Unannounced Meals in the Artificial Pancreas: Detection Using Continuous Glucose Monitoring
Abstract
:1. Introduction
2. Materials and Methods
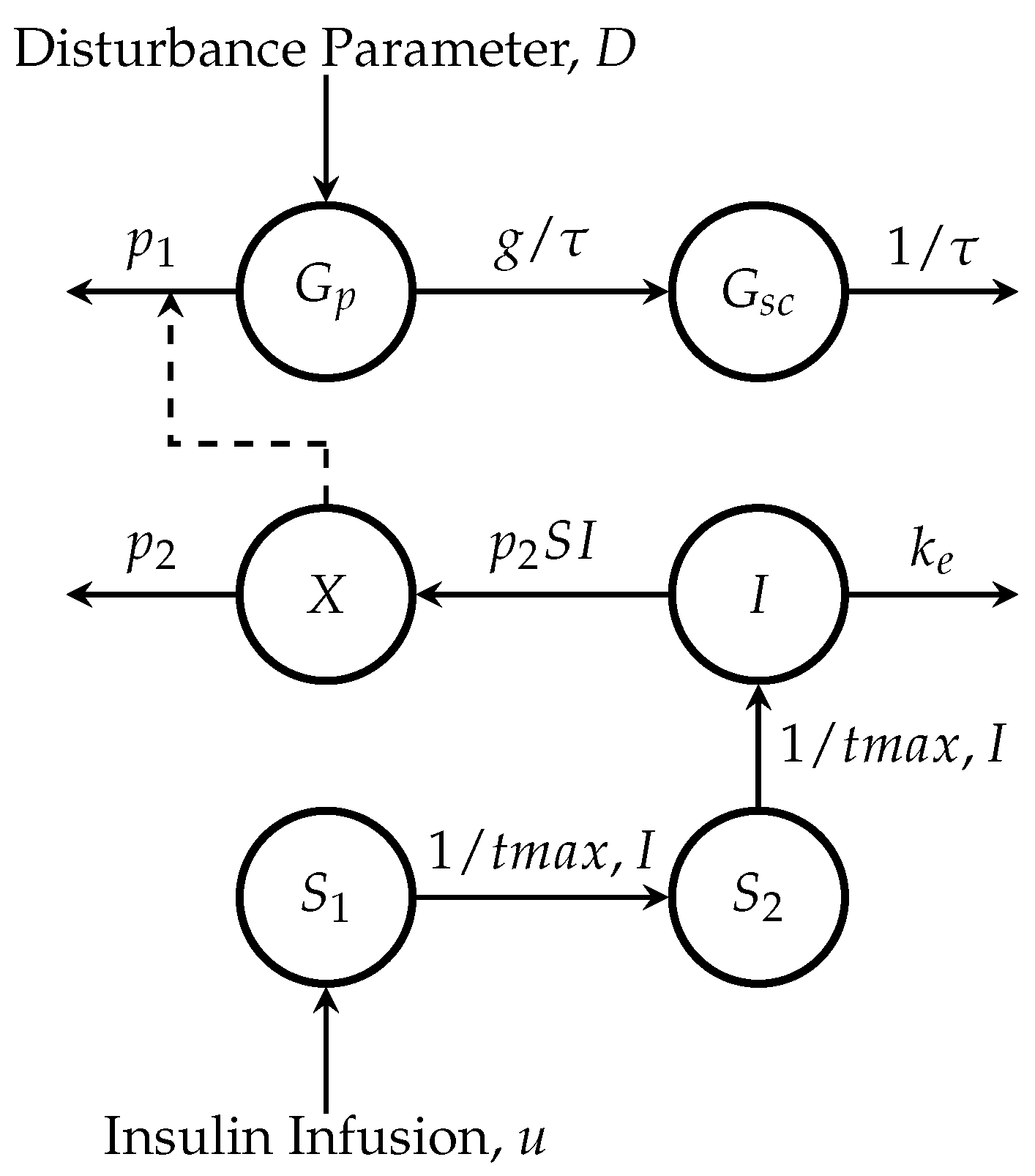
2.1. Minimal Model
2.1.1. Glucose Subsystem
2.1.2. Insulin Subsystem
2.2. Unscented Kalman Filter
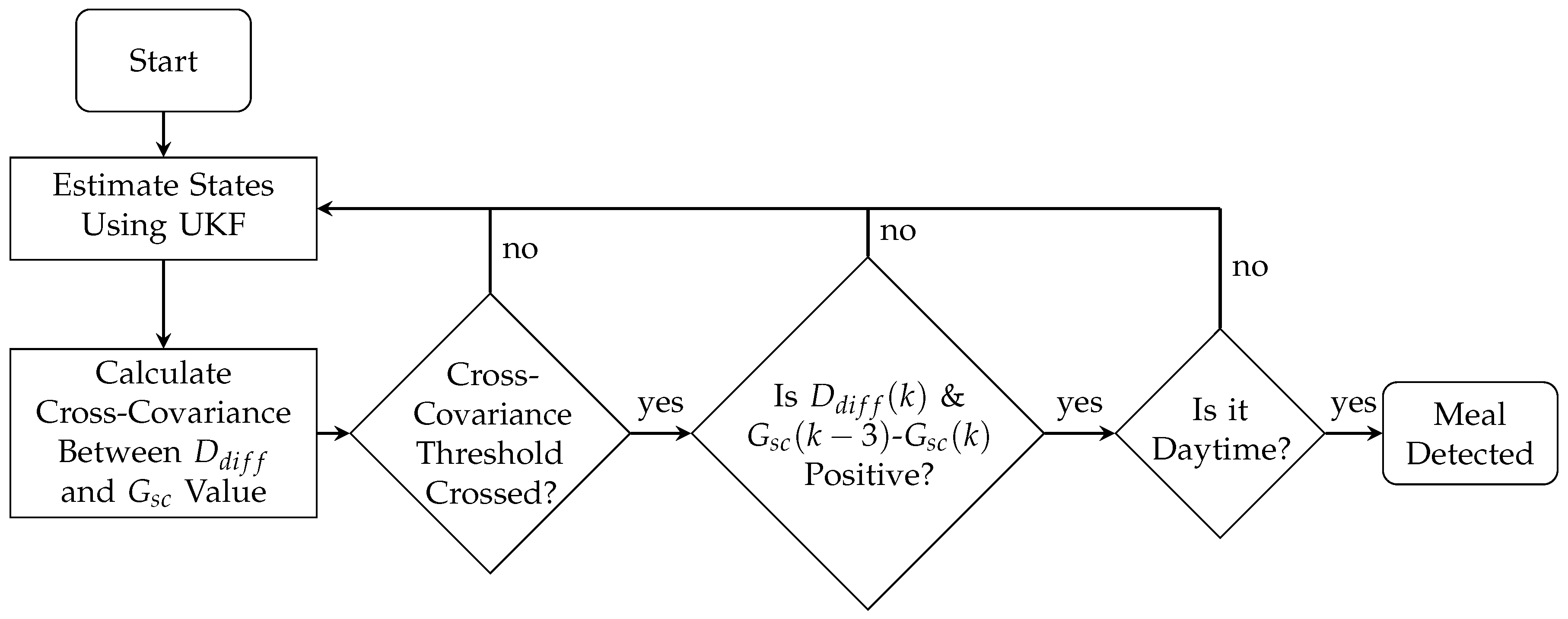
2.3. Meal Detection
2.4. Performance Metrics
2.5. Diabetes Simulation Scenario
2.5.1. Meal Detection Tuning and Validation Scenario
2.5.2. Meal Detection Sensitivity Analysis Scenario
3. Results
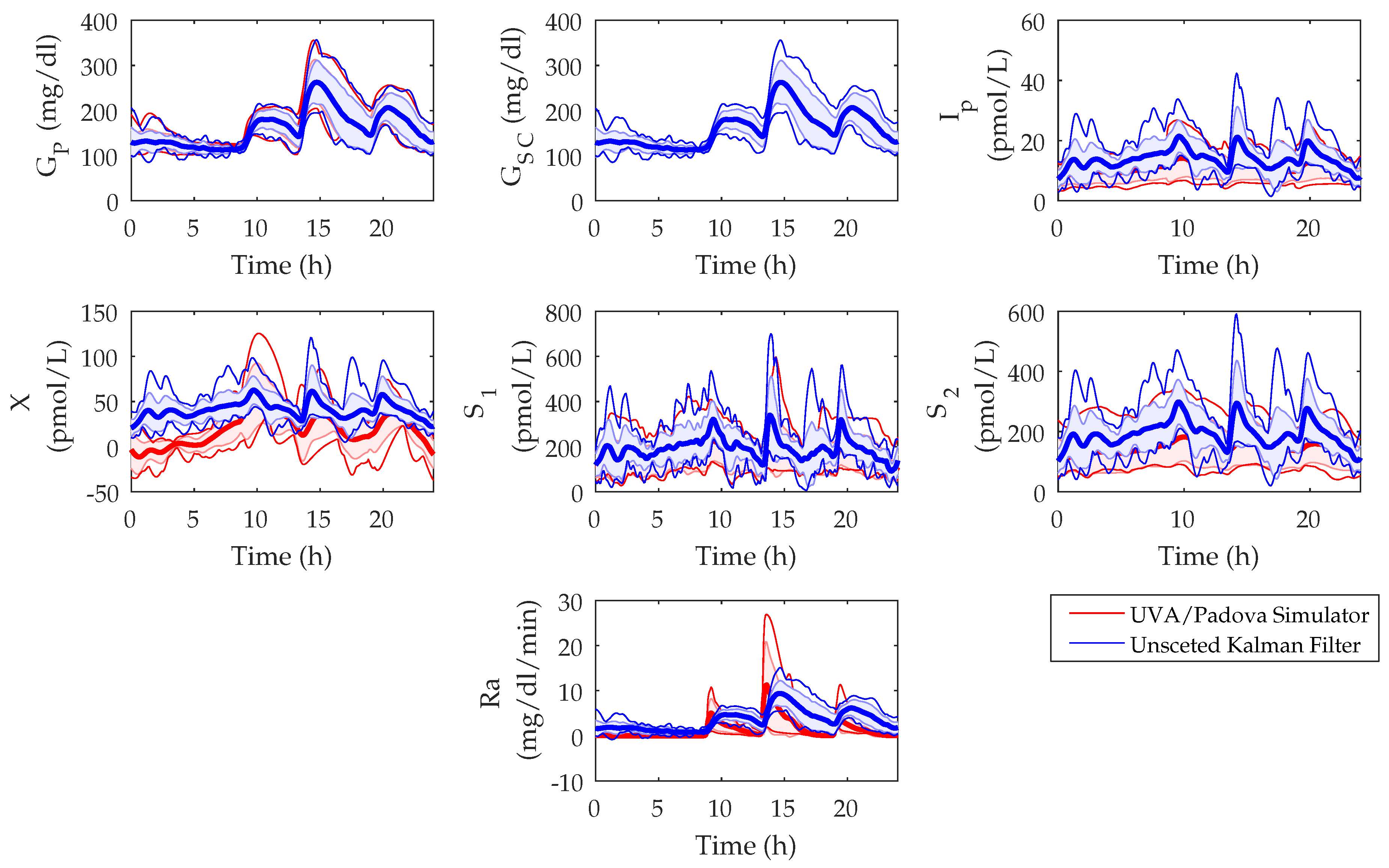
3.1. UKF State Estimations
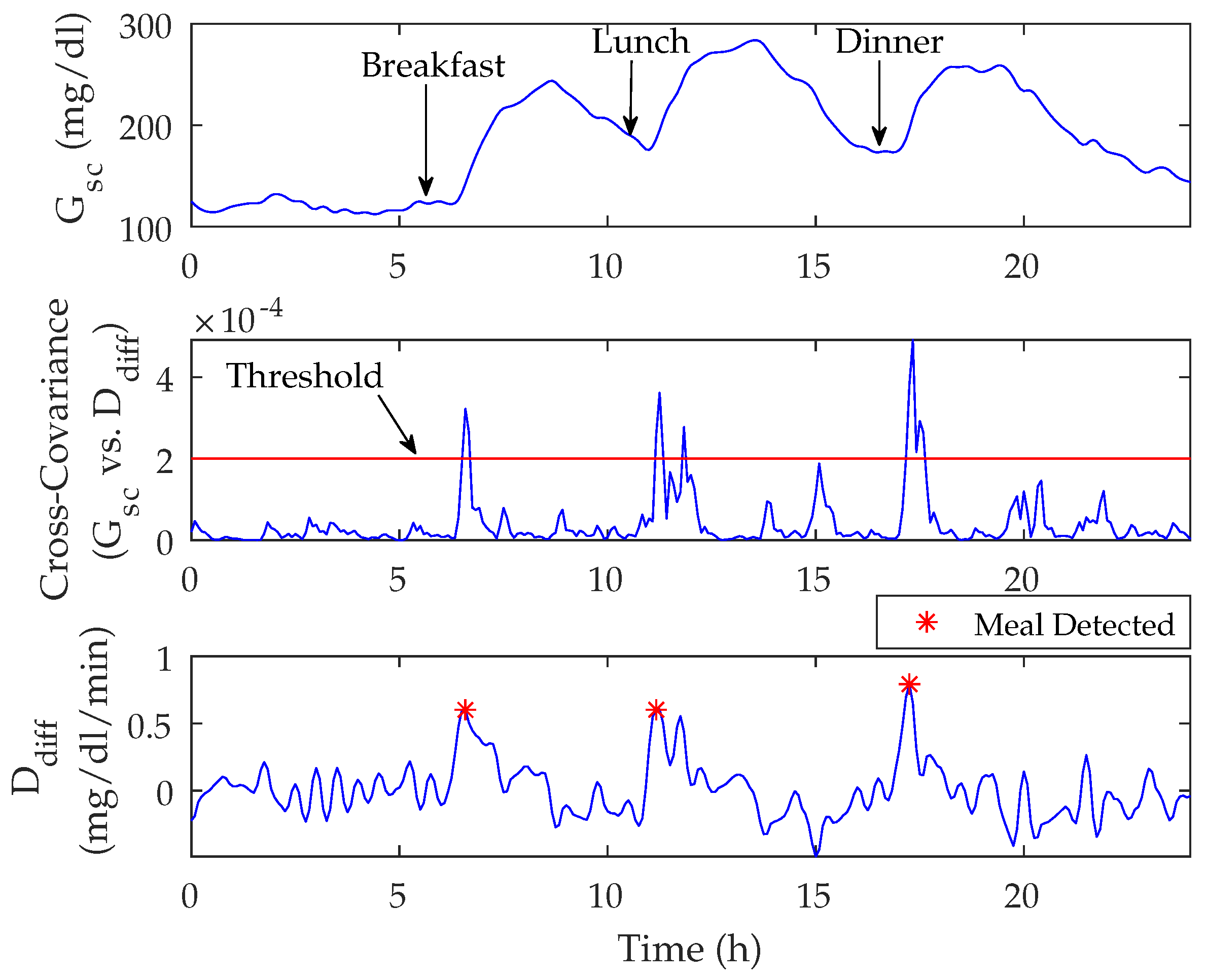
3.2. Meal Detection Algorithm
3.2.1. Meal Detection Algorithm: Tuning
3.2.2. Meal Detection Algorithm: Sensitivity Analysis
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| AP | Artificial Pancreas |
| BG | Blood Glucose |
| CGM | Continuous Glucose Monitor |
| FN | False Negative |
| FP | False Positive |
| HbA1c | Glycated Hemoglobin |
| Ra | Rate of Glucose Appearance |
| RMSE | Root Mean Squared Error |
| T1D | Type 1 Diabetes |
| TN | True Negative |
| TP | True Positive |
| UKF | Unscented Kalman Filter |
| UVA/Padova | University of Virginia/Padova |
References
- Burdick, J.; Chase, H.P.; Slover, R.H.; Knievel, K.; Scrimgeour, L.; Maniatis, A.K.; Klingensmith, G.J. Missed insulin meal boluses and elevated hemoglobin A1c levels in children receiving insulin pump therapy. Pediatrics 2004, 113, e221-4. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, M.; Donath, S.; Cameron, F. Poor adherence to integral daily tasks limits the efficacy of CSII in youth. Pediatr. Diabetes 2011, 12, 556–559. [Google Scholar] [CrossRef] [PubMed]
- Driscoll, K.a.; Johnson, S.B.; Hogan, J.; Gill, E.; Wright, N.; Deeb, L.C. Insulin bolusing software: The potential to optimize health outcomes in type 1 diabetes mellitus. J. Diabetes Sci. Technol. 2013, 7, 646–652. [Google Scholar] [CrossRef] [PubMed]
