1. Introduction
Biometric identity recognition systems using human body measurement data are vital for the security of financial transactions, access control, travel, etc. The various biometrics that are currently being adopted in identity recognition scenarios such as fingerprint, iris, face and speech recognition have operational trade-offs in terms of performance, measurability, circumvention and liveness detection [
1]. These limitations have led to the development of smart solutions such as: (i) improving voice recognition by sophisticated pre-processing, followed by heuristical selection of significant features in time and frequency domains [
2]; (ii) integrating face- and body-related soft biometric traits [
3]; (iii) providing security and flexibility by simultaneous application of numeric password data and biometric fingerprint information [
4]; (iv) assuring liveness detection via optical sensors, which establish the presence of pulse, the variations of optical characteristics caused by pressure changes and skin reaction to illumination with different wavelengths [
5].
For more than a decade the electrocardiogram (ECG) has been intensively investigated in the context of biometric applications. The attention is attracted by its ability to provide an unambiguous sign of liveness and to exhibit several personalized traits, reflecting the underlying physiological properties of the heart such as relative timings of the P, QRS, T waves and differences in beat geometry. An additional precondition for the use of biomedical data for identification purposes is the rapid development of multidimensional sensors making it possible to efficiently acquire such data [
6,
7]. Recently, the strategic application of biometric systems has been extended towards:
- -
remote healthcare monitoring [
8,
9] with biosensors integrated into mobile devices [
10];
- -
secure wireless body area sensor networks [
11,
12];
- -
continuous authentication applications with adaptive strategies to follow individual beat variations in 24 h ECG recordings [
13];
- -
continuous human recognition in driving settings [
14].
Similar to any other biometric system, the operation of those, embedding ECG analysis, could be spitted into two phases [
15]:
- -
Enrolment or registration phase, when a reference database from subjects with known identity is collected;
- -
Matching phase with two possible scenarios:
Person verification, when the system accepts or rejects someone’s identity based on a single comparison to the data for a given ID in the reference database (one-to-one scenario);
Person identification, when the system is finding the identity of an individual based on a search in a reference database for the subject whose data best matches the input data (one-to-many scenario).
In terms of feature extraction methods, identification techniques based on ECG analysis could be assigned into three categories:
- -
Fiducial based approaches, which analyse:
Temporal, amplitude, area and angle features over single lead [
9] or 12-lead ECG [
16];
Only temporal features measured in a single lead that are hypothesized to be invariant to sensor placement [
12,
17,
18].
The accuracy of these methods strongly depends on the correct localization of the waves’ boundaries within the P-QRS-T segment.
- -
Non-fiducial methods based on calculation of:
Autocorrelation functions and their post-processing via discrete cosine transform [
14,
19];
Linear discriminant analysis [
20] for feature space reduction and further identification;
Assessment of correlation coefficients between beats with known and unknown identity [
21,
22,
23];
Pattern matching and estimation of normalized Euclidian distance between the tested and the target heartbeat [
24];
Features describing the dynamic characteristics and chaotic behaviour in the ECG time series [
25].
- -
Hybrid human identification methods combining both fiducial and non-fiducial strategies, which include:
Hierarchical identification scheme for temporal and amplitude measurements based on fiducial points detection and estimation of waveforms‘ similarity via Euclidean distance [
26];
Fiducial-based measurements for reduction of the matching candidates’ number followed by wavelet transform and application of the coefficients for template matching [
8].
Numerous studies are focused on general-purpose practical applications limited to the use of a single ECG lead, usually from the limbs [
10,
17,
18,
23,
24]. Others aim at effective channel combination schemes by involving 12-lead ECG [
16,
20,
21,
22] into the analysis in view of feasible applications in healthcare scenarios. The merit of multi-lead ECG is controversial, considering: on the one hand the identification accuracy improvement by increasing the analysed ECG leads in a fiducial independent application [
22] and on the other hand the same level of identification accuracy for all 12 leads, only limb, only chest and only lead I in a fiducial-based application [
16].
Despite that the majority of the studies have used public [
9,
10,
19,
20,
23,
26] or proprietary [
10,
16,
17,
18,
21,
22,
23,
24] clinical databases, some authors have addressed the application of different non-invasive portable single-channel ECG sensors which support the acquisition of ECG signals for human identification purposes. The reported identification accuracies are:
- -
~99% for 30 ECG signals acquired from the individual via a biosensor integrated into mobile devices [
10];
- -
~90% for 30 ECG test patterns acquired from the palms of 9 subjects via a two poles self-developed ECG signal acquisition system [
25];
- -
~94% achieved for ECG recordings acquired from the fingers of 16 individuals via self-developed ECG sensor [
27].
