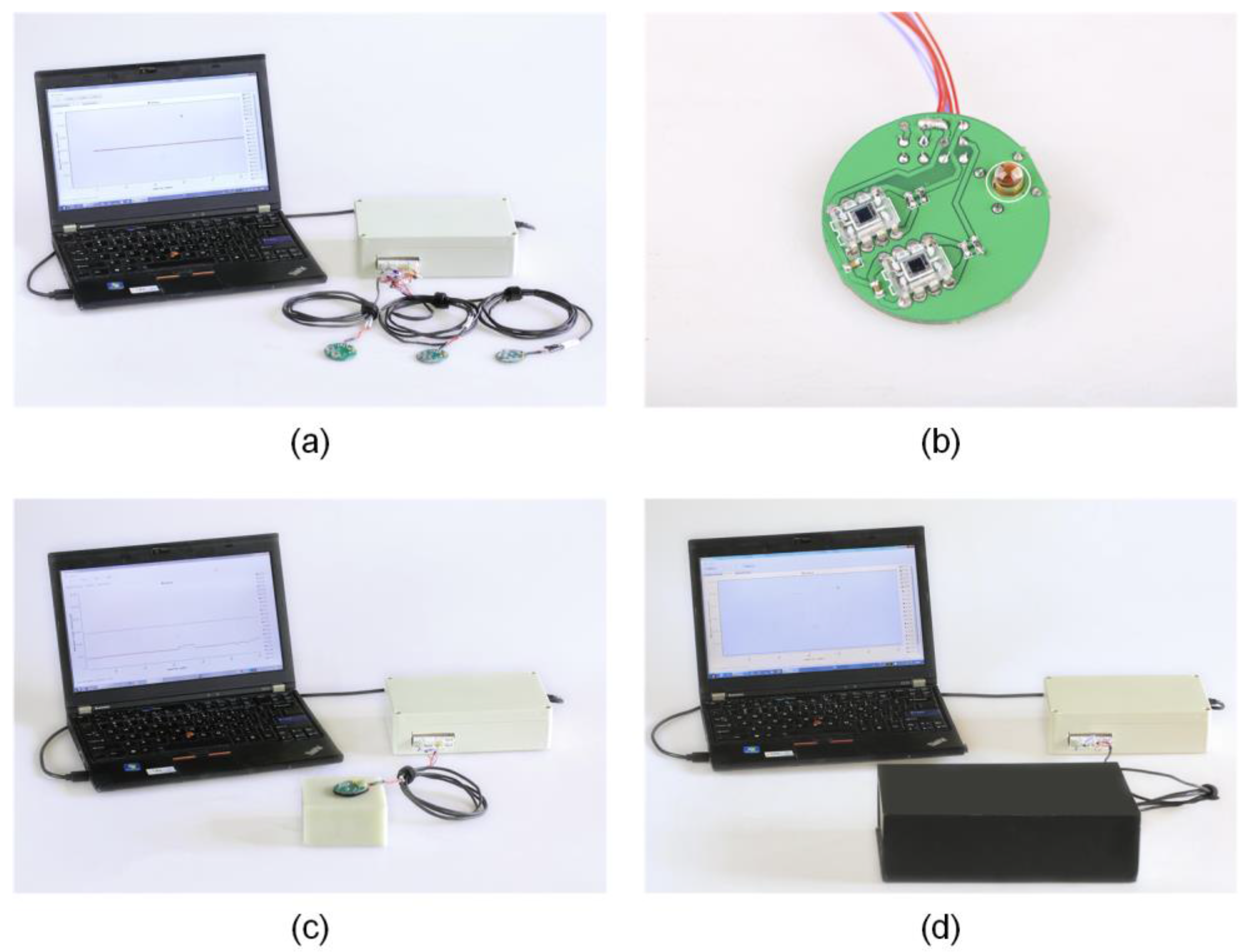
2.3. Data Acquisition System
2.4. Baseline Extraction and Removal
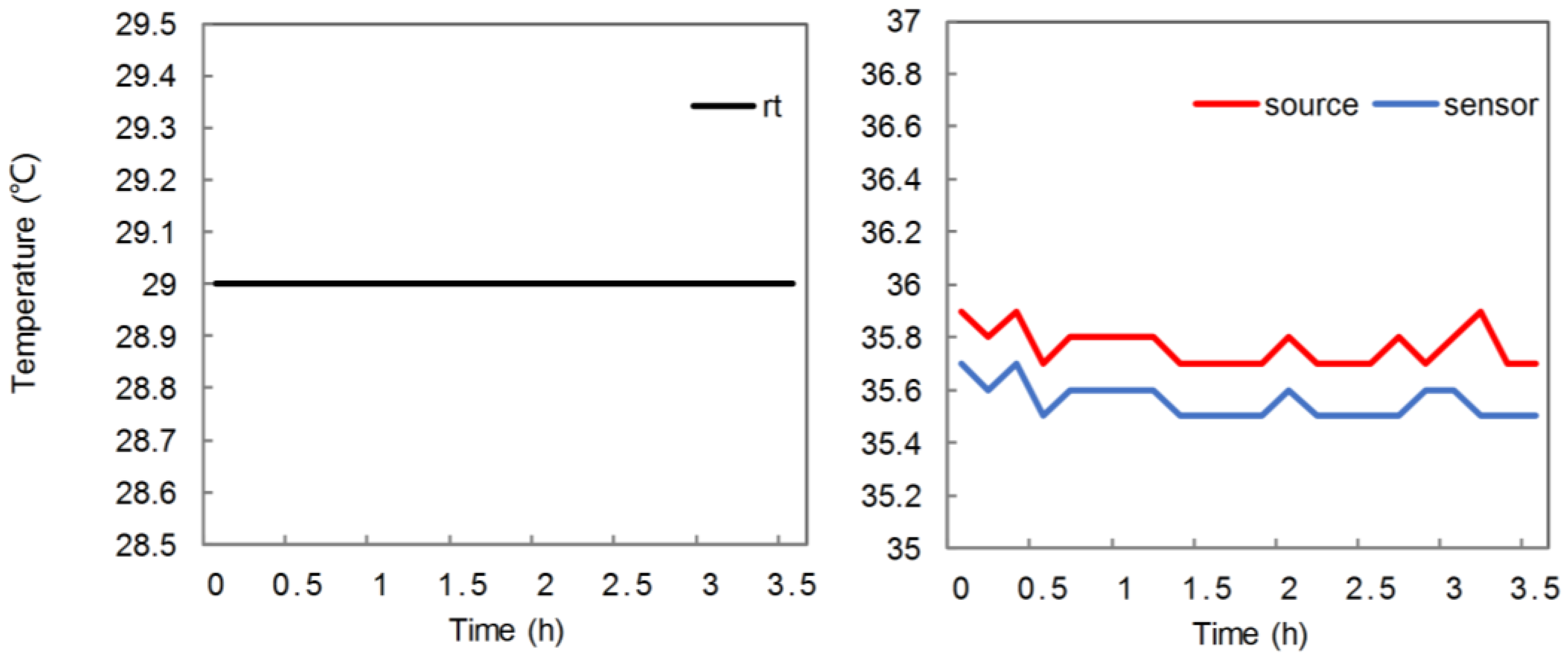
2.5. Experiment on Removal Effect
3. Results
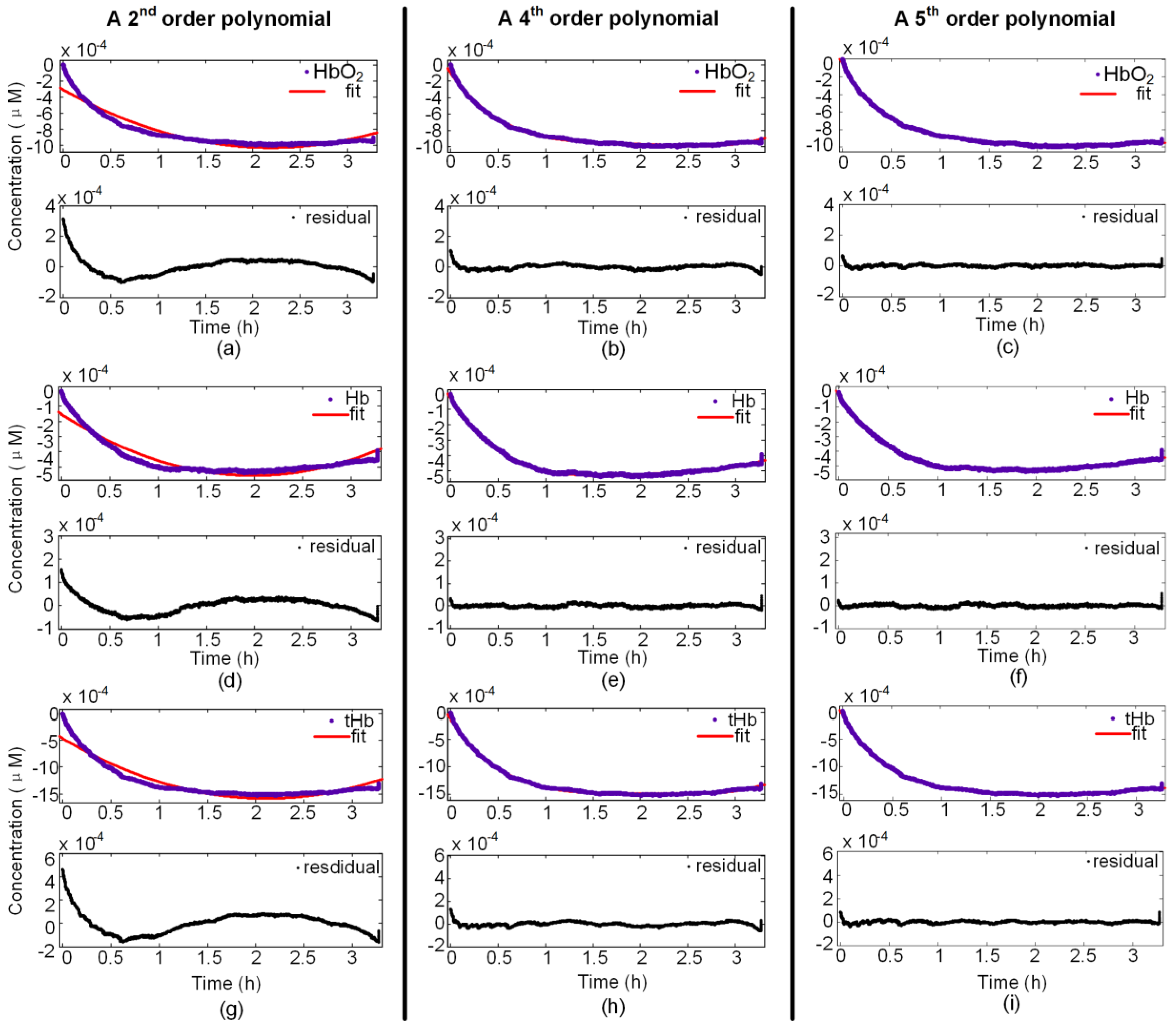
3.1. Fourth-Order Polynomial Function
3.2. Evaluating the Level of Fit
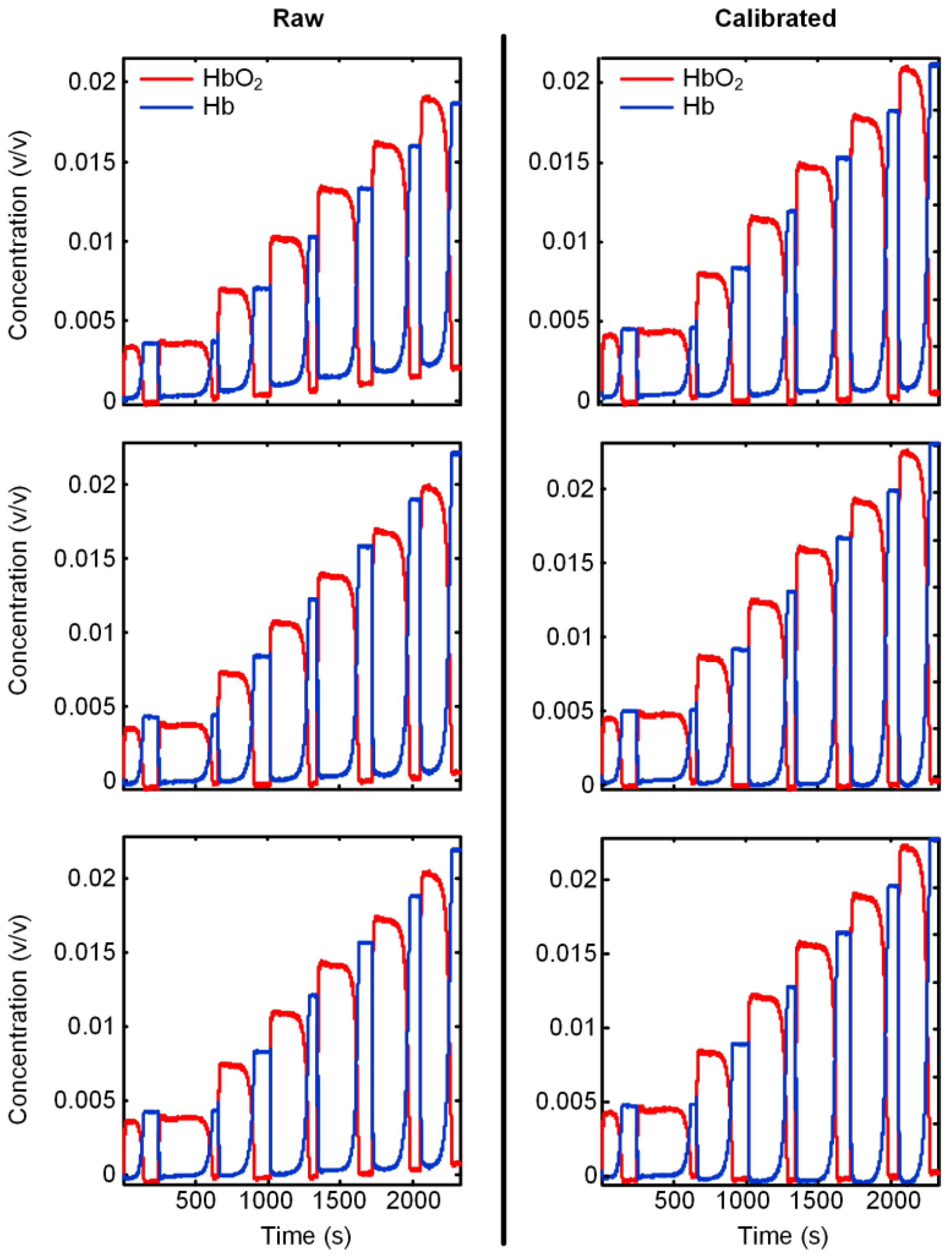
3.3. Verification of the Calibration
4. Conclusions and Discussion
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Geladi, P.; Dåbakk, E. An overview of chemometrics applications in near infrared spectrometry. J. Near Infrared Spectrosc. 1995, 3, 119–132. [Google Scholar] [CrossRef]
- Williams, P.; Norris, K. Near-Infrared Technology in the Agricultural and Food Industries; American Association of Cereal Chemists, Inc.: Eagan, MN, USA, 1987. [Google Scholar]
- Jobsis, F.F. Noninvasive, infrared monitoring of cerebral and myocardial oxygen sufficiency and circulatory parameters. Science 1977, 198, 1264–1267. [Google Scholar] [CrossRef] [PubMed]
- Noman, N.; Keum-Shik, H. Corrigendum “fnirs-based brain-computer interfaces: A review”. Front. Hum. Neurosci. 2015, 9, 172. [Google Scholar]
- Naseer, N.; Hong, K.S. Classification of functional near-infrared spectroscopy signals corresponding to the right- and left-wrist motor imagery for development of a brain-computer interface. Neurosci. Lett. 2013, 553, 84–89. [Google Scholar] [CrossRef] [PubMed]