- Patton, S.R.; Clements, M.A.; Fridlington, A.; Cohoon, C.; Turpin, A.L.; DeLurgio, S.A. Frequency of Mealtime Insulin Bolus as a Proxy Measure of Adherence for Children and Youths with Type 1 Diabetes Mellitus. Diabetes Technol. Ther. 2013, 15, 124–128. [Google Scholar] [CrossRef] [PubMed]
- Ludvik, B.; Thomaseth, K.; Nolan, J.J.; Clodi, M.; Prager, R.; Pacini, G. Inverse relation between amylin and glucagon secretion in healthy and diabetic human subjects. Eur. J. Clin. Investig. 2003, 33, 316–322. [Google Scholar] [CrossRef]
- Haidar, A.; Elleri, D.; Kumareswaran, K.; Leelarathna, L.; Allen, J.M.; Caldwell, K.; Murphy, H.R.; Wilinska, M.E.; Acerini, C.L.; Evans, M.L.; et al. Pharmacokinetics of Insulin Aspart in Pump-Treated Subjects with Type 1 Diabetes: Reproducibility and Effect of Age, Weight, and Duration of Diabetes. Diabetes Care 2013, 36, e173–e174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Basu, A.; Dube, S.; Veettil, S.; Slama, M.; Kudva, Y.C.; Peyser, T.; Carter, R.E.; Cobelli, C.; Basu, R. Time lag of glucose from intravascular to interstitial compartment in type 1 diabetes. J. Diabetes Sci. Technol. 2015, 9, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Biagi, L.; Ramkissoon, C.M.; Facchinetti, A.; Leal, Y.; Vehi, J. Modeling the error of the medtronic paradigm veo enlite sensor. Sensors 2017, 17, 1361. [Google Scholar] [CrossRef] [PubMed]
- Reddy, M.; Herrero, P.; El Sharkawy, M.; Pesl, P.; Jugnee, N.; Pavitt, D.; Godsland, I.F.; Alberti, G.; Toumazou, C.; Johnston, D.G. Metabolic Control With the Bio-inspired Artificial Pancreas in Adults with Type 1 Diabetes A 24-Hour Randomized Controlled Crossover Study. J. Diabetes Sci. Technol. 2015, 10, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Dassau, E.; Zisser, H.; Harvey, R.A.; Percival, M.W.; Grosman, B.; Bevier, W.; Atlas, E.; Miller, S.; Nimri, R.; Jovanovic, L.; et al. Clinical evaluation of a personalized artificial pancreas. Diabetes Care 2013, 36, 801–809. [Google Scholar] [CrossRef] [PubMed]