The use of such ECG sensors facilitates the practical application of ECG biometrics in areas such as cyber-security [
25,
27], cloud data security or remote healthcare systems [
10].
The majority of the cited methods report high identification accuracy. However, certain limitations are observed such as:
- -
Small population size (<30 individuals) [
16,
17,
18,
19,
20,
21,
22,
25,
26,
27] with ECG recordings acquired in a very short temporal interval [
16,
21,
22] or even during the same session [
9,
17,
18,
20,
26];
- -
Application of one and the same database for both training and testing.
This is far from the realm of biometrics. Significant accuracy degradation could be expected [
28] with these methods, due to the influence of different unpredictable factors such as long-term ECG changes [
13], noisy environment [
14], electrode misplacement, overtraining of the identification model, etc.
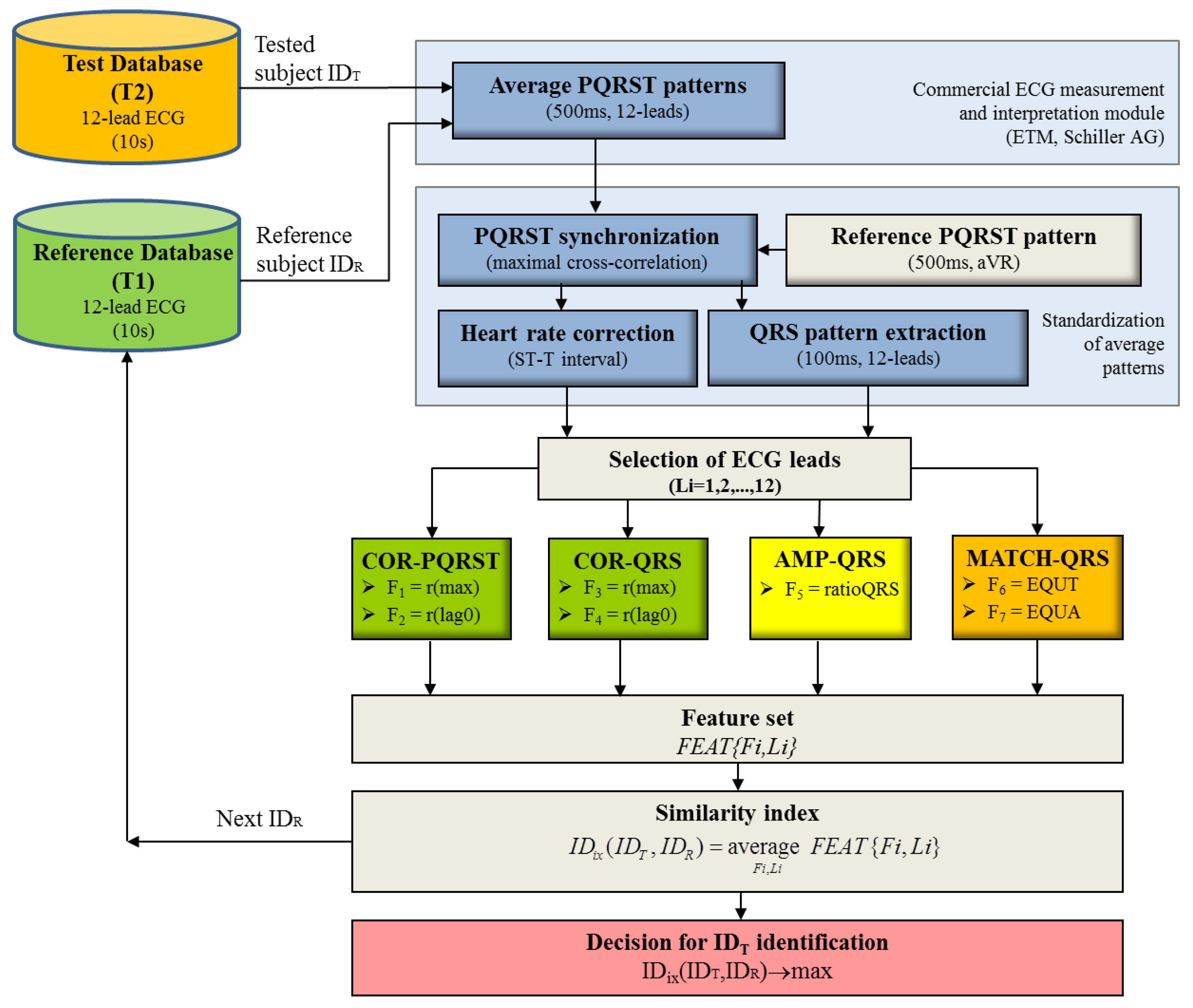
The aim of this paper is to present the 12-lead ECG as a biometric modality by describing three methods for comparison between input-to-template waveforms of personalized beat patterns based on cross-correlation, amplitude relations and a new method for binary pattern matching. We further investigate their potential for human identification within an uncommonly large population with one-year distant collection of the reference and test datasets. Unbiased statistical analysis on independent validation set is presented for different perspectives of the human identification problem, considering: (i) the choice of the optimal single and multi-lead ECG set; (ii) the influence of the reference database size, providing evidence about the reasonable size of the population that could be tolerated in ECG biometric studies.