- Scholkmann, F.; Kleiser, S.; Metz, A.J.; Zimmermann, R.; Pavia, J.M.; Wolf, U.; Wolf, M. A review on continuous wave functional near-infrared spectroscopy and imaging instrumentation and methodology. Neuroimage 2014, 85, 6–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrari, M.; Quaresima, V. A brief review on the history of human functional near-infrared spectroscopy (fnirs) development and fields of application. Neuroimage 2012, 63, 921–935. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Lin, Y.; Li, T. Effect of human brain edema on light propagation: A monte carlo modeling based on the visible chinese human dataset. IEEE Photonics J. 2017, 9. [Google Scholar] [CrossRef]
- Li, T.; Yu, L.; Yu, S.; Lian, H.; Chong, H.; Szabunio, M.; Yu, G. Simultaneous measurement of deep tissue blood flow and oxygenation using noncontact diffuse correlation spectroscopy flow-oximeter. Sci. Rep. 2013, 3, 1358. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Xue, C.; Wang, P.; Li, Y.; Wu, L. Photon penetration depth in human brain for light stimulation and treatment: A realistic monte carlo simulation study. J. Innov. Opt. Health Sci. 2017, 10. [Google Scholar] [CrossRef]
- Esquerre, C.; Gowen, A.A.; Burger, J.; Downey, G.; O’Donnell, C.P. Suppressing sample morphology effects in near infrared spectral imaging using chemometric data pre-treatments. Chemom. Intell. Lab. Syst. 2012, 117, 129–137. [Google Scholar] [CrossRef]
- Liu, J.M. Photonic Devices; Cambridge University Press: Cambridge, UK, 2005. [Google Scholar]
- Rinnan, Å.; Berg, F.V.D.; Engelsen, S.B. Review of the most common pre-processing techniques for near-infrared spectra. TrAC Trends Anal. Chem. 2009, 28, 1201–1222. [Google Scholar] [CrossRef]
- Wold, S.; Antti, H.; Lindgren, F.; Öhman, J. Orthogonal signal correction of near-infrared spectra. Chemom. Intell. Lab. Syst. 1998, 44, 175–185. [Google Scholar] [CrossRef]
- Savitzky, A.; Golay, M.J.E. Smoothing and differentiation of data by simplified least squares procedures. Anal. Chem. 1964, 36, 1627–1639. [Google Scholar] [CrossRef]
- Dhanoa, M.S.; Barnes, R.J.; Lister, S.J. Standard normal variate transformation and de-trending of near-infrared diffuse reflectance spectra. Appl. Spectrosc. 2016, 43, 772–777. [Google Scholar]
- Lipponen, J.A.; Geest, K.B.V.D.; Tarvainen, M.P.; Leinonen, A.; Lahtinen, M.; Karjalainen, P.A. Baseline removal from near infrared spectroscopy measurements for lactate concentration estimation. In IFMBE Proceedings; Springer: Berlin, Germany, 2009; Volume 25, pp. 2042–2045. [Google Scholar]