- Dassau, E.; Bequette, B.W.; Buckingham, B.A.; Doyle, F.J., III. Detection of a Meal Using Continuous Glucose Monitoring. Diabetes Care 2008, 31, 295–300. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Bequette, B.W. A closed-loop artificial pancreas based on model predictive control: Human-friendly identification and automatic meal disturbance rejection. Biomed. Signal Process. Control 2009, 4, 347–354. [Google Scholar] [CrossRef]
- Lee, H.; Buckingham, B.A.; Wilson, D.M.; Bequette, B.W. A closed-loop artificial pancreas using model predictive control and a sliding meal size estimator. J. Diabetes Sci. Technol. 2009, 3, 1082–1090. [Google Scholar] [CrossRef] [PubMed]
- Cameron, F.; Niemeyer, G.; Buckingham, B.A. Probabilistic evolving meal detection and estimation of meal total glucose appearance. J. Diabetes Sci. Technol. 2009, 3, 1022–1030. [Google Scholar] [CrossRef] [PubMed]
- Cameron, F.; Niemeyer, G. Predicting Blood Glucose Levels Around Meals for Patients With Type I Diabetes. In Proceedings of the ASME 2010 Dynamic Systems and Control Conference, Cambridge, MA, USA, 12–15 September 2010; Volume 1, pp. 289–296. [Google Scholar]
- Chen, S.; Weimer, J.; Rickels, M.; Peleckis, A.; Lee, I. Towards a Model-based Meal Detector for Type I Diabetics. In Proceedings of the 6th Medical Cyber-Physical Systems Workshop, Seattle, WA, USA, 10–13 April 2015. [Google Scholar]
- Weimer, J.; Chen, S.; Peleckis, A.; Rickels, M.R.; Lee, I. Physiology-Invariant Meal Detection for Type 1 Diabetes. Diabetes Technol. Ther. 2016, 18, 616–624. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Wang, Q. Meal Detection and Meal Size Estimation for Type 1 Diabetes Treatment: A Variable State Dimension Approach. In Proceedings of the ASME 2015 Dynamic Systems and Control Conference, Columbus, OH, USA, 28–30 October 2015; Volume 1, p. V001T15A003. [Google Scholar]
