Label-Free Quantification of Anti-TNF-α in Patients Treated with Adalimumab Using an Optical Biosensor
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Rreagents
2.2. Plasma Ccollection
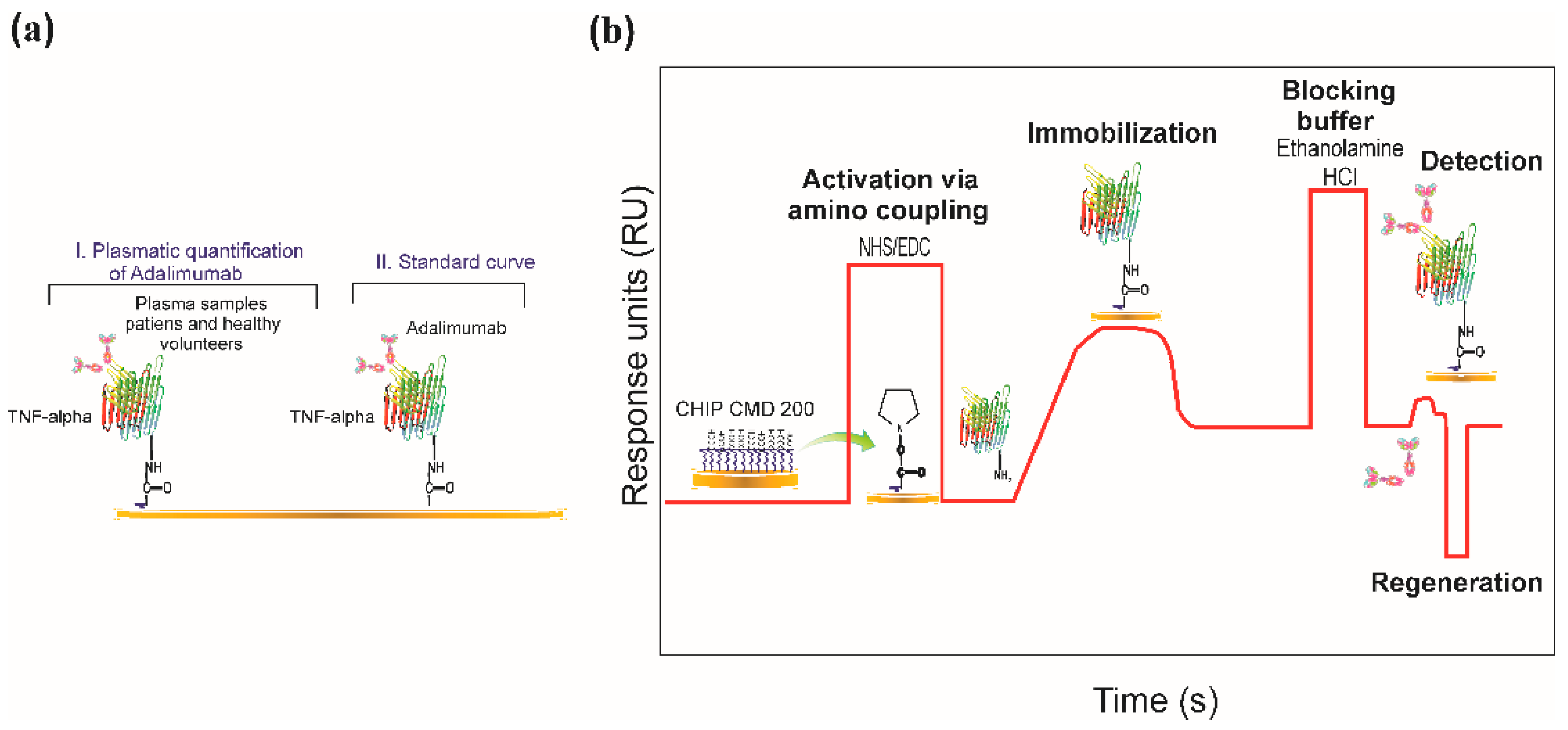
2.3. Biosensor System
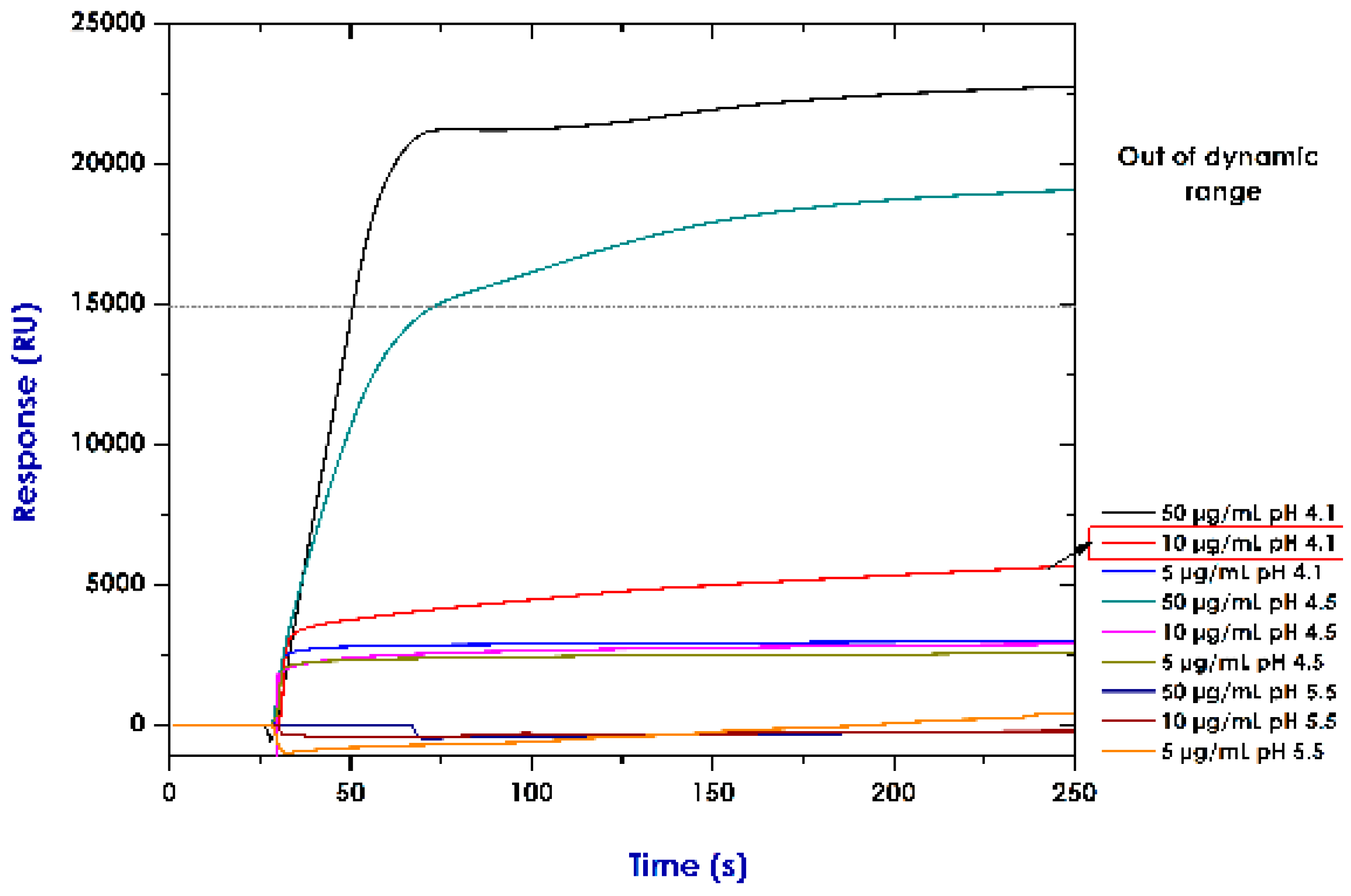
2.4. Establishing Conditions for Immobilising Recombinant Proteins (Pre-Concentration Assays)
2.5. Immobilising TNF-α Recombinant Protein
2.6. Regeneration Assays
2.7. SPR-Based Real-Time Quantification
2.8. ELISA Assays
2.9. Statistical Analysis
3. Results
