A New Strategy Involving the Use of Peptides and Graphene Oxide for Fluorescence Turn-on Detection of Proteins
Abstract
:1. Introduction
2. Materials and Methods
2.1. Apparatus and Materials
2.2. Protein Expression And Purification
2.3. Fabrication of Peptide-GO Sensor and Fluorescence Detection
2.4. Preparation of Human Serum Samples
2.5. Construction of Bcl-xL Overexpression Cell Line
2.6. Western Blotting
2.7. Cytotoxicity Assay
2.8. Cell Imaging
3. Results and Discussion
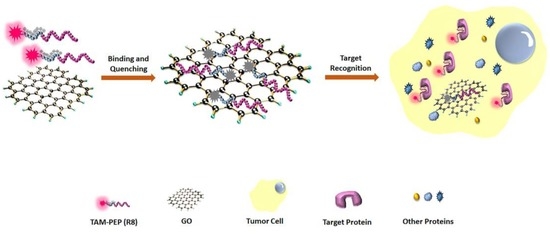
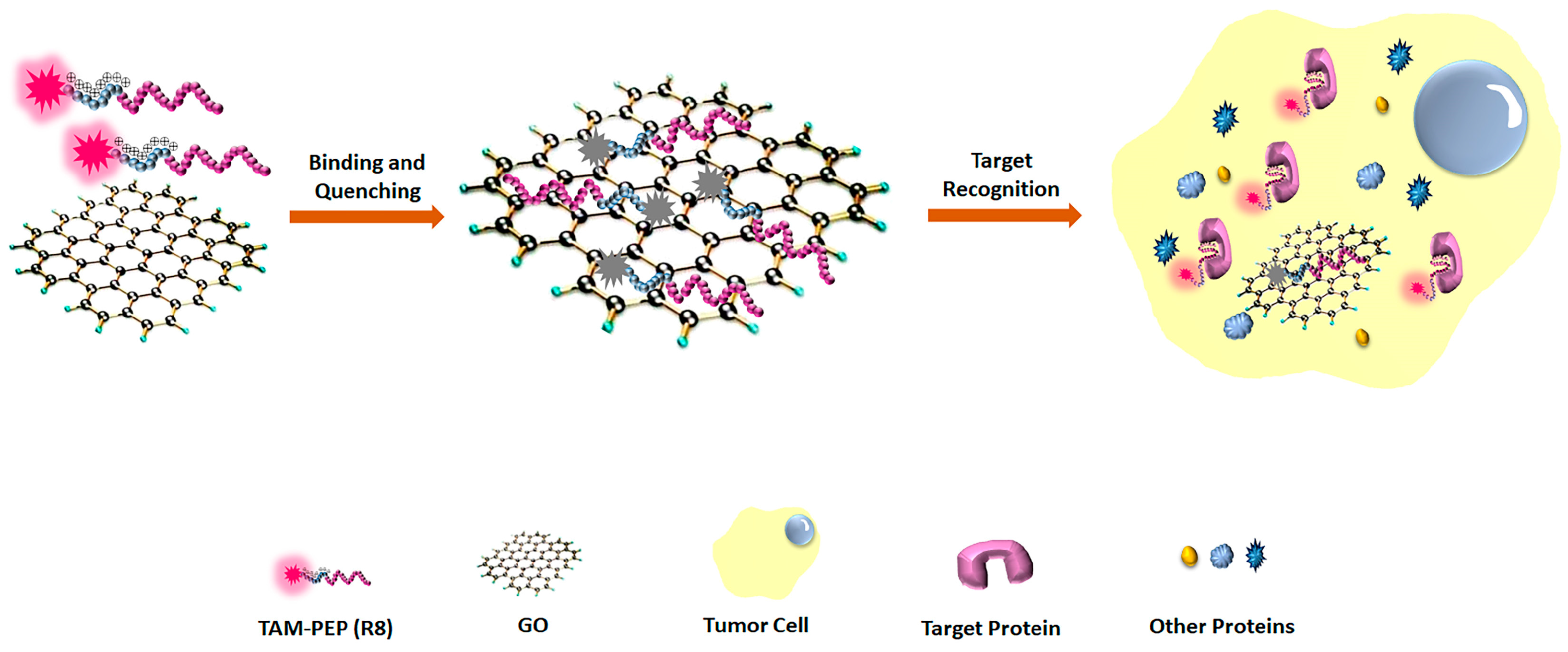
3.1. The Principle of the Strategy
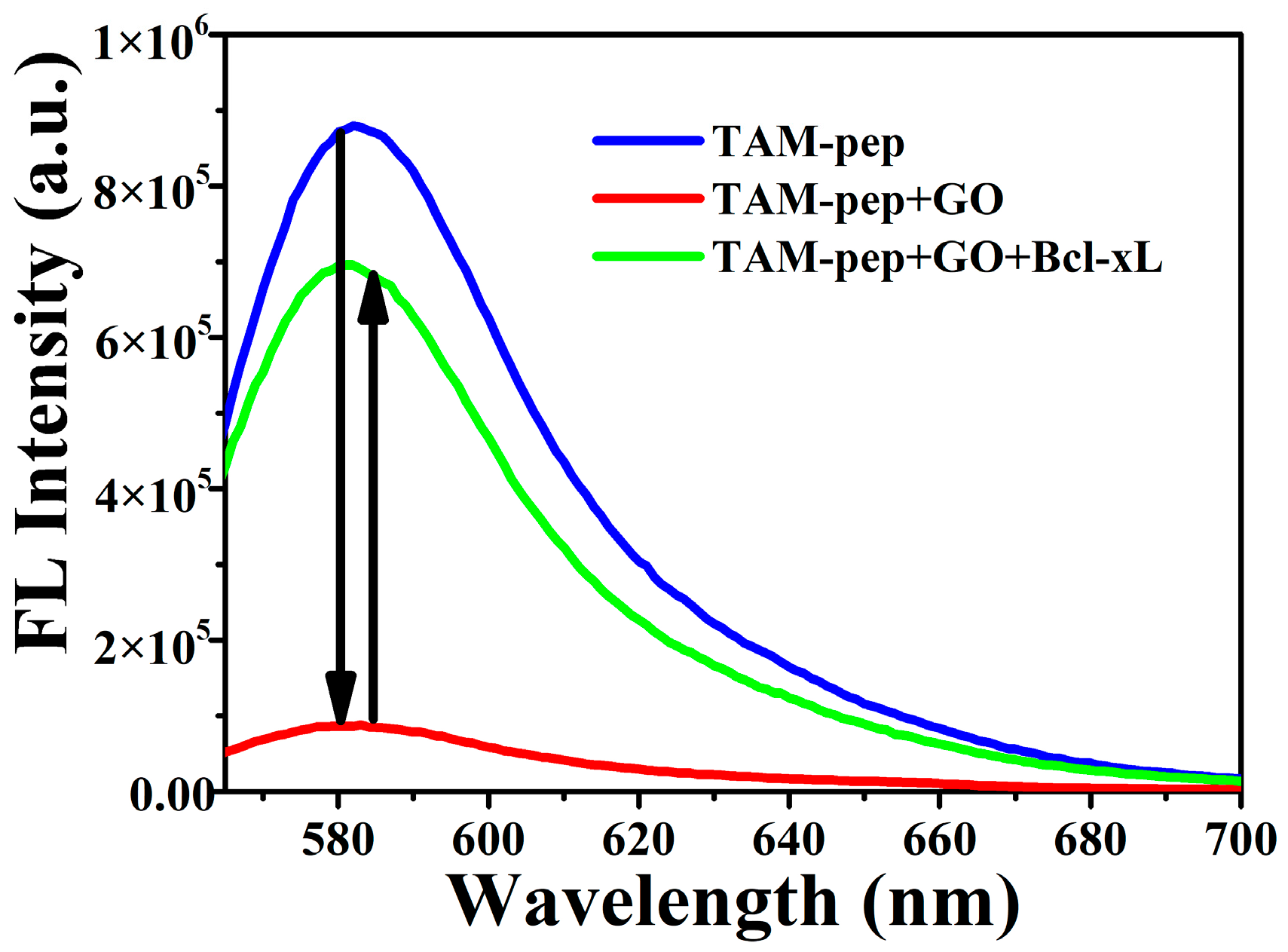
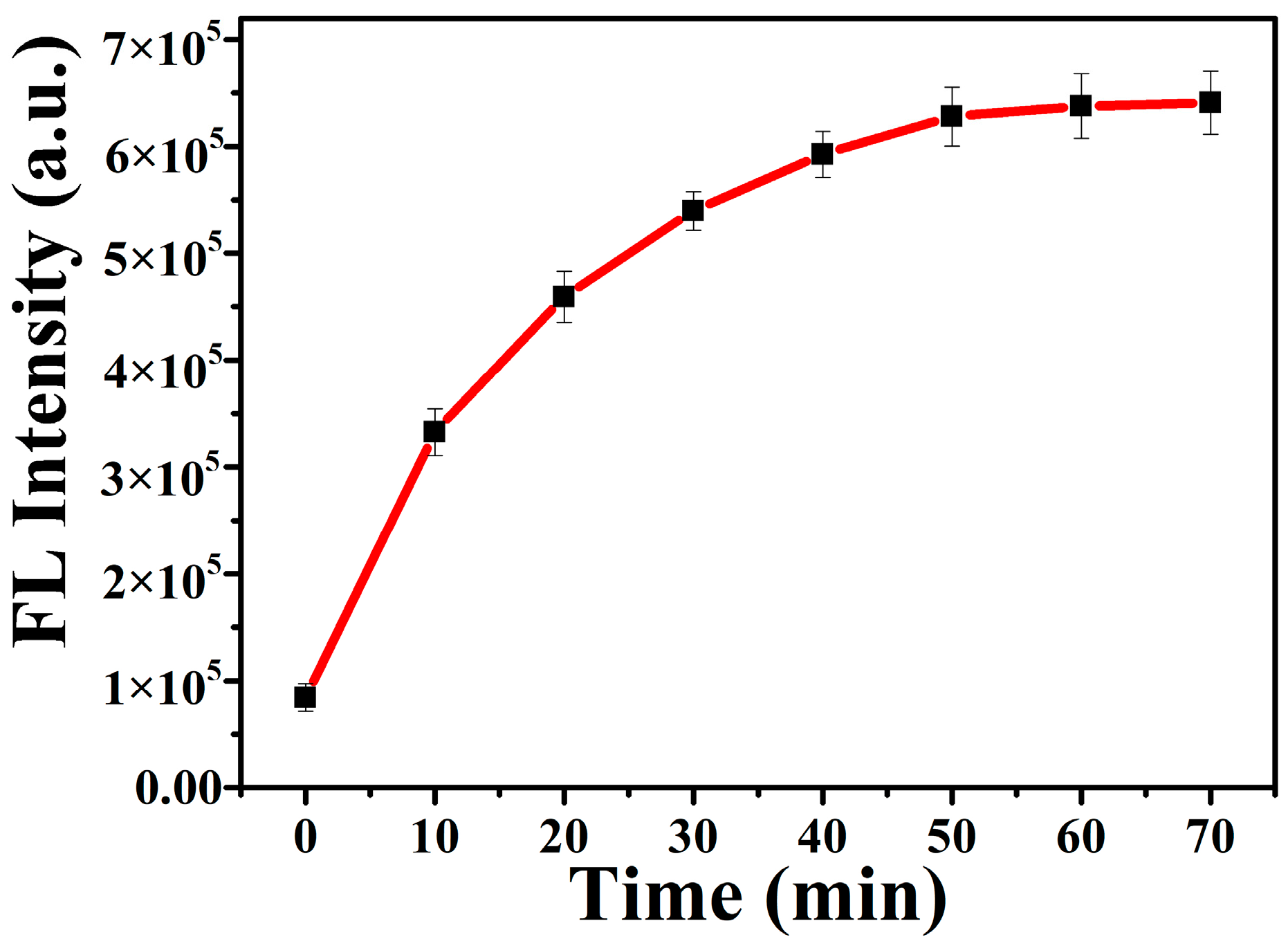
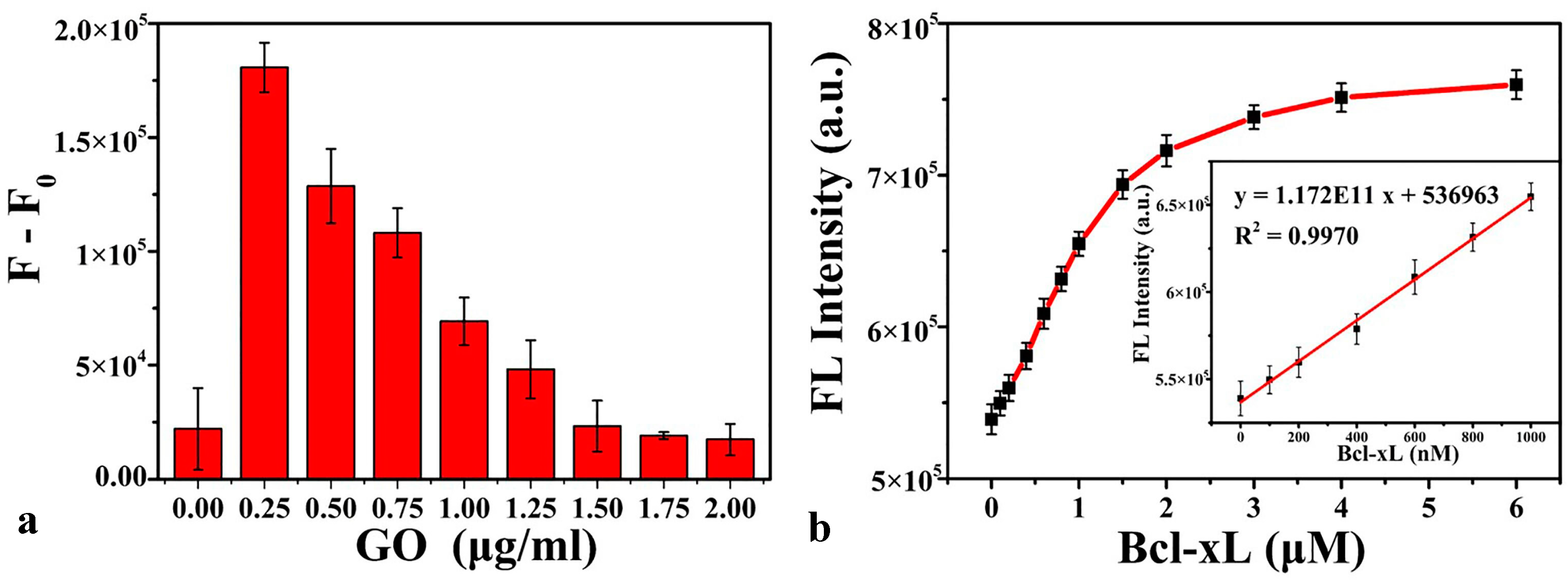
3.2. Optimization of the Detection Conditions
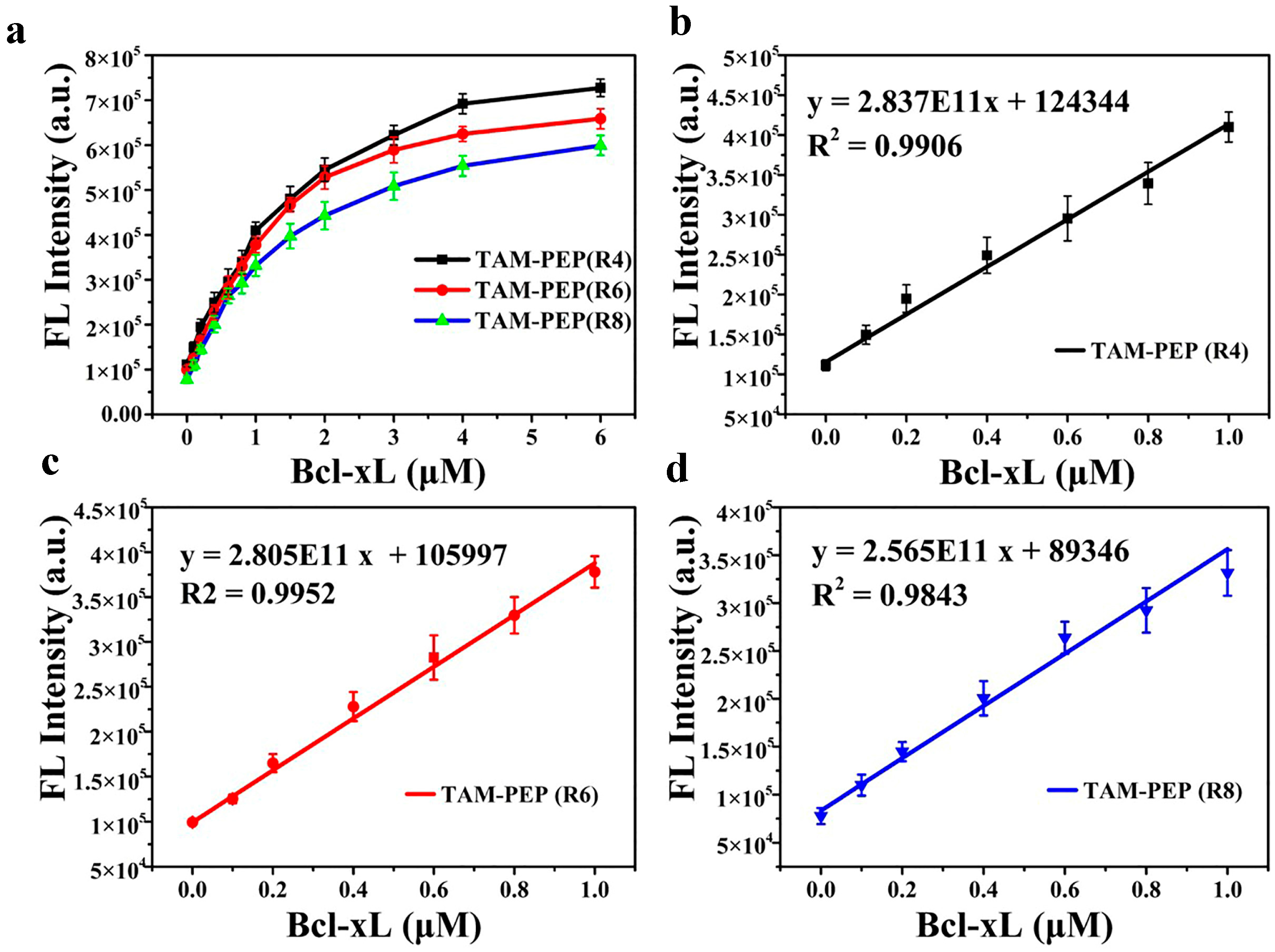
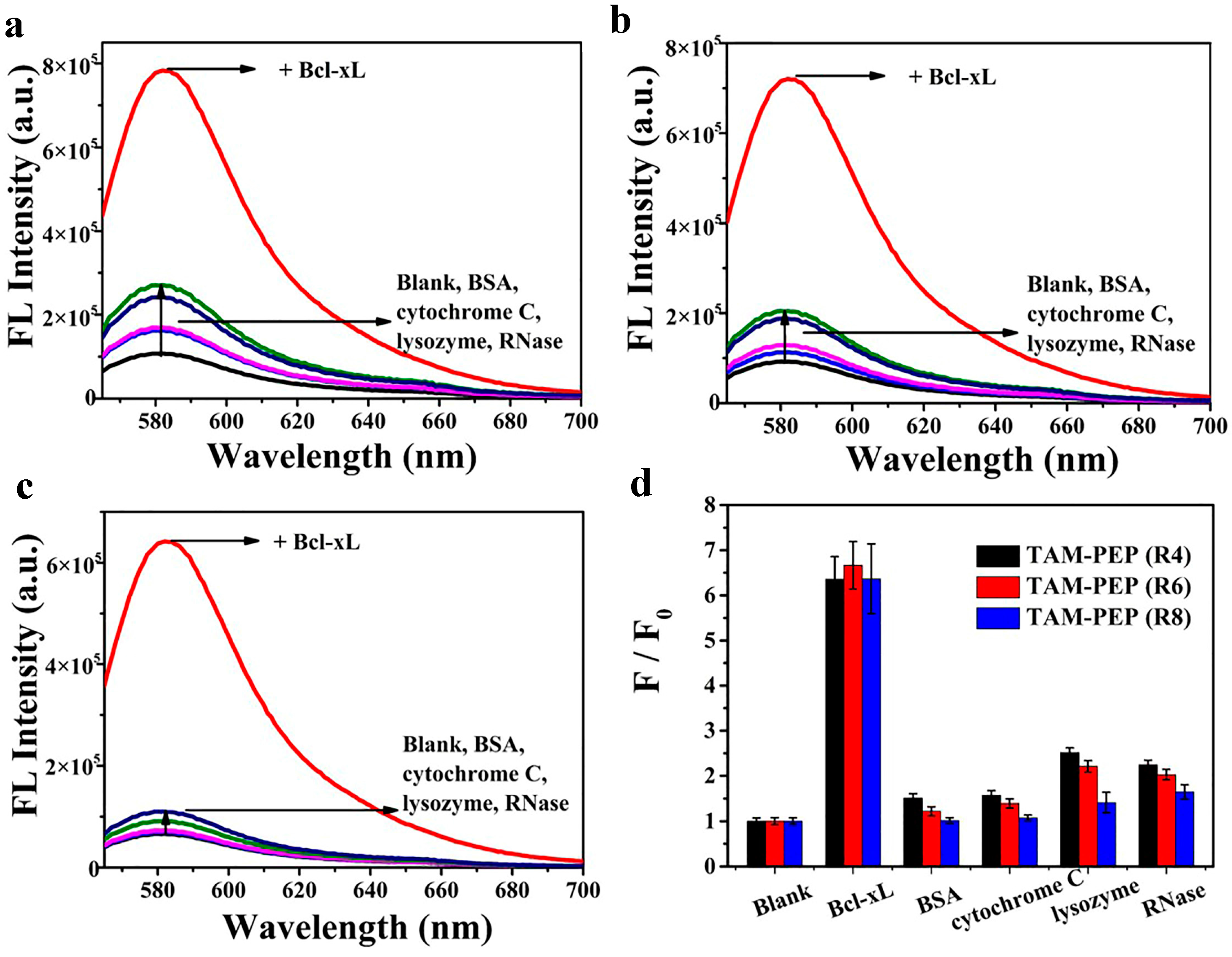
3.3. The Sensitivity and Specificity of the System
3.4. Bcl-xL Detection in Serum
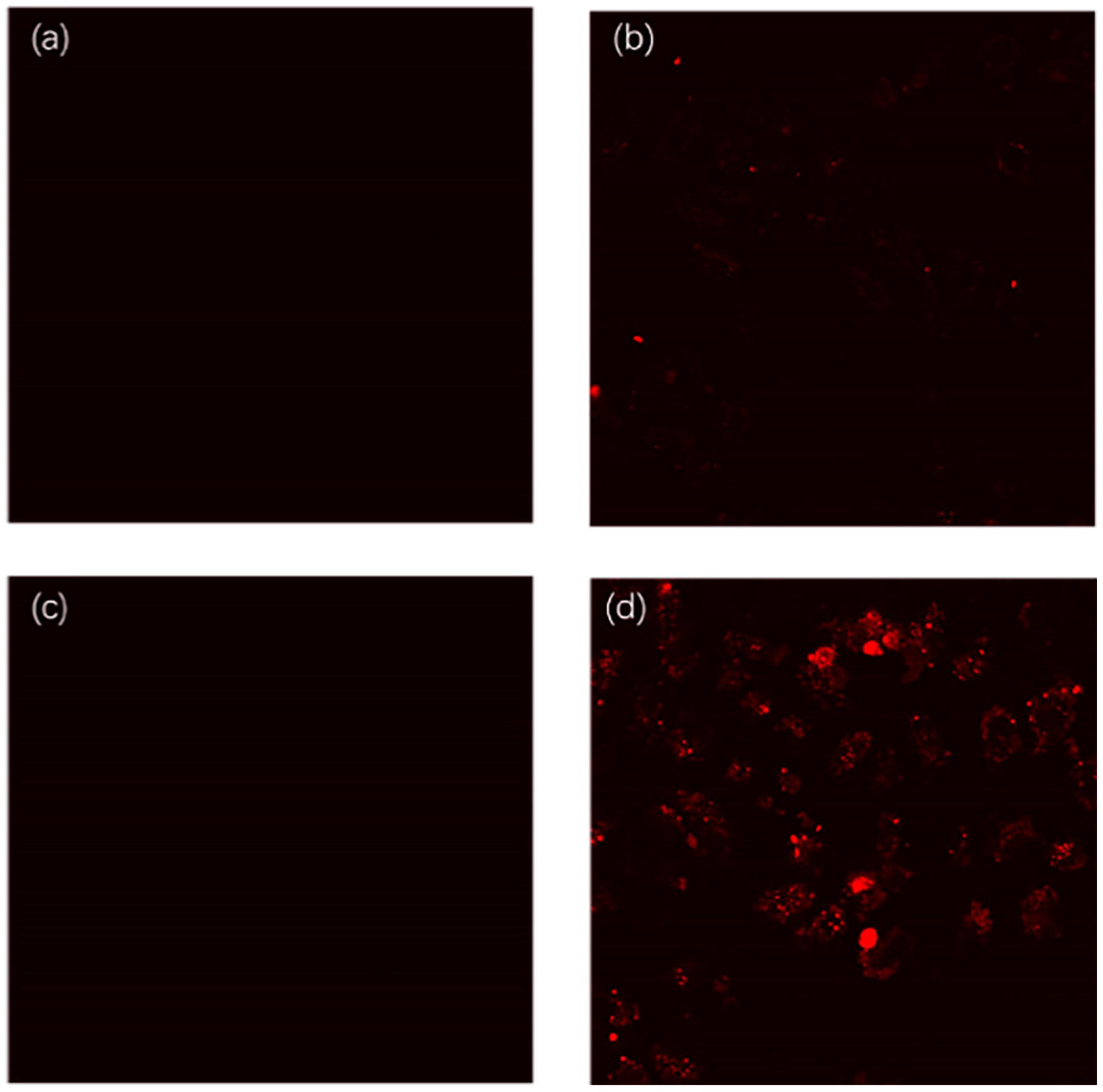
3.5. Cell Imaging