- Turksoy, K.; Samadi, S.; Feng, J.; Littlejohn, E.; Quinn, L.; Cinar, A. Meal detection in patients with type 1 diabetes: A new module for the multivariable adaptive artificial pancreas control system. IEEE J. Biomed. Health Inform. 2016, 20, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Mahmoudi, Z.; Nørgaard, K.; Poulsen, N.K.; Madsen, H.; Jørgensen, J.B. Fault and meal detection by redundant continuous glucose monitors and the unscented Kalman filter. Biomed. Signal Process. Control 2017, 38, 86–99. [Google Scholar] [CrossRef]
- Turksoy, K.; Hajizadeh, I.; Samadi, S.; Feng, J.; Sevil, M.; Park, M.; Quinn, L.; Littlejohn, E.; Cinar, A. Real-time insulin bolusing for unannounced meals with artificial pancreas. Control Eng. Pract. 2017, 59, 159–164. [Google Scholar] [CrossRef]
- Cameron, F.; Ly, T.T.; Buckingham, B.A.; Maahs, D.M.; Forlenza, G.P.; Levy, C.J.; Lam, D.; Clinton, P.; Messer, L.H.; Westfall, E.; et al. Closed-Loop Control Without Meal Announcement in Type 1 Diabetes. Diabetes Technol. Ther. 2017, 19, 527–532. [Google Scholar] [CrossRef] [PubMed]
- Bergman, R.N.; Phillips, L.S.; Cobelli, C. Physiologic evaluation of factors controlling glucose tolerance in man: measurement of insulin sensitivity and beta-cell glucose sensitivity from the response to intravenous glucose. J. Clin. Investig. 1981, 68, 1456–1467. [Google Scholar] [CrossRef] [PubMed]
- Facchinetti, A.; Sparacino, G.; Cobelli, C. Enhanced Accuracy of Continuous Glucose Monitoring by Online Extended Kalman Filtering. Diabetes Technol. Ther. 2010, 12, 353–363. [Google Scholar] [CrossRef] [PubMed]
- Hovorka, R.; Canonico, V.; Chassin, L.J.; Haueter, U.; Massi-benedetti, M.; Federici, M.O.; Pieber, T.R.; Schaller, H.C.; Schaupp, L.; Vering, T.; et al. Nonlinear model predictive control of glucose concentration in subjects with type 1 diabetes. Physiol. Meas. 2004, 25, 905–920. [Google Scholar] [CrossRef] [PubMed]
- Herrero, P.; Calm, R.; Vehí, J.; Armengol, J.; Georgiou, P.; Oliver, N.; Tomazou, C. Robust fault detection system for insulin pump therapy using continuous glucose monitoring. J. Diabetes Sci. Technol. 2012, 6, 1131–1141. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Parker, R.S. Dynamic modeling of exercise effects on plasma glucose and insulin levels. J. Diabetes Sci. Technol. 2007, 1, 338–347. [Google Scholar] [CrossRef] [PubMed]