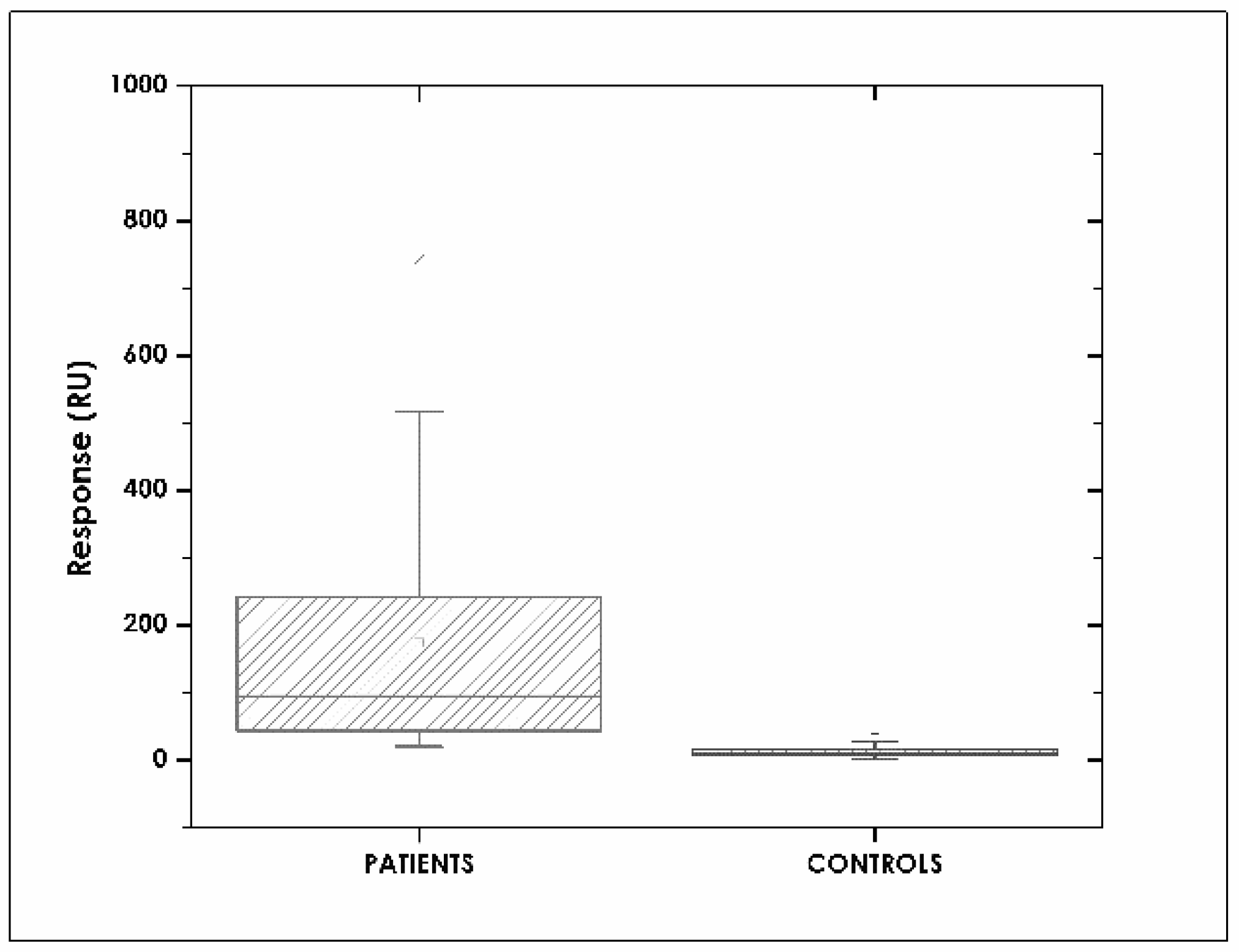
3.1. A Description of the Patients
3.2. Establishing Conditions for Immobilising Rrecombinant Pproteins (Pre-Concentration Assays)
3.3. Immobilising TNF-α Recombinant Protein
3.4. Regeneration Assays
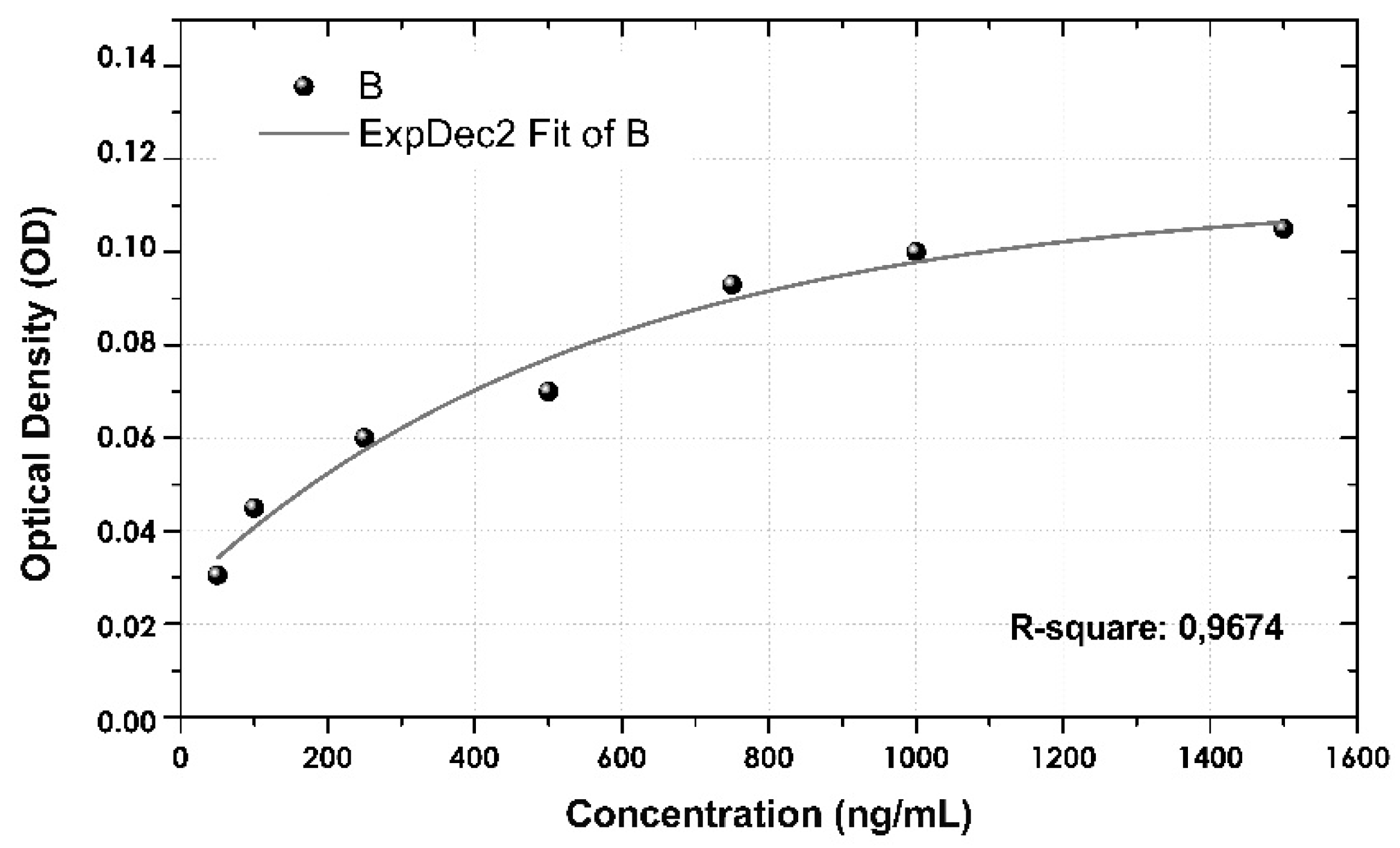
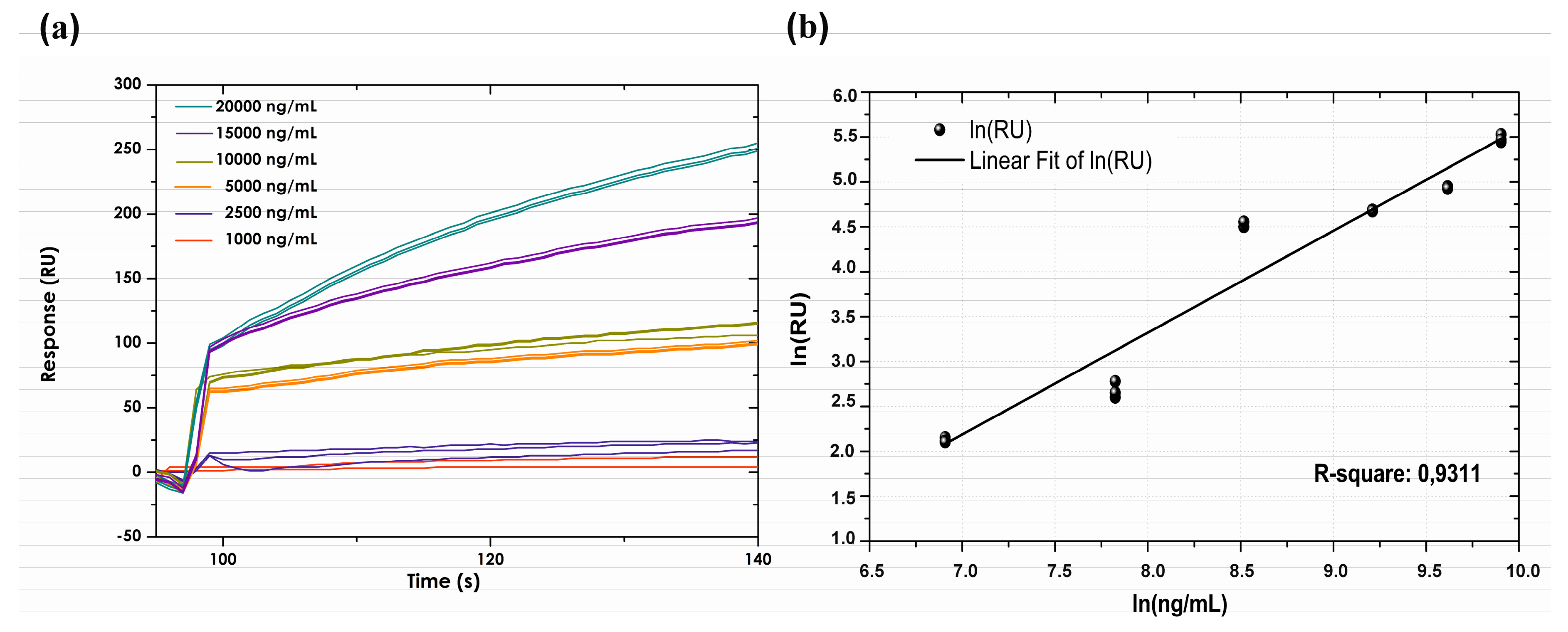
3.5. Calibration Curve
3.6. Assay Specificity
3.7. Correlation between SPR and ELISA Assays
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Appendix A
References