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Tompa, P. The principle of conformational signaling. Chem. Soc. Rev. 2016, 45, 4252–4284. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.T.; Sun, M.; Lu, M.X.; Du, Y.Z. Expression patterns of five heat shock proteins in Sesamia inferens (Lepidoptera: Noctuidae) during heat stress. J. Asia Pac. Entomol. 2015, 18, 529–533. [Google Scholar] [CrossRef]
- Jia, X.; Song, T.; Liu, Y.; Meng, L.; Mao, X. An immunochromatographic assay for carcinoembryonic antigen on cotton thread using a composite of carbon nanotubes and gold nanoparticles as reporters. Anal. Chim. Acta 2017, 969, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Pihikova, D.; Kasak, P.; Kubanikova, P.; Sokol, R.; Tkac, J. Aberrant sialylation of a prostate-specific antigen: Electrochemical label-free glycoprofiling in prostate cancer serum samples. Anal. Chim. Acta 2016, 934, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Shi, Z.; Fang, C.; Gao, A.; Li, C.M.; Yu, L. Versatile microfluidic complement fixation test for disease biomarker detection. Anal. Chim. Acta 2016, 916, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Qu, X. Cancer biomarker detection: Recent achievements and challenges. Chem. Soc. Rev. 2015, 44, 2963–2997. [Google Scholar] [CrossRef] [PubMed]