4. Discussion
Although being a part of one general biometric task, the aims of human “identification” and human “verification” are quite different, i.e. to recognize an individual in a database by comparing a single input sample to many stored templates and to verify/reject person’s identity by comparing a single input sample to a single stored template. The same features for comparison between input-to-template waveforms could be used in both sub-tasks, however, the decision-making process is different. Usually, the decision for “identification” is performed via the best similarity assessment approach, while the “verification” relies on threshold based techniques. In our recent study [
29] we solve the “verification” problem via 6 cross-correlation and 2 amplitude features of QRS and PQRST patterns, involved in a linear discriminant threshold decision function. In this study, we use the same large population to solve the “identification” task adopting 4 cross-correlation and 1 amplitude features and adding 2 more QRS pattern features that measure the similarity in the time and amplitude scales. They are calculated with a brand-new method for binary template matching of short-duration QRS patterns (100 ms). The presented identification model applies a normalized scale for representation of all 7 features (0–100%), where 0% corresponds to the least matching and 100% to the best matching between input and template waveforms. The optimization of the feature selection scheme involved in the calculation of the common similarity index is presented by iterative maximization of the identification accuracy
AccID.
The questions of greatest concern for the human identification task are: “Is there an individual whose personalized beat pattern would be more similar to another than to his own in a long-term basis?”, “What is the probability to find such an individual within an increasing population of reference subjects?”
A quantitative answer to these questions is presented in this study following a straightforward list of training and validation tests which aim to reveal specific aspects of the current human ID investigation.
(1) Is there a statistical justification for using the defined features for waveform similarity?
Table 1 presents the statistics of the features’ distributions in the training dataset, proving that all of them exhibit significantly higher similarity of the waveforms measured across the same individuals in a long-term basis (>1 year) than what can be seen between different individuals (mean value in the range about 82–96% vs. 60–86%,
p < 0.0001). This proves that the similarity index ID
ix defined for finding the best match between subjects could include all features on the same basis.