- Barbin, D.F.; Kaminishikawahara, C.M.; Soares, A.L.; Mizubuti, I.Y.; Grespan, M.; Shimokomaki, M.; Hirooka, E.Y. Prediction of chicken quality attributes by near infrared spectroscopy. Food Chem. 2015, 168, 554–560. [Google Scholar] [CrossRef] [PubMed]
- Laxalde, J.; Ruckebusch, C.; Devos, O.; Caillol, N.; Wahl, F.; Duponchel, L. Characterisation of heavy oils using near-infrared spectroscopy: Optimisation of pre-processing methods and variable selection. Anal. Chim. Acta 2011, 705, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yang, M.; Wei, G.; Hu, R.; Luo, Z.; Li, G. Improved pls regression based on svm classification for rapid analysis of coal properties by near-infrared reflectance spectroscopy. Sens. Actuators B Chem. 2014, 193, 723–729. [Google Scholar] [CrossRef]
- Pierce, K.M.; Kehimkar, B.; Marney, L.C.; Hoggard, J.C.; Synovec, R.E. Review of chemometric analysis techniques for comprehensive two dimensional separations data. J. Chromatogr. A 2012, 1255, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Hong, K.S.; Naseer, N. Reduction of delay in detecting initial dips from functional near-infrared spectroscopy signals using vector-based phase analysis. Int. J. Neural Syst. 2016, 26, 1650012. [Google Scholar] [CrossRef] [PubMed]