- Feagan, B.G.; Choquette, D.; Ghosh, S.; Gladman, D.D.; Ho, V.; Meibohm, B.; Zou, G.; Xu, Z.; Shankar, G.; Sealey, D.C.; et al. The challenge of indication extrapolation for infliximab biosimilars. Biologicals 2014, 42, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Bendtzen, K.; Svenson, M. Enzyme Immunoassays and Radioimmunoassays for Quantification of Anti-TNF Biopharmaceuticals and Anti-Drug Antibodies. In Detection and Quantification of Antibodies to Biopharmaceuticals; John Wiley & Sons: Hoboken, NJ, USA, 2011. [Google Scholar]
- European Medicines Agency. Guideline on Immunogenicity Assessment of Biotechnology-Derived Therapeutic Proteins. 2015. Available online: http://www.ema.europa.eu/docs/en_GB/document_library/Scientific_guideline/2015/10/WC500194507.pdf (accessed on 3 November 2017).
- US Food and Drug Administration. Biosimilars Guidances; US FDA: New Hampshire, MD, USA, 2017.
- WHO (World Health Organization). Guidelines of Evaluation for Similar Biotherapeutics Products (SBPs). 2009. Available online: http://www.who.int/biologicals/areas/biological_therapeutics/BIOTHERAPEUTICS_FOR_WEB_22APRIL2010.pdf (accessed on 21 February 2018).