(2) Which are the most reliable QRS and PQRST features for human ID?
This aspect is investigated by training optimal ID models with different configurations of features using a large dataset with 230 individuals. The simplest model (one-feature in one-lead,
Table 2) distinguishes
r(
lag0) in leads (aVL, aVF, III) with maximal
AccID in the range of 35 to 41%. It is about 10% points better than the other distinguished feature
r(max) where
AccID ranges between 24 and 35%. The advance of both correlation coefficients is due to different spatial dynamics of the cardiac vector between different subjects. This dissimilarity is enhanced in the zero-lag correlation which additionally represents the temporal desynchronization of the depolarization-repolarization process in different individuals. We suggest that the latter effect is compensated in aVR by using this lead for synchronization of each subject’s average beat to the reference pattern and this explains the remarkably low
AccID in aVR (11–15%) in respect to the other limb leads. The joint analysis of
r(
lag0) and
r(max) in 12-lead ECG increases
AccID by about 40% reaching 73.5% (COR-QRS) and 77.4% (COR-PQRST)—see
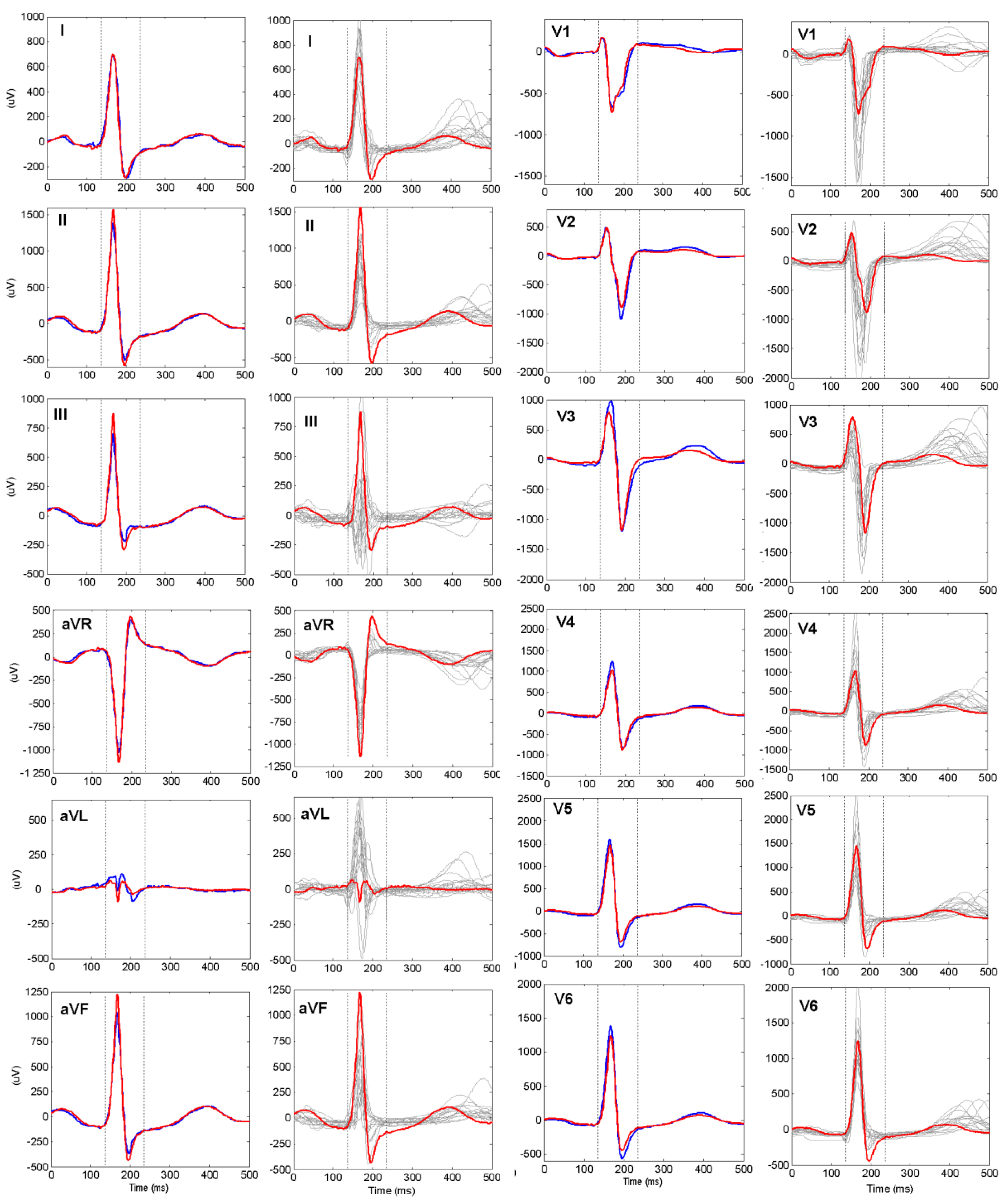
Table 3. These results indicate that the QRS pattern (100 ms) carry the essential subject-specific information, while P and T waves in the PQRST pattern (500 ms) contribute only for a slight ID accuracy improvement by about 4 percentage points.
The analysis of AMP-QRS shows definitively that the QRS peak-to-peak range across individuals is an indistinguishable feature. It provides
AccID < 1% for all single leads (
Table 2), which increases up to 24% for the optimal ID model (
Table 3) including the 7 ECG leads that are the most distinguishable in amplitude (I, II, III, V1, V2, V3, V5).
The MATCH-QRS method is not powerful when its features are analysed in single leads (maximal accuracy of 26.5% for
EQUA in aVF,
Table 2). This is suggested by the methodological approach for QRS waveform approximation in
binQRS matrix [100 × 80], which gives indistinguishably high waveform similarity for many subjects whose QRS waveforms are matching within the pre-set time and amplitude tolerances of ±1 ms and ±5%, respectively. The accumulated similarity over 12 leads, however stays high for the same identity subjects and decreases for different identity subjects. Thus, 15 MATCH-QRS measures are selected in the best ID model (
AccID = 83.5%,
Table 3), which represents the best similarity for 10 leads in the amplitude scale (I, II, III, avR, avF, V1, V2, V4, V5, V6) and for 5 leads in the time scale (I, aVR, aVL, V1, V3).