- Saita, T.; Yamamoto, Y.; Hosoya, K.; Yamamoto, Y.; Kimura, S.; Narisawa, Y.; Shin, M. An ultra-specific and sensitive sandwich ELISA for imatinib using two anti-imatinib antibodies. Anal. Chim. Acta 2017, 969, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Wang, Y.; Duan, X. Biofunctional polyelectrolytes assembling on biosensors—A versatile surface coating method for protein detections. Anal. Chim. Acta 2017, 964, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Hitchins, V.M.; Wickramasekara, S. Rapid detection of bacterial endotoxins in ophthalmic viscosurgical device materials by direct analysis in real time mass spectrometry. Anal. Chim. Acta 2016, 943, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Ma, H.; Lv, X.; Yan, T.; Li, N.; Cao, W.; Wei, Q. A novel electrochemical immunosensor using β-cyclodextrins functionalized silver supported adamantine-modified glucose oxidase as labels for ultrasensitive detection of alpha-fetoprotein. Anal. Chim. Acta 2015, 893, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Peng, W.P.; Chou, S.W.; Patil, A.A. Measuring masses of large biomolecules and bioparticles using mass spectrometric techniques. Analyst 2014, 139, 3507–3523. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Zhuo, Y.; Chai, Y.; Xiang, Y.; Liao, N.; Gui, G.; Yuan, R. Dual signal amplification strategy for the fabrication of an ultrasensitive electrochemiluminescenct aptasensor. Analyst 2013, 138, 6639–6644. [Google Scholar] [CrossRef] [PubMed]
- Gao, N.; Gao, F.; He, S.; Zhu, Q.; Huang, J.; Tanaka, H.; Wang, Q. Graphene oxide directed in-situ deposition of electroactive silver nanoparticles and its electrochemical sensing application for DNA analysis. Anal. Chim. Acta 2017, 951, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Chullasat, K.; Nurerk, P.; Kanatharana, P.; Kueseng, P.; Sukchuay, T.; Bunkoed, O. Hybrid monolith sorbent of polypyrrole-coated graphene oxide incorporated into a polyvinyl alcohol cryogel for extraction and enrichment of sulfonamides from water samples. Anal. Chim. Acta 2017, 961, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Aphale, A.N.; Macwan, I.G.; Patra, P.K.; Gonzalez, W.G.; Miksovska, J.; Leblanc, R.M. Graphene Oxide as a Quencher for Fluorescent Assay of Amino Acids, Peptides, and Proteins. ACS Appl. Mater. Interfaces 2012, 4, 7069–7075. [Google Scholar] [CrossRef] [PubMed]