- Yeganeh, H.Z.; Toronov, V.; Elliott, J.T.; Diop, M.; Lee, T.Y.; Lawrence, K.S. Broadband continuous-wave technique to measure baseline values and changes in thetissue chromophore concentrations. Biomed. Opt. Express 2012, 3, 2761–2770. [Google Scholar] [CrossRef] [PubMed]
- Lapborisuth, P.; Zhang, X.; Noah, A.; Hirsch, J. Neurofeedback-based functional near-infrared spectroscopy upregulates motor cortex activity in imagined motor tasks. Neurophotonics 2017, 4, 021107. [Google Scholar] [CrossRef] [PubMed]
- Huppert, T.J.; Diamond, S.G.; Boas, D.A. Quantitative estimation of cerebral hemodynamic changes through the multimodality fusion of bold and diffuse optical tomography. In Proceedings of the Biomedical Topical Meeting, Fort Lauderdale, FL, USA, 19–22 March 2006. [Google Scholar]
- Scholkmann, F.; Spichtig, S.; Muehlemann, T.; Wolf, M. How to detect and reduce movement artifacts in near-infrared imaging using moving standard deviation and spline interpolation. Physiol. Meas. 2010, 31, 649–662. [Google Scholar] [CrossRef] [PubMed]
- Hebden, J.C.; Price, B.D.; Gibson, A.P.; Royle, G. A soft deformable tissue-equivalent phantom for diffuse optical tomography. Phys. Med. Biol. 2006, 51, 5581–5590. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Duan, M.; Li, K.; Yu, G.; Ruan, Z. Bedside monitoring of patients with shock using a portable spatially-resolved near-infrared spectroscopy. Biomed. Opt. Express 2015, 6, 3431–3436. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Li, Y.; Lin, Y.; Li, K. Significant and sustaining elevation of blood oxygen induced by Chinese cupping therapy as assessed by near-infrared spectroscopy. Biomed. Opt. Express 2017, 8, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Zhong, F.; Pan, B.; Li, Z.; Chong, H.; Deng, Z. A brief review of opt101 sensor application in near-infrared spectroscopy instrumentation for intensive care unit clinics. Sensors 2017, 17, 1701. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Xu, G.; Li, T. A portable high-density absolute-measure nirs imager for detecting prefrontal lobe response to driving fatigue. Microelectron. Reliab. 2017, in press. [Google Scholar]
- Afseth, N.K.; Kohler, A. Extended multiplicative signal correction in vibrational spectroscopy, a tutorial. Chemom. Intell. Lab. Syst. 2012, 117, 92–99. [Google Scholar] [CrossRef]
- Jiang, S.; Pogue, B.W.; Mcbride, T.O.; Paulsen, K.D. Quantitative analysis of near-infrared tomography: Sensitivity to the tissue-simulating precalibration phantom. J. Biomed. Opt. 2003, 8, 308–315. [Google Scholar] [CrossRef] [PubMed]
- MathWorks. Evaluating Goodness of Fit. Available online: http://cn.mathworks.com/help/curvefit/evaluating-goodness-of-fit.html?requestedDomain=www.mathworks.com (accessed on 7 October 2017).