(3) Which are the most reliable leads for human ID?
This aspect is investigated by training optimal ID models with different configurations of leads on a large dataset with 230 individuals (
Table 4). In a single lead ID scenario, we highlight limb leads I and II (maximal
AccID of 58%) followed by aVF (55%) and aVL (50%) including 3 to 5 features from all feature extraction methods. Compared to the best limb leads (I and II), the single chest leads are from 14 to 30% less powerful for the purposes of human ID. They are ranked in the following order: V1 (maximal
AccID of 44%) followed by V6, V5, V2 (37–35%) and V3, V4 (27–30%). Such a drop in accuracy, especially in anterior leads, could be explained by V1–V6 proximity to the heart, so that small electrode misplacements across different recording sessions of the same individual result in considerable ECG morphology changes seen in chest leads.
Multi-lead ID models gain considerable raise in accuracy (about 20%) comparing 6 limb leads to the best II (77% vs. 58%) and 6 chest leads to the best V1 (64% vs. 44%). The optimal ID models are built with a prevalent selection of features from leads I, II (6 features), V1, V6 (5 features), aVR, V5 (4 features). The complete ID model, selecting features from all 12-leads, reaches maximal AccID of 91.3%. This is better than 6 limb, 6 chest leads and the best single lead (II) with about 14%, 27% and 33% points, respectively.
(4) What is the tolerated reference database size for searching human ID?
This question arises from our previous experience on human identification [
23] showing that the time elapsed between the collection of the reference and the test dataset as well as the number of enrolled subjects in the reference dataset have a severe impact on the ID performance. There,
AccID has been found to drop by 15% (from 93% to 78%) while the population increases by 35 subjects (from 14 to 49). The last is in agreement with [
28], reporting
AccID drop by 10% (from 99% to 89%) for population increase by 40 subjects (from 10 to 50). Naturally,
AccID worsening could be expected for reference database with larger size, since the more the subjects there are, the higher the random match probability is, i.e. to find a subject with better similarity than his own. According to a comparative study [