- Ternant, D.; Paintaud, G. Pharmacokinetics and concentration-effect relationships of therapeutic monoclonal antibodies and fusion proteins. Expert Opin. Biol. Ther. 2005, 5, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, Y.; Takeuchi, T. Targeted antibody therapy and relevant novel biomarkers for precision medicine for rheumatoid arthritis. Int. Immunol. 2017, 29, 511–517. [Google Scholar] [CrossRef] [PubMed]
- Schmier, J.; Ogden, K.; Nickman, N.; Halpern, M.T.; Cifaldi, M.; Ganguli, A.; Bao, Y.; Garg, V. Costs of Providing Infusion Therapy for Rheumatoid Arthritis in a Hospital-based Infusion Center Setting. Clin. Ther. 2017, 39, 1600–1617. [Google Scholar] [CrossRef] [PubMed]
- Einodshofer, M.T.; Duren, L.N. Cost Management through Care Management, Part 2: The Importance of Managing Specialty Drug Utilization in the Medical Benefit. Am. Health Drug Benefits 2012, 5, 359–364. [Google Scholar] [PubMed]
- Batticciotto, A.; Ravasio, R.; Riva, M.; Sarzi-Puttini, P. Efficacy and Treatment Costs of Monotherapy with bDMARDs in the Treatment of Rheumatoid Arthritis in Patients Intolerant to or Inappropriate to Continue Treatment with Methotrexate. Adv. Ther. 2016, 33, 1360–1373. [Google Scholar] [CrossRef] [PubMed]
- Wong, U.; Cross, R.K. Primary and secondary nonresponse to infliximab: Mechanisms and countermeasures. Expert Opin. Drug Metab. Toxicol. 2017, 13, 1039–1046. [Google Scholar] [CrossRef] [PubMed]
- Ordas, I.; Mould, D.R.; Feagan, B.G.; Sandborn, W.J. Anti-TNF monoclonal antibodies in inflammatory bowel disease: Pharmacokinetics-based dosing paradigms. Clin. Pharmacol. Ther. 2012, 91, 635–646. [Google Scholar] [CrossRef] [PubMed]
- Kamath, A.V. Translational pharmacokinetics and pharmacodynamics of monoclonal antibodies. Drug Discov. Today Technol. 2016, 21–22, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Deng, R.; Jin, F.; Prabhu, S.; Iyer, S. Monoclonal antibodies: What are the pharmacokinetic and pharmacodynamic considerations for drug development? Expert Opin. Drug Metab. Toxicol. 2012, 8, 141–160. [Google Scholar] [CrossRef] [PubMed]
- Lobo, E.D.; Hansen, R.J.; Balthasar, J.P. Antibody pharmacokinetics and pharmacodynamics. J. Pharm. Sci. 2004, 93, 2645–2668. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wang, E.Q.; Balthasar, J.P. Monoclonal antibody pharmacokinetics and pharmacodynamics. Clin. Pharmacol. Ther. 2008, 84, 548–558. [Google Scholar] [CrossRef] [PubMed]
- Yanai, H.; Lichtenstein, L.; Assa, A.; Mazor, Y.; Weiss, B.; Levine, A.; Ron, Y.; Kopylov, U.; Bujanover, Y.; Rosenbach, Y.; et al. Levels of drug and antidrug antibodies are associated with outcome of interventions after loss of response to infliximab or adalimumab. Clin. Gastroenterol. Hepatol. 2015, 13, 522–530. [Google Scholar] [CrossRef] [PubMed]
- Van Schie, K.A.; Wolbink, G.J.; Rispens, T. Cross-reactive and pre-existing antibodies to therapeutic antibodies—Effects on treatment and immunogenicity. MAbs 2015, 7, 662–671. [Google Scholar] [CrossRef] [PubMed]
- Krishna, M.; Nadler, S.G. Immunogenicity to Biotherapeutics—The Role of Anti-drug Immune Complexes. Front. Immunol. 2016, 7, 21. [Google Scholar] [CrossRef] [PubMed]
- Jani, M.; Isaacs, J.D.; Morgan, A.W.; Wilson, A.G.; Plant, D.; Hyrich, K.L.; Chinoy, H.; Barton, A.; BRAGGSS. High frequency of antidrug antibodies and association of random drug levels with efficacy in certolizumab pegol-treated patients with rheumatoid arthritis: Results from the BRAGGSS cohort. Ann. Rheum. Dis. 2017, 76, 208–213. [Google Scholar] [CrossRef] [PubMed]
- Vincent, F.B.; Morand, E.F.; Murphy, K.; Mackay, F.; Mariette, X.; Marcelli, C. Antidrug antibodies (ADAb) to tumour necrosis factor (TNF)-specific neutralising agents in chronic inflammatory diseases: A real issue, a clinical perspective. Ann. Rheum. Dis. 2013, 72, 165–178. [Google Scholar] [CrossRef] [PubMed]
- Wolbink, G.J.; Vis, M.; Lems, W.; Voskuyl, A.E.; de Groot, E.; Nurmohamed, M.T.; Stapel, S.; Tak, P.P.; Aarden, L.; Dijkmans, B. Development of antiinfliximab antibodies and relationship to clinical response in patients with rheumatoid arthritis. Arthritis Rheum. 2006, 54, 711–715. [Google Scholar] [CrossRef] [PubMed]
- Prado, M.S.; Bendtzen, K.; Andrade, L.E.C. Biological anti-TNF drugs: Immunogenicity underlying treatment failure and adverse events. Expert Opin. Drug Metab. Toxicol. 2017, 13, 985–995. [Google Scholar] [CrossRef] [PubMed]
- Bartelds, G.M.; Krieckaert, C.L.; Nurmohamed, M.T.; van Schouwenburg, P.A.; Lems, W.F.; Twisk, J.W.; Dijkmans, B.A.; Aarden, L.; Wolbink, G.J. Development of antidrug antibodies against adalimumab and association with disease activity and treatment failure during long-term follow-up. JAMA 2011, 305, 1460–1468. [Google Scholar] [CrossRef] [PubMed]
- Seitz, K.; Zhou, H. Pharmacokinetic drug-drug interaction potentials for therapeutic monoclonal antibodies: Reality check. J. Clin. Pharmacol. 2007, 47, 1104–1118. [Google Scholar] [CrossRef] [PubMed]