- Feng, D.; Zhang, Y.; Feng, T.; Shi, W.; Li, X.; Ma, H. A graphene oxide-peptide fluorescence sensor tailor-made for simple and sensitive detection of matrix metalloproteinase 2. Chem. Commun. 2011, 47, 10680–10682. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, Q.; Chu, X.; Chen, T.; Ge, J.; Yu, R. Graphene oxide-peptide conjugate as an intracellular protease sensor for caspase-3 activation imaging in live cells. Angew. Chem. 2011, 50, 7065–7069. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Gao, Y.; Qi, Y.; Chen, L.; Ma, Y.; Li, Y. Peptide-based cancer therapy: Opportunity and challenge. Cancer Lett. 2014, 351, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Yin, B.C.; Wang, X.F.; Ye, B.C. Interaction of peptides with graphene oxide and its application for real-time monitoring of protease activity. Chem. Commun. 2011, 47, 2399–2401. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Yang, G.; Zhang, G.; Zhang, D.; Zhu, D. A new fluorescence turn-on assay for trypsin and inhibitor screening based on graphene oxide. ACS Appl. Mater. Interfaces 2011, 3, 1175–1179. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Müller, M.B.; Gilje, S.; Kaner, R.B.; Wallace, G.G. Processable aqueous dispersions of graphene nanosheets. Nat. Nanotechnol. 2008, 3, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Lerf, A.; He, H.; Forster, M.; Klinowski, J. Structure of Graphite Oxide RevisitedII. J. Phys. Chem. B 1998, 102, 4477–4482. [Google Scholar] [CrossRef]
- Li, X.; Ding, X.; Fan, J. Nicking endonuclease-assisted signal amplification of a split molecular aptamer beacon for biomolecule detection using graphene oxide as a sensing platform. Analyst 2015, 140, 7918–7925. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Tao, M.; Jin, Y. An enzyme-aided amplification strategy for sensitive detection of DNA utilizing graphene oxide (GO) as a fluorescence quencher. Analyst 2014, 139, 3455–3459. [Google Scholar] [CrossRef] [PubMed]
- Dho, S.H.; Deverman, B.E.; Lapid, C.; Manson, S.R.; Gan, L.; Riehm, J.J.; Aurora, R.; Kwon, K.S.; Weintraub, S.J. Control of cellular Bcl-xL levels by deamidation-regulated degradation. PLoS Biol. 2013, 11, e1001588–e1001603. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Liang, Z.; Hu, T.; Xu, S.; Ding, J.; Chen, K.; Jiang, H.; Liu, D. A conserved hydrophobic core at Bcl-xL mediates its structural stability and binding affinity with BH3-domain peptide of pro-apoptotic protein. Arch. Biochem. Biophys. 2009, 484, 46–54. [Google Scholar]
- Sun, Z.; Lu, W.; Tang, Y.; Zhang, J.; Chen, J.; Deng, H.; Li, X.; Liu, J. Expression, purification and characterization of human urodilatin in E. coli. Protein Expr. Purif. 2007, 55, 312–318. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Tan, C.; Xue, P.; Cao, J.; Liu, F.; Tan, Y.; Jiang, Y. Proteolysis targeting peptide (PROTAP) strategy for protein ubiquitination and degradation. Biochem. Biophys. Res. Commun. 2016, 470, 936–940. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Wu, J.; Wu, P.; Chen, Y.Z.; Tan, Y.; Tan, C.; Jiang, Y. A sensitive polymeric dark quencher-based sensing platform for fluorescence “turn on” detection of proteins. RSC Adv. 2016, 6, 42443–42446. [Google Scholar] [CrossRef]
- He, Y.; Lin, Y.; Tang, H.; Pang, D. A graphene oxide-based fluorescent aptasensor for the turn-on detection of epithelial tumor marker mucin 1. Nanoscale 2012, 4, 2054–2059. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Feng, J.; Liu, H.; Li, Q.; Tong, L.; Tang, B. Two-color Imaging of MicroRNA with Enzyme-Free Signal Amplification via Hybridization Chain Reactions in Living Cells. Chem. Sci. 2016, 7, 1940–1945. [Google Scholar] [CrossRef]
- Jiang, Y.; Fang, X.; Bai, C. Signaling Aptamer/Protein Binding by a Molecular Light Switch Complex. Anal. Chem. 2004, 76, 5230–5235. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, H.; Zhang, B.; Liu, S.; Tan, C.; Tan, Y.; Jiang, Y. A New Strategy Involving the Use of Peptides and Graphene Oxide for Fluorescence Turn-on Detection of Proteins. Sensors 2018, 18, 385. https://doi.org/10.3390/s18020385
Shi H, Zhang B, Liu S, Tan C, Tan Y, Jiang Y. A New Strategy Involving the Use of Peptides and Graphene Oxide for Fluorescence Turn-on Detection of Proteins. Sensors. 2018; 18(2):385. https://doi.org/10.3390/s18020385
Chicago/Turabian StyleShi, Huan, Bibo Zhang, Shuwen Liu, Chunyan Tan, Ying Tan, and Yuyang Jiang. 2018. "A New Strategy Involving the Use of Peptides and Graphene Oxide for Fluorescence Turn-on Detection of Proteins" Sensors 18, no. 2: 385. https://doi.org/10.3390/s18020385