1], the majority of the published investigations on human ID have been conducted on small populations (about a few dozens of subjects). Considering the missing research information about the influence of the reference database size on the ID accuracy, we present consistent validation of our optimal ID models on an independent dataset by increasing its size from 10 to 230 subjects. Our validation results definitively confirm the expected trend for
AccID drop with the increase of the number of the reference subjects which however is non-linear and depends on the particular ID model (
Figure 4 and
Figure 5). We consider as good models those with higher
AccID and lower
AccID drop in respect of the number of reference subjects.
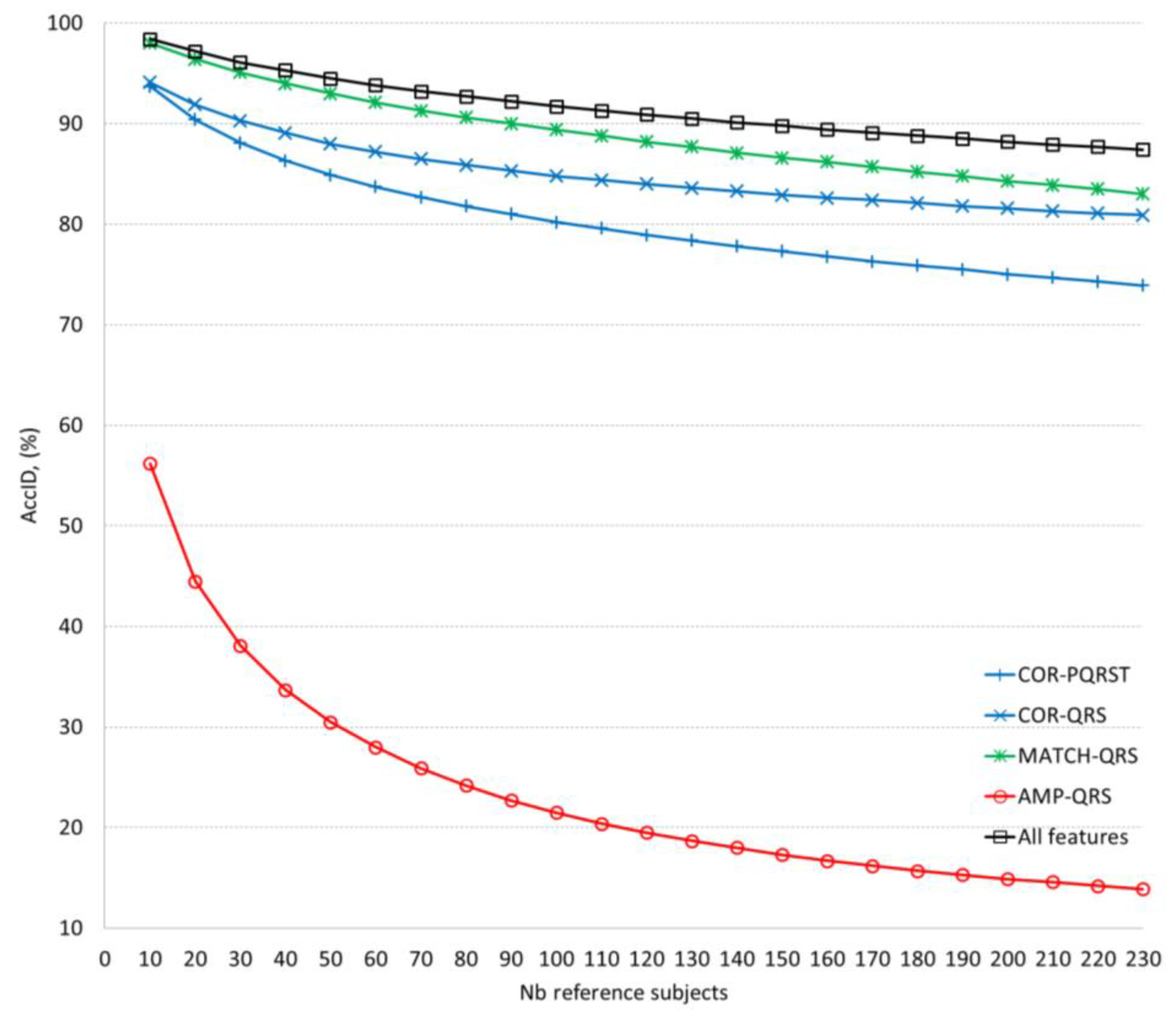
The validation of the ID models applying different feature extraction methods (
Table 5,
Figure 4) closely reproduces the
AccID ranges on the training dataset with 230 reference subjects (
Table 3). The observed
AccID differences (training vs. validation) are: COR-PQRST (77% vs. 74%), COR-QRS (74% vs. 81%), AMP-QRS (24% vs. 14%), MATCH-QRS (84% vs. 83%), all features (91% vs. 87%). The differences are relatively small which is a sign of confident training of the ID models with 12-lead ECG capable of adequately evaluating independent database without bias. We observed that the minimal set of 2 MATCH-QRS features is capable of providing human ID with almost the same accuracy (
AccID > 90%) as the maximal set of all 7 features. However, this is valid for limited reference datasets of up to 100 subjects. More subjects are best identified by the complete ID model including all features which maintains relatively stable performance with a drop of 11% (from 98% to 87%) for population increase by 220 subjects (from 10 to 230). These results outperform the cited in [
23,
28] which report the same level of
AccID drop (10–15%) for about 6 times smaller population increase (35–40 subjects). The best performance of our complete ID model could be explained by the fact that larger datasets increase the probability to find similar characteristics across different subjects and thus, more and more robust features are required to distinguish their individuality.