- Darwish, M.; Burke, J.M.; Hellriegel, E.; Robertson, P., Jr.; Phillips, L.; Ludwig, E.; Munteanu, M.C.; Bond, M. An evaluation of the potential for drug-drug interactions between bendamustine and rituximab in indolent non-Hodgkin lymphoma and mantle cell lymphoma. Cancer Chemother. Pharmacol. 2014, 73, 1119–1127. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Sharma, A. Therapeutic protein-drug interactions: Plausible mechanisms and assessment strategies. Expert Opin. Drug Metab. Toxicol. 2016, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Matucci, A.; Nencini, F.; Pratesi, S.; Maggi, E.; Vultaggio, A. An overview on safety of monoclonal antibodies. Curr. Opin. Allergy Clin. Immunol. 2016, 16, 576–581. [Google Scholar] [CrossRef] [PubMed]
- Doessegger, L.; Banholzer, M.L. Clinical development methodology for infusion-related reactions with monoclonal antibodies. Clin. Transl. Immunol. 2015, 4, e39. [Google Scholar] [CrossRef] [PubMed]
- Vultaggio, A.; Maggi, E.; Matucci, A. Immediate adverse reactions to biologicals: From pathogenic mechanisms to prophylactic management. Curr. Opin. Allergy Clin. Immunol. 2011, 11, 262–268. [Google Scholar] [CrossRef] [PubMed]
- Hansel, T.T.; Kropshofer, H.; Singer, T.; Mitchell, J.A.; George, A.J. The safety and side effects of monoclonal antibodies. Nat. Rev. Drug Discov. 2010, 9, 325–338. [Google Scholar] [CrossRef] [PubMed]
- Vultaggio, A.; Matucci, A.; Nencini, F.; Pratesi, S.; Parronchi, P.; Rossi, O.; Romagnani, S.; Maggi, E. Anti-infliximab IgE and non-IgE antibodies and induction of infusion-related severe anaphylactic reactions. Allergy 2010, 65, 657–661. [Google Scholar] [CrossRef] [PubMed]
- Suntharalingam, G.; Perry, M.R.; Ward, S.; Brett, S.J.; Castello-Cortes, A.; Brunner, M.D.; Panoskaltsis, N. Cytokine storm in a phase 1 trial of the anti-CD28 monoclonal antibody TGN1412. N. Engl. J. Med. 2006, 355, 1018–1028. [Google Scholar] [CrossRef] [PubMed]
- Wing, M. Monoclonal antibody first dose cytokine release syndromes-mechanisms and prediction. J. Immunotoxicol. 2008, 5, 11–15. [Google Scholar] [CrossRef] [PubMed]
- Williams, M.; Khalid, T.; Hughes, S.; Bonney, D.; Wynn, R. Rituximab-induced Cytokine Storm in the Absence of Overt Lymphoproliferative Disease. J. Pediatr. Hematol. Oncol. 2016, 38, 29–31. [Google Scholar] [CrossRef] [PubMed]
- Seymour, H.E.; Worsley, A.; Smith, J.M.; Thomas, S.H. Anti-TNF agents for rheumatoid arthritis. Br. J. Clin. Pharmacol. 2001, 51, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Caminati, M.; Senna, G.; Stefanizzi, G.; Bellamoli, R.; Longhi, S.; Chieco-Bianchi, F.; Guarnieri, G.; Tognella, S.; Olivieri, M.; Micheletto, C. Drop-out rate among patients treated with omalizumab for severe asthma: Literature review and real-life experience. BMC Pulm. Med. 2016, 16, 128. [Google Scholar] [CrossRef] [PubMed]
- Nowatzke, W.L.; Rogers, K.; Wells, E.; Bowsher, R.R.; Ray, C.; Unger, S. Unique challenges of providing bioanalytical support for biological therapeutic pharmacokinetic programs. Bioanalysis 2011, 3, 509–521. [Google Scholar] [CrossRef] [PubMed]
- Van Bezooijen, J.S.; Koch, B.C.; van Doorn, M.B.; Prens, E.P.; van Gelder, T.; Schreurs, M.W. Comparison of Three Assays to Quantify Infliximab, Adalimumab, and Etanercept Serum Concentrations. Ther. Drug Monit. 2016, 38, 432–438. [Google Scholar] [CrossRef] [PubMed]
- Freeman, K.; Connock, M.; Auguste, P.; Taylor-Phillips, S.; Mistry, H.; Shyangdan, D.; Court, R.; Arasaradnam, R.; Sutcliffe, P.; Clarke, A. Clinical effectiveness and cost-effectiveness of use of therapeutic monitoring of tumour necrosis factor alpha (TNF-alpha) inhibitors [LISA-TRACKER(R) enzyme-linked immunosorbent assay (ELISA) kits, TNF-alpha-Blocker ELISA kits and Promonitor(R) ELISA kits] versus standard care in patients with Crohn’s disease: Systematic reviews and economic modelling. Health Technol. Assess. 2016, 20, 1–288. [Google Scholar] [PubMed]
- Chellaraj, V.; Cicero, K.; Abuarjah, K.; Lundberg, L.; Desai, H.; Gadkari, S.; Hantash, J.; Scott, G.; Beaver, C. Comparison Between Two UV ELISA Kits and an Electrochemiluminescence ELISA Method for the Quantification of HUMIRA® (Adalimumab) in Human Serum. 2012. Available online: http://www.inventivhealthclinical.com/497801ee-21a9-4235-91b1-2d3c9e279b38/download.htm (accessed on 5 November 2017).
- Bendtzen, K.; Geborek, P.; Svenson, M.; Larsson, L.; Kapetanovic, M.C.; Saxne, T. Individualized monitoring of drug bioavailability and immunogenicity in rheumatoid arthritis patients treated with the tumor necrosis factor alpha inhibitor infliximab. Arthritis Rheum. 2006, 54, 3782–3789. [Google Scholar] [CrossRef] [PubMed]
- Svenson, M.; Geborek, P.; Saxne, T.; Bendtzen, K. Monitoring patients treated with anti-TNF-alpha biopharmaceuticals: Assessing serum infliximab and anti-infliximab antibodies. Rheumatology 2007, 46, 1828–1834. [Google Scholar] [CrossRef] [PubMed]
- Radstake, T.R.; Svenson, M.; Eijsbouts, A.M.; van den Hoogen, F.H.; Enevold, C.; van Riel, P.L.; Bendtzen, K. Formation of antibodies against infliximab and adalimumab strongly correlates with functional drug levels and clinical responses in rheumatoid arthritis. Ann. Rheum. Dis. 2009, 68, 1739–1745. [Google Scholar] [CrossRef] [PubMed]
- The United States Pharmacopeial Convention. Available online: https://www.google.com.hk/url?sa=t&rct=j&q=&esrc=s&source=web&cd=1&ved=0ahUKEwiL9riFmLvZAhXMEbwKHdWOAwMQFgglMAA&url=https%3A%2F%2Fwww.ipqpubs.com%2Fwp-content%2Fuploads%2F2010%2F06%2FUSP_1032.pdf&usg=AOvVaw2leNseZklkroyS3UtgspjB (accessed on 20 January 2018).