- Julier, S.J.; Uhlmann, J.K. Unscented filtering and nonlinear estimation. Proc. IEEE 2004, 92, 401–422. [Google Scholar] [CrossRef]
- Uhlmann, J.K. Simultaneous Map Building and Localization for Real Time Applications; Technical Report; University of Oxford: Oxford, UK, 1994. [Google Scholar]
- Larsen, J. Correlation Functions and Power Spectra, Section for Cognitive Systems, Informatics and Mathematical Modelling; Technical University of Denmark: Lyngby, Denmark, 2009. [Google Scholar]
- Dalla Man, C.; Micheletto, F.; Lv, D.; Breton, M.; Kovatchev, B.; Cobelli, C. The UVA/PADOVA Type 1 Diabetes Simulator: New Features. J. Diabetes Sci. Technol. 2014, 8, 26–34. [Google Scholar]
- Dalla Man, C.; Camilleri, M.; Cobelli, C. A System Model of Oral Glucose Absorption: Validation on Gold Standard Data. IEEE Trans. Biomed. Eng. 2006, 53, 2472–2478. [Google Scholar] [PubMed]
- Bell, K.J.; Smart, C.E.; Steil, G.M.; Brand-Miller, J.C.; King, B.; Wolpert, H.A. Impact of Fat, Protein, and Glycemic Index on Postprandial Glucose Control in Type 1 Diabetes: Implication for Intensive Diabetes Management in the Continuous Glucose Monitoring Era. Diabetes Care 2015, 38, 1008–1015. [Google Scholar] [CrossRef] [PubMed]
| Symbol | Quantity | Value | Units | Reference |
|---|---|---|---|---|
| Glucose removal rate from the plasma space independent of the influence of insulin | 0.035 | 1/min | [27] | |
| Disappearance rate of remote insulin from the remote insulin compartment | 0.05 | 1/min | [27] | |
| Appearance rate of remote insulin into the remote insulin compartment | 0.000028 | mL/U · min | [27] | |
| Basal plasma glucose | 100 | mg/dL | [23] | |
| Volume distribution of glucose compartment | 1.6 | dL/kg | [25] | |
| Volume distribution of insulin compartment | 120 | mL/kg | [25] | |
| First-order decay rate of insulin in plasma | 0.138 | 1/min | [25] | |
| Time-to-maximum insulin absorption | 55 | min | [25] | |