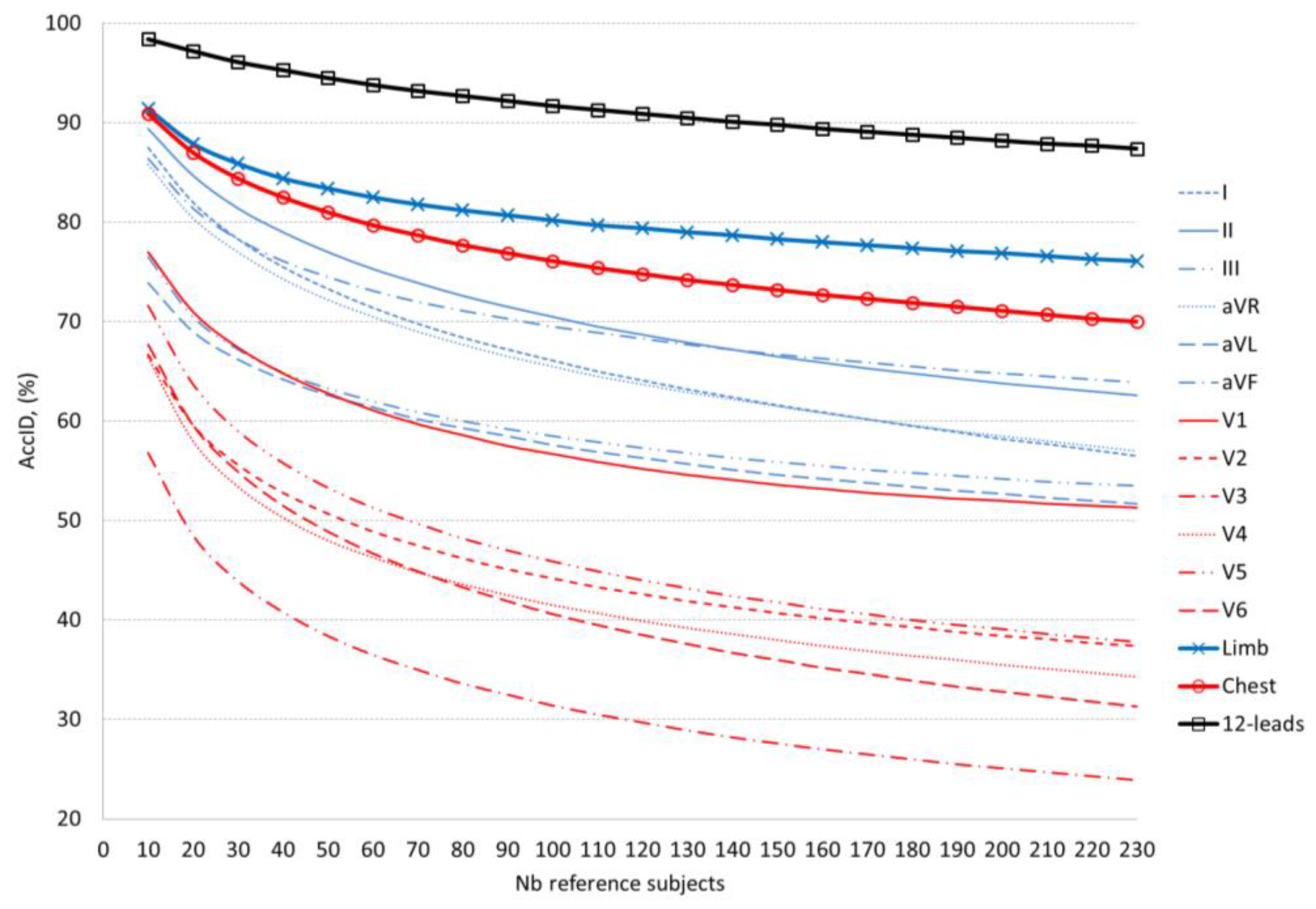
In a single lead ID scenario (
Figure 5,
Table 6) we can distinguish two best ID models based on analysis of lead II and aVF for small (10–140) and large (140–230) number of reference subjects, respectively. Overall, even for the best limb leads (II, aVF), we observe a limited ID capability, not exceeding 70% for more than 100 reference subjects and 80% for more than 30 reference subjects. Single chest leads are not usable for human ID. The majority of them (V2–V6) do not exceed 50% for more than 50 subjects. Only V1 can be distinguished with
AccID above 70% for less than 20 subjects. We recommend multi-lead ID scenarios for management of human search in large populations. Both ID models based on 6 chest and 6 limb leads pass the
AccID > 70% threshold for the maximal reference dataset (230 subjects) and
AccID > 80% for up to 100 subjects (limb leads) and up to 50 subjects (chest leads). As expected, 12-lead ECG provides the most robust beat patterns for human ID with
AccID > 90% for up to 140 subjects, reaching 98.4% for 10 subjects.
All published studies do not report training and validation of their ID models on independent datasets (as used in this study), therefore biased
AccID is probable due to overtraining. For example, such an effect is suspected for studies with accuracy close to 100%, especially on small sized databases [
9,
16,
18,
19,
20,
26]. Overall, the ID studies with enrolment and test phase in a single day report higher
AccID [
9,
10,
18,
26,
28]. However, they could not consider longitudinal ECG morphology changes due to physiological or technical sources (e.g. electrode displacement across different sessions). There are two ID studies using multiple sessions in different days on large populations with 74 [
24] and 89 subjects [
10] that report less than 5 percentage points better performance than our model. This might be due either to superior model algorithm or to the lack of independent validation, considering that the differences between our training and validation accuracies are in the same range.