- Zhao, Y.; Qiu, L.; Sun, Y.; Huang, C.; Li, T. Optimal hemoglobin extinction coefficient data set for near-infrared spectroscopy. Biomed. Opt. Express 2017, 8, 5151–5159. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Sun, Y.; Chen, X.; Zhao, Y.; Ren, R. Noninvasive diagnosis and therapeutic effect evaluation of deep vein thrombosis in clinics by near-infrared spectroscopy. J. Biomed. Opt. 2015, 20, 010502. [Google Scholar] [CrossRef] [PubMed]
| P | C | HP | Second Order | Third Order | Fourth Order | Fifth Order | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| SSE | R-Square | SSE | R-Square | SSE | R-Square | SSE | R-Square | |||
| 1 | 1 | HbO2 | 4.67 × 10−4 | 0.977 | 4.25 × 10−4 | 0.98 | 2.07 × 10−4 | 0.99 | 8.3 × 10−5 | 0.996 |
| Hb | 1.65 × 10−4 | 0.979 | 1.27 × 10−4 | 0.981 | 7.81 × 10−5 | 0.99 | 3.05 × 10−5 | 0.996 | ||
| tHb | 1.17 × 10−3 | 0.978 | 9.63 × 10−4 | 0.979 | 5.34 × 10−4 | 0.99 | 2.08 × 10−4 | 0.996 | ||
| 2 | HbO2 | 1.66 × 10−3 | 0.979 | 1.54 × 10−4 | 0.983 | 5.90 × 10−4 | 0.993 | 2.26 × 10−4 | 0.997 | |
| Hb | 1.87 × 10−4 | 0.975 | 1.59 × 10−4 | 0.981 | 3.26 × 10−5 | 0.996 | 1.71 × 10−5 | 0.998 | ||
| tHb | 8.01 × 10−4 | 0.979 | 7.29 × 10−4 | 0.982 | 3.77 × 10−4 | 0.99 | 1.47 × 10−4 | 0.996 | ||
| 2 | 1 | HbO2 | 3.87 × 10−4 | 0.957 | 3.38 × 10−4 | 0.958 | 3.52 × 10−5 | 0.996 | 8.08 × 10–6 | 0.999 |
| Hb | 6.97 × 10−4 | 0.957 | 6.37 × 10−4 | 0.961 | 5.60 × 10−5 | 0.997 | 1.26 × 10−5 | 0.999 | ||
| tHb | 4.57 × 10−5 | 0.955 | 3.34 × 10−5 | 0.963 | 4.38 × 10−7 | 0.998 | 5.43 × 10−7 | 0.999 | ||
| 2 | HbO2 | 1.60 × 10−3 | 0.977 | 1.29 × 10−3 | 0.979 | 1.61 × 10−4 | 0.998 | 3.78 × 10−5 | 0.999 | |
| Hb | 2.96 × 10−4 | 0.961 | 2.43 × 10−4 | 0.967 | 1.87 × 10−5 | 0.998 | 5.52 × 10–6 | 0.999 | ||
| tHb | 5.43 × 10−4 | 0.982 | 4.03 × 10−4 | 0.986 | 8.97 × 10−5 | 0.997 | 3.45 × 10−5 | 0.999 | ||
| 3 | 1 | HbO2 | 4.67 × 10−4 | 0.977 | 2.21 × 10−4 | 0.927 | 2.07 × 10−4 | 0.99 | 8.3 × 10−5 | 0.996 |
| Hb | 1.65 × 10−4 | 0.979 | 6.29 × 10−5 | 0.913 | 7.81 × 10−5 | 0.99 | 3.05 × 10−5 | 0.996 | ||
| tHb | 1.17 × 10−3 | 0.978 | 5.23 × 10−4 | 0.917 | 5.34 × 10−4 | 0.99 | 2.08 × 10−4 | 0.996 | ||
| 2 | HbO2 | 1.66 × 10−3 | 0.979 | 3.34 × 10−4 | 0.959 | 5.90 × 10−4 | 0.993 | 2.26 × 10−4 | 0.997 | |
| Hb | 1.87 × 10−4 | 0.975 | 1.43 × 10−4 | 0.968 | 3.26 × 10−5 | 0.996 | 1.71 × 10−5 | 0.998 | ||
| tHb | 8.01 × 10−4 | 0.979 | 4.03 × 10−4 | 0.95 | 3.77 × 10−4 | 0.99 | 1.47 × 10−4 | 0.996 | ||
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, K.; Ji, Y.; Li, Y.; Li, T. Online Removal of Baseline Shift with a Polynomial Function for Hemodynamic Monitoring Using Near-Infrared Spectroscopy. Sensors 2018, 18, 312. https://doi.org/10.3390/s18010312
Zhao K, Ji Y, Li Y, Li T. Online Removal of Baseline Shift with a Polynomial Function for Hemodynamic Monitoring Using Near-Infrared Spectroscopy. Sensors. 2018; 18(1):312. https://doi.org/10.3390/s18010312
Chicago/Turabian StyleZhao, Ke, Yaoyao Ji, Yan Li, and Ting Li. 2018. "Online Removal of Baseline Shift with a Polynomial Function for Hemodynamic Monitoring Using Near-Infrared Spectroscopy" Sensors 18, no. 1: 312. https://doi.org/10.3390/s18010312