| Time constant of the system | 8.2237 | min | [24] | |
| g | Static gain of the system | 1 | unitless | [24] |
| Sensitivity (%) | Glucose (mg/dL) | Detection Time (min) | TP | FP | FN | FP/day | |
|---|---|---|---|---|---|---|---|
| Highest Sensitivity (window = 3; threshold = 0.000039) | |||||||
| Tuning | 99 ± 2 | 7 ± 7 | 28 ± 10 | 41 ± 1 | 17 ± 5 | 1 ± 1 | 1 ± 1 |
| 99 (95, 100) | 6 (−1, 17) | 25 (15, 45) | 42 (40, 42) | 19 (9, 25) | 1 (0, 2) | 1 (0, 3) | |
| Validation | 98 ± 4 | 6 ± 8 | 28 ± 9 | 41 ± 2 | 18 ± 5 | 1 ± 2 | 1 ± 1 |
| 98 (90, 100) | 5 (−2, 13) | 25 (15, 40) | 42 (38, 42) | 20 (11, 26) | 0 (0, 4) | 1 (0, 3) | |
| Trade-Off (window = 9; threshold = 0.00019) | |||||||
| Tuning | 93 ± 5 | 19 ± 5 | 37 ± 9 | 39 ± 2 | 3 ± 2 | 3 ± 2 | 0.2 ± 0.5 |
| 93 (86, 100) | 19 (11, 28) | 35 (25, 55) | 39 (36, 42) | 3 (0, 7) | 3 (0, 6) | 0 (0, 1) | |
| Validation | 93 ± 6 | 19 ± 5 | 37 ± 83 | 39 ± 3 | 3 ± 3 | 3 ± 3 | 0 ± 0 |
| 94 (83, 100) | 18.43 (11, 26) | 35 (25, 50) | 40 (35, 42) | 4 (1, 9) | 3 (0, 7) | 0 (0, 0) | |
| Lowest False Positive (window = 12; threshold = 0.00082) | |||||||
| Tuning | 47 ± 12 | 45 ± 7 | 46 ± 8 | 20± 5 | 0 ± 0 | 22 ± 5 | 0 ± 0 |
| 50 (26, 64) | 45 (36, 54) | 45 (35, 60) | 21 (11, 27) | 0 (0, 0) | 21 (15, 31) | 0 (0, 0) | |
| Validation | 46 ± 16 | 43 ± 7 | 48 ± 8 | 19 ± 7 | 0.2 ± 0.4 | 23 ± 7 | 0.01 ± 0.1 |
| 45 (29, 71) | 45 (35, 56) | 50 (35, 60) | 19 (12, 30) | 0 (0, 1) | 23 (12, 30) | 0 (0, 0) | |
| Sensitivity (%) | Glucose (mg/dL) | Detection Time (min) | TP | FP | FN | FP/day |
|---|---|---|---|---|---|---|
| Highest Sensitivity (window = 3; threshold = 0.000039) | ||||||
| 92 ± 3 | 6 ± 13 | 31 ± 16 | 1375 ± 38 | 719 ± 111 | 126 ± 38 | 1 ± 1 |
| 92 (87, 96) | 5 (−8, 17) | 25 (10, 60) | 1374 (1306, 1433) | 716 (561, 897) | 126 (67, 194) | 1 (0, 3) |
| Trade-Off (window = 9; threshold = 0.00019) | ||||||
| 82 ± 4 | 19 ± 9 | 38 ± 14 | 1232 ± 56 | 100 ± 23 | 268 ± 56 | 0.2 ± 0.5 |
| 83 (76, 87) | 19 (6, 31) | 35 (25, 65) | 1242 (1139, 1310) | 96 (72, 154) | 259 (190, 361) | 0 (0, 1) |