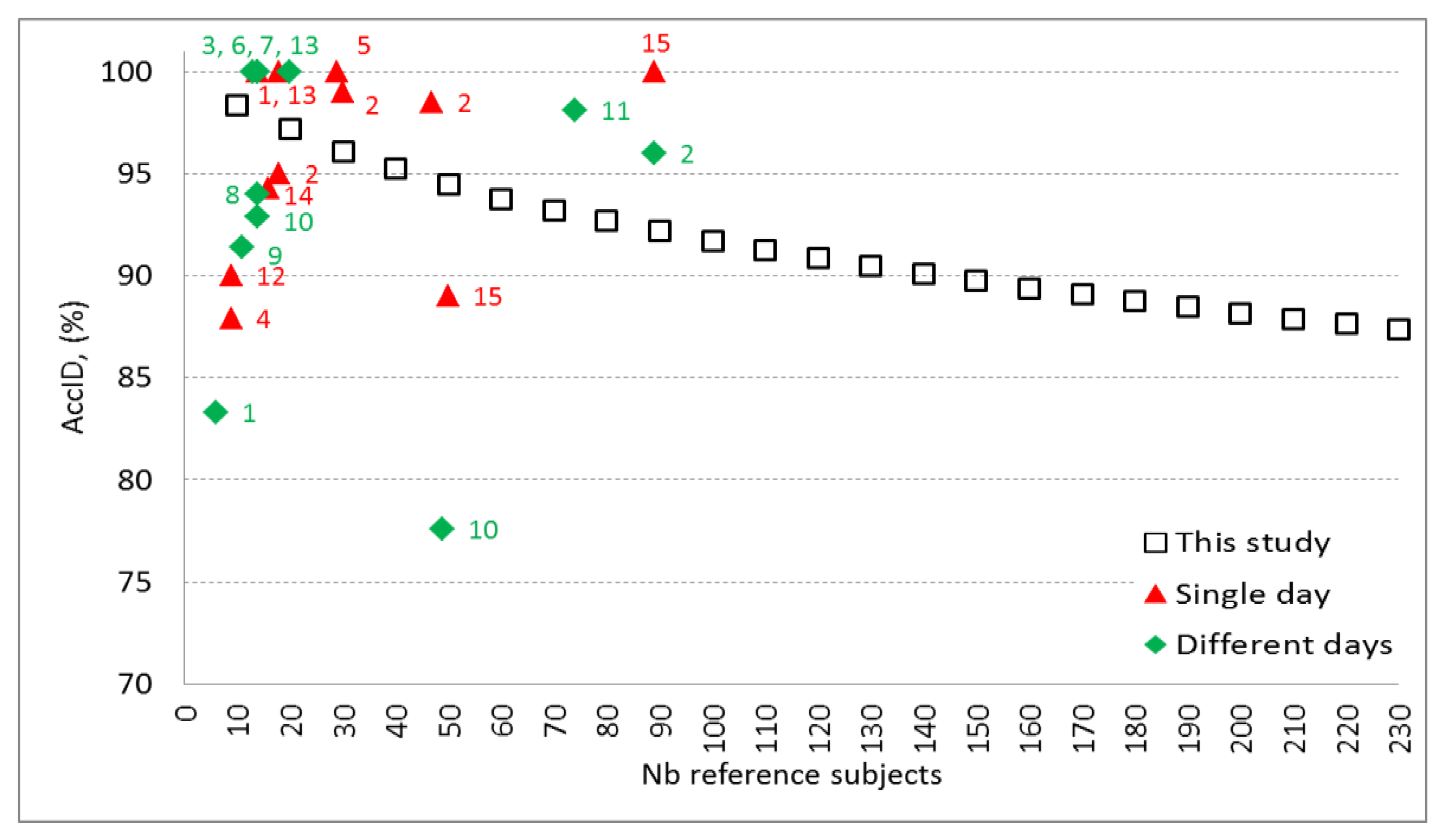
Figure 6 illustrates validation results of this study which extends the knowledge on the ID model performance over the largest span from 10 to 230 reference subjects. The observed decrease of
AccID could be explained with the appearance of new subjects in the reference dataset who present PQRST waveforms similar to already existing ones. Thus, some of the subjects that match exactly their own pattern in the reference dataset of e.g. 100 subjects show higher correlation and pattern matching with different subject in the reference dataset that contains 230 individuals. This could be due to time-related ECG changes and possible small differences in the electrode placement during the acquisition of the test dataset. Possible approach to keep high
AccID for larger reference datasets is to include additional robust features in the identification model. However, this should be done very carefully and the designed models should be validated on independent dataset in order to avoid overtraining. Another option to prevent from the assignment of wrong identity to the tested subject is to keep the size of the reference dataset as small as possible for the particular application and to update it with actual PQRST patterns on a regular basis.