- Moberg, A.; Lager, A.; Hamalainen, M.D.; Jarhede, T. Increased sensitivity of SPR assays in plasma through efficient parallel assay optimization. J. Pharm. Biomed. Anal. 2013, 78–79, 224–232. [Google Scholar] [CrossRef] [PubMed]
- Swanson, S.; Mytych, D. The use of Surface Plasmon Resonance for the Detection and Characterization of Antibodies. In Detection and Quantification of Antibodies to Biopharmaceuticals; John Wiley & Sons: Hoboken, NJ, USA, 2011. [Google Scholar]
- Mire-Sluis, A.R.; Barrett, Y.C.; Devanarayan, V.; Koren, E.; Liu, H.; Maia, M.; Parish, T.; Scott, G.; Shankar, G.; Shores, E. Recommendations for the design and optimization of immunoassays used in the detection of host antibodies against biotechnology products. J. Immunol. Methods 2004, 289, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Bhalla, N.; Jolly, P.; Formisano, N.; Estrela, P. Introduction to biosensors. Essays Biochem. 2016, 60, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Damborsky, P.; Svitel, J.; Katrlik, J. Optical biosensors. Essays Biochem. 2016, 60, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.; Nanda, R.; Sahoo, S.; Mohapatra, E. Biosensors in Health Care: The Milestones Achieved in Their Development towards Lab-on-Chip-Analysis. Biochem. Res. Int. 2016, 2016, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.H.; Park, J.; Kang, S.; Kim, M. Surface plasmon resonance: A versatile technique for biosensor applications. Sensors 2015, 15, 10481–10510. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Gallego, R.; Bosch, J.; Such-Sanmartin, G.; Segura, J. Surface plasmon resonance immunoassays—A perspective. Growth Horm. IGF Res. 2009, 19, 388–398. [Google Scholar] [CrossRef] [PubMed]
- Malmborg, A.C.; Michaelsson, A.; Ohlin, M.; Jansson, B.; Borrebaeck, C.A. Real time analysis of antibody-antigen reaction kinetics. Scand. J. Immunol. 1992, 35, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Mytych, D.T.; La, S.; Barger, T.; Ferbas, J.; Swanson, S.J. The development and validation of a sensitive, dual-flow cell, SPR-based biosensor immunoassay for the detection, semi-quantitation, and characterization of antibodies to darbepoetin alfa and epoetin alfa in human serum. J. Pharm. Biomed. Anal. 2009, 49, 415–426. [Google Scholar] [CrossRef] [PubMed]
- Wadhwa, M.; Bird, C.; Dilger, P.; Gaines-Das, R.; Thorpe, R. Strategies for detection, measurement and characterization of unwanted antibodies induced by therapeutic biologicals. J. Immunol. Methods 2003, 278, 1–17. [Google Scholar] [CrossRef]
- Centers for Medicare & Medicaid Services. Clinical Laboratory Improvement Amendments (CLIA). 2017. Available online: https://www.cms.gov/Regulations-and-Guidance/Legislation/CLIA/index.html?redirect=/CLIA (accessed on 21 February 2018).
- Van der Merwe, A. Surface Plasmon Resonance. 2011. Available online: http://www.biophysics.bioc.cam.ac.uk/wp-content/uploads/2011/02/spr1.pdf (accessed on 14 November 2017).
- O’Shannessy, D.J.; Brigham-Burke, M.; Peck, K. Immobilization chemistries suitable for use in the BIAcore surface plasmon resonance detector. Anal. Biochem. 1992, 205, 132–136. [Google Scholar] [CrossRef]
- Trilling, A.K.; Beekwilder, J.; Zuilhof, H. Antibody orientation on biosensor surfaces: A minireview. Analyst 2013, 138, 1619–1627. [Google Scholar] [CrossRef] [PubMed]
- Andersson, K.; Areskoug, D.; Hardenborg, E. Exploring buffer space for molecular interactions. J. Mol. Recognit. 1999, 12, 310–315. [Google Scholar] [CrossRef]
- GE Healthcare. BiacoreTM Assay Handbook. 2012. Available online: http://proteins.gelifesciences.com/~/media/protein-purification-ib/documents/handbooks/biacore_assay_handbook.pdf?la=en (accessed on 15 November 2017).
- Bustos, R.H.; Suesca, E.; Millan, D.; Gonzalez, J.M.; Fontanilla, M.R. Real-time quantification of proteins secreted by artificial connective tissue made from uni- or multidirectional collagen I scaffolds and oral mucosa fibroblasts. Anal. Chem. 2014, 86, 2421–2428. [Google Scholar] [CrossRef] [PubMed]
- Marquart, J.A. Surface Plasmon Resonance and Biomolecular Interaction Analysis Theory and Practice. 2014. Available online: http://www.lulu.com/shop/ja-marquart/surface-plasmon-resonance-and-biomolecular-interaction-analysis/hardcover/product-21404460.html (accessed on 20 February 2018).