| Low False Positive (window = 12; threshold = 0.00082) | ||||||
| 54 ± 9 | 42 ± 18 | 43 ± 11 | 813 ± 130 | 11 ± 9 | 687 ± 130 | 0.02 ± 0.2 |
| 52 (44, 71) | 45 (10, 56) | 40 (30, 65) | 776 (663, 1060) | 10 (5, 34) | 724 (440, 837) | 0 (0, 0) |
| Carbohydrates (grams) | |||||
|---|---|---|---|---|---|
| 20–40 | 40–80 | 80–120 | |||
| High Sensitivity window = 3 threshold = 0.000039 | Detection Time (min) | Mean | 39 ± 22 | 31 ± 16 | 25 ± 13 |
| Median | 30 (10, 90) | 30 (10, 60) | 25 (0, 50) | ||
| Glucose (mg/dL) | Mean | 5 ± 14 | 5 ± 10 | 7 ± 15 | |
| Median | 5 (−13, 23) | 5 (−7, 16) | 6 (−6, 15) | ||
| Sensitivity (%) | Mean | 74 ± 6 | 93 ± 3 | 99 ± 1 | |
| Median | 72 (67, 87) | 94 (87,96) | 99 (97, 100) | ||
| Trade-Off window = 9 threshold = 0.00019 | Detection Time (min) | Mean | 46 ± 18 | 40 ± 16 | 32± 15 |
| Median | 45 (25, 80) | 35 (25, 65) | 30 (0, 55) | ||
| Glucose (mg/dL) | Mean | 18 ± 11 | 17 ± 9 | 20 ± 9 | |
| Median | 19 (0, 32) | 18 (0, 28) | 20 (8, 32) | ||
| Sensitivity (%) | Mean | 49 ± 9 | 84 ± 4 | 96 ± 2 | |
| Median | 47 (39, 64) | 86 (76, 89) | 97 (94, 98) | ||
| Low False Positive window = 12 threshold = 0.00082 | Detection Time (min) | Mean | 45 ± 11 | 44 ± 8 | 41 ± 11 |
| Median | 50 (14, 71) | 45 (30, 65) | 40 (30, 60) | ||
| Glucose (mg/dL) | Mean | 29 ± 42 | 42 ± 18 | 42 ± 16 | |
| Median | 43 (−46, 57) | 45 (12, 57) | 44 (16, 56) | ||
| Sensitivity (%) | Mean | 8 ± 7 | 49 ± 12 | 83 ± 7 | |
| Median | 6 (2, 26) | 45 (36, 71) | 82 (73, 93) | ||
| Rate of Glucose Appearance | |||||
|---|---|---|---|---|---|
| Slow | Medium | Fast | |||
| High Sensitivity window = 3 threshold = 0.000039 | Detection Time (min) | Mean | 47 ± 22 | 38 ± 20 | 27 ± 11 |
| Median | 45 (10, 90) | 35 (10, 80) | 25 (10, 45) | ||
| Glucose (mg/dL) | Mean | 3 ± 15 | 4 ± 13 | 6 ± 13 | |
| Median | 4 (−22, 25) | 4 (−14, 23) | 6 (−5, 15) | ||
| Sensitivity (%) | Mean | 77 ± 9 | 87 ± 4 | 95 ± 1 | |
| Median | 77 (62, 92) | 88 (81, 95) | 95 (92, 97) | ||
| Trade-Off window = 9 threshold = 0.00019 | Detection Time (min) | Mean | 57 ± 18 | 47 ± 16 | 34 ± 10 |
| Median | 55 (35, 90) | 45 (30, 80) | 30 (25, 50) | ||
| Glucose (mg/dL) | Mean | 16 ± 14 | 18 ± 12 | 20 ± 7 | |
| Median | 18 (−5, 36) | 19 (1, 33) | 19 (9, 30) | ||
| Sensitivity (%) | Mean | 54 ± 10 | 73 ± 6 | 89 ± 3 | |
| Median | 57 (38, 71) | 73 (65, 81) | 89 (84, 92) | ||
| Low False Positive window = 12 threshold = 0.00082 | Detection Time (min) | Mean | 63 ± 14 | 52 ± 13 | 40 ± 9 |
| Median | 65 (45, 80) | 50 (35, 75) | 40 (30, 55) | ||
| Glucose (mg/dL) | Mean | 37 ± 44 | 43 ± 24 | 41 ± 15 | |
| Median | 48 (−39, 72) | 47 (−2, 63) | 44 (13, 55) | ||
| Sensitivity (%) | Mean | 10 ± 8 | 33 ± 12 | 68 ± 7 | |
| Median | 6 (2, 29) | 29 (18, 57) | 67 (59, 81) | ||
| Reference | Sensitivity (%) | Glucose (mg/dL) | Detection Time (min) | TP | FP | FN | FP/day |
|---|---|---|---|---|---|---|---|
| Dassau et al. [11] | − | − | 30 | − | − | − | − |
| Lee et al. [13] | 82 | − | 31 | 656 | 54 | 144 | − |
| Chen et al. [16] | 99.6 | − | − | − | − | − | − |
| Weimer et al. [17] | 86.9 | − | − | − | − | − | 2.01 |
| Xie and Wang [18] | 95 | − | − | − | − | − | − |
| Turksoy et al. [19] | 97 ± 6 | 16 ± 9 | − | 7 ± 2 | 0.1 ± 0.3 | 0.2 ± 0.4 | − |
| Mahmoudi et al. [20] | 99.5 | 46.3 ± 21.2 | 58.4 ± 18.7 | − | − | − | − |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramkissoon, C.M.; Herrero, P.; Bondia, J.; Vehi, J. Unannounced Meals in the Artificial Pancreas: Detection Using Continuous Glucose Monitoring. Sensors 2018, 18, 884. https://doi.org/10.3390/s18030884
Ramkissoon CM, Herrero P, Bondia J, Vehi J. Unannounced Meals in the Artificial Pancreas: Detection Using Continuous Glucose Monitoring. Sensors. 2018; 18(3):884. https://doi.org/10.3390/s18030884
Chicago/Turabian StyleRamkissoon, Charrise M., Pau Herrero, Jorge Bondia, and Josep Vehi. 2018. "Unannounced Meals in the Artificial Pancreas: Detection Using Continuous Glucose Monitoring" Sensors 18, no. 3: 884. https://doi.org/10.3390/s18030884