- Homola, J.; Piliarik, M. Surface Plasmon Resonance (SPR) Sensors; Springer Series on Chemical Sensors and Biosensors; Springer: Berlin/Heidelberg, Germany, 2006. [Google Scholar]
- Jang, D.; Chae, G.; Shin, S. Analysis of Surface Plasmon Resonance Curves with a Novel Sigmoid-Asymmetric Fitting Algorithm. Sensors 2015, 15, 25385–25398. [Google Scholar] [CrossRef] [PubMed]
- Lundström, I. Real-time biospecific interaction analysis. Biosens. Bioelectron. 1994, 9, 725–736. [Google Scholar] [CrossRef]
- Sanquin. Adalimumab Level Elisa M1885. 2015. Available online: https://www.sanquin.nl/repository/reagentia/ifu/Product_flyer_Adalimumab_ELISA_kit.pdf (accessed on 15 November 2017).
- Bian, S.; Stappen, T.V.; Baert, F.; Compernolle, G.; Brouwers, E.; Tops, S.; Vries, A.; Rispens, T.; Lammertyn, J.; Vermeire, S.; et al. Generation and characterization of a unique panel of anti-adalimumab specific antibodies and their application in therapeutic drug monitoring assays. J. Pharm. Biomed. Anal. 2016, 125, 62–67. [Google Scholar] [CrossRef] [PubMed]
- Van der Neut Kolfschoten, M.; Schuurman, J.; Losen, M.; Bleeker, W.K.; Martinez-Martinez, P.; Vermeulen, E.; den Bleker, T.H.; Wiegman, L.; Vink, T.; Aarden, L.A. Anti-inflammatory activity of human IgG4 antibodies by dynamic Fab arm exchange. Science 2007, 317, 1554–1557. [Google Scholar] [CrossRef] [PubMed]
- Gizeli, E.; Lowe, C. Biomolecular Sensors; Taylor & Francis: London, UK, 2002. [Google Scholar]
- Lu, J.; Van Stappen, T.; Spasic, D.; Delport, F.; Vermeire, S.; Gils, A.; Lammertyn, J. Fiber optic-SPR platform for fast and sensitive infliximab detection in serum of inflammatory bowel disease patients. Biosens. Bioelectron. 2016, 79, 173–179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bian, S.; Lu, J.; Delport, F.; Vermeire, S.; Spasic, D.; Lammertyn, J.; Gils, A. Development and validation of an optical biosensor for rapid monitoring of adalimumab in serum of patients with Crohn’s disease. Drug Test. Anal. 2017. [Google Scholar] [CrossRef] [PubMed]
- Real Fernandez, F.; Di Pisa, M.; Rossi, G.; Auberger, N.; Lequin, O.; Larregola, M.; Benchohra, A.; Mansuy, C.; Chassaing, G.; Lolli, F. Antibody recognition in multiple sclerosis and Rett syndrome using a collection of linear and cyclic N-glucosylated antigenic probes. Biopolymers 2015, 104, 560–576. [Google Scholar] [CrossRef] [PubMed]
- Real-Fernandez, F.; Cimaz, R.; Rossi, G.; Simonini, G.; Giani, T.; Pagnini, I.; Papini, A.M.; Rovero, P. Surface plasmon resonance-based methodology for anti-adalimumab antibody identification and kinetic characterization. Anal. Bioanal. Chem. 2015, 407, 7477–7485. [Google Scholar] [CrossRef] [PubMed]
- Tam, Y.J.; Zeenathul, N.A.; Rezaei, M.A.; Mustafa, N.H.; Azmi, M.L.M.; Bahaman, A.R.; Lo, S.C.; Tan, J.S.; Hani, H.; Rasedee, A. Wide dynamic range of surface-plasmon-resonance-based assay for hepatitis B surface antigen antibody optimal detection in comparison with ELISA. Biotechnol. Appl. Biochem. 2017, 64, 735–744. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, E.M.; van de Kerkhof, D.; Hamann, D.; van Dongen, J.L.; Kuijper, P.H.; Brunsveld, L.; Scharnhorst, V.; Broeren, M.A. Therapeutic drug monitoring of infliximab: Performance evaluation of three commercial ELISA kits. Clin. Chem. Lab. Med. 2016, 54, 1211–1219. [Google Scholar] [CrossRef] [PubMed]
- Sanquin. Adalimumab Level ELISA. 2009. Available online: http://78.41.76.137/antibodyshop/datasheet/sanquin/M1885.pdf (accessed on 21 January 2018).



© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bustos, R.H.; Zapata, C.; Esteban, E.; García, J.-C.; Jáuregui, E.; Jaimes, D. Label-Free Quantification of Anti-TNF-α in Patients Treated with Adalimumab Using an Optical Biosensor. Sensors 2018, 18, 691. https://doi.org/10.3390/s18030691
Bustos RH, Zapata C, Esteban E, García J-C, Jáuregui E, Jaimes D. Label-Free Quantification of Anti-TNF-α in Patients Treated with Adalimumab Using an Optical Biosensor. Sensors. 2018; 18(3):691. https://doi.org/10.3390/s18030691
Chicago/Turabian StyleBustos, Rosa Helena, Carlos Zapata, Efraín Esteban, Julio-César García, Edwin Jáuregui, and Diego Jaimes. 2018. "Label-Free Quantification of Anti-TNF-α in Patients Treated with Adalimumab Using an Optical Biosensor" Sensors 18, no. 3: 691. https://doi.org/10.3390/s18030691