A Urea Biosensor from Stacked Sol-Gel Films with Immobilized Nile Blue Chromoionophore and Urease Enzyme
Abstract
:1. Introduction
2. Results and Discussion
2.1 The pH sensor based on sol-gel Nile blue chromoionophore film
2.2 Effect of buffer capacity on the optical biosensor response
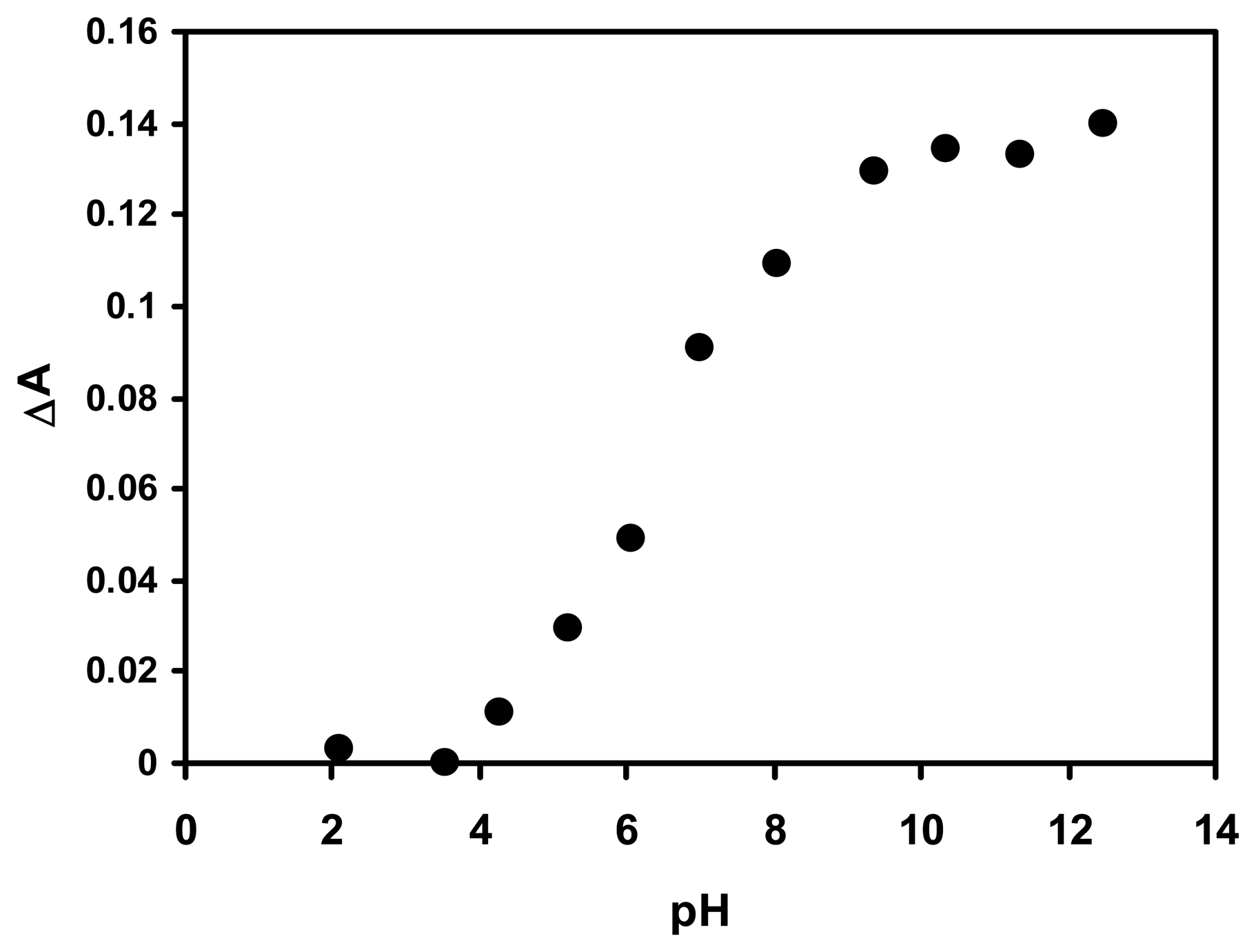
2.3 Effect of pH on the response of the optical biosensor
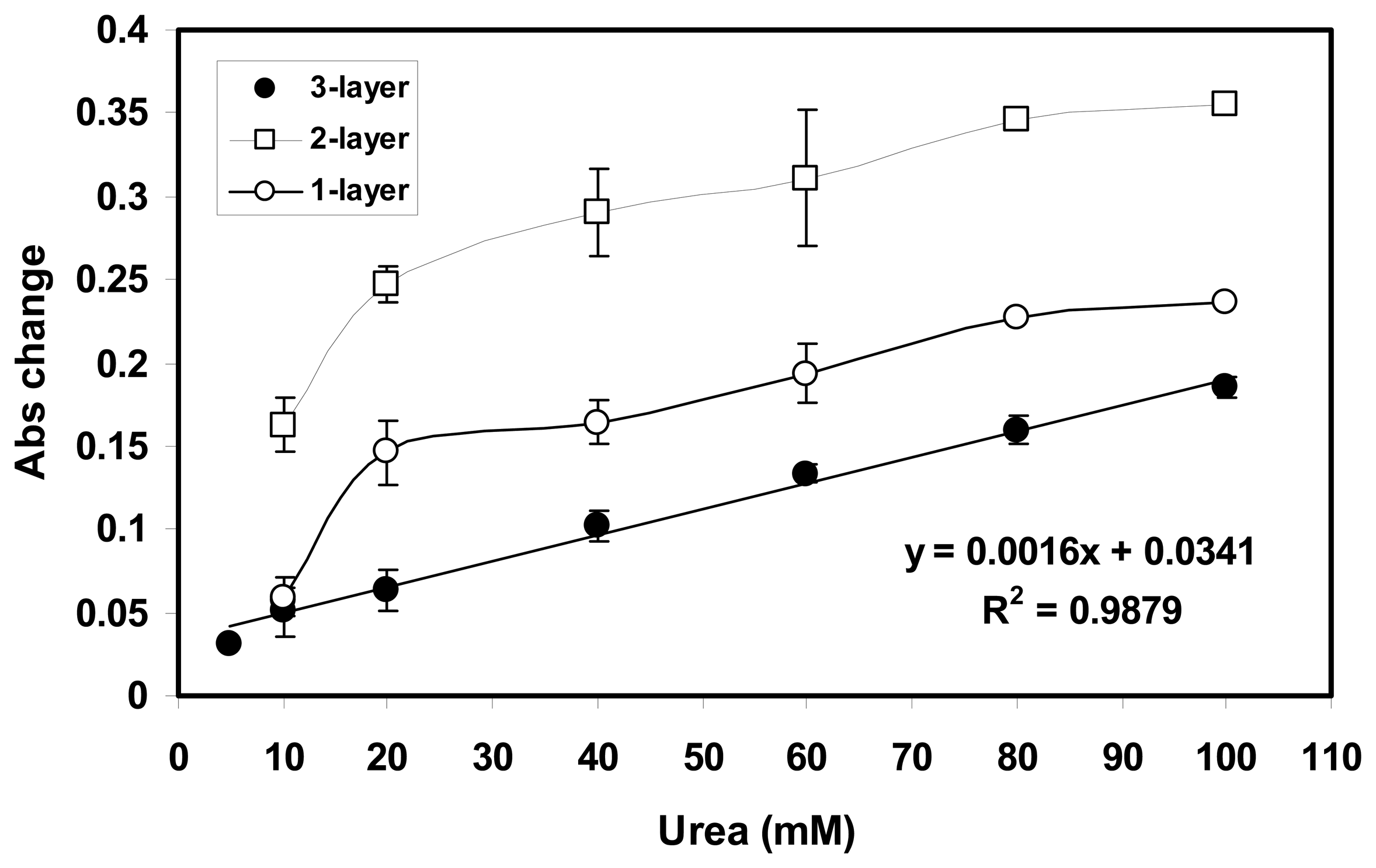
2.4 Effect on enzyme loading on the biosensor response
2.5 The reproducibility and repeatability characteristics of the urea biosensor
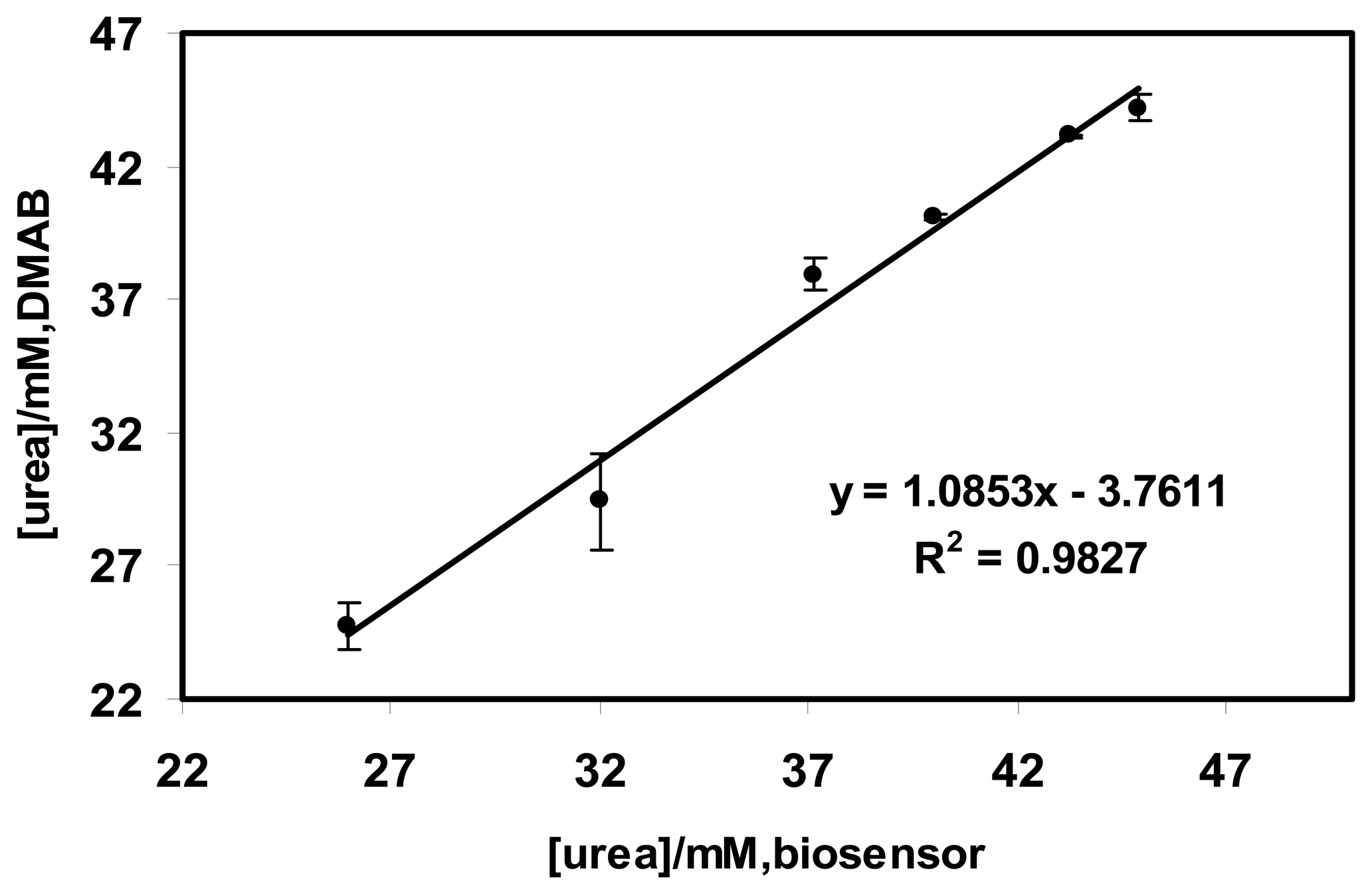
2.6 The validation results of the optical urea biosensor with urine samples
2.7 Recovery characteristics of urea biosensor in urine samples
2.8 Stability of the urea biosensor
2.9 Comparison with other urea biosensors
3. Experimental
3.1 Materials
3.2 Preparation of optical pH sensor and evaluation of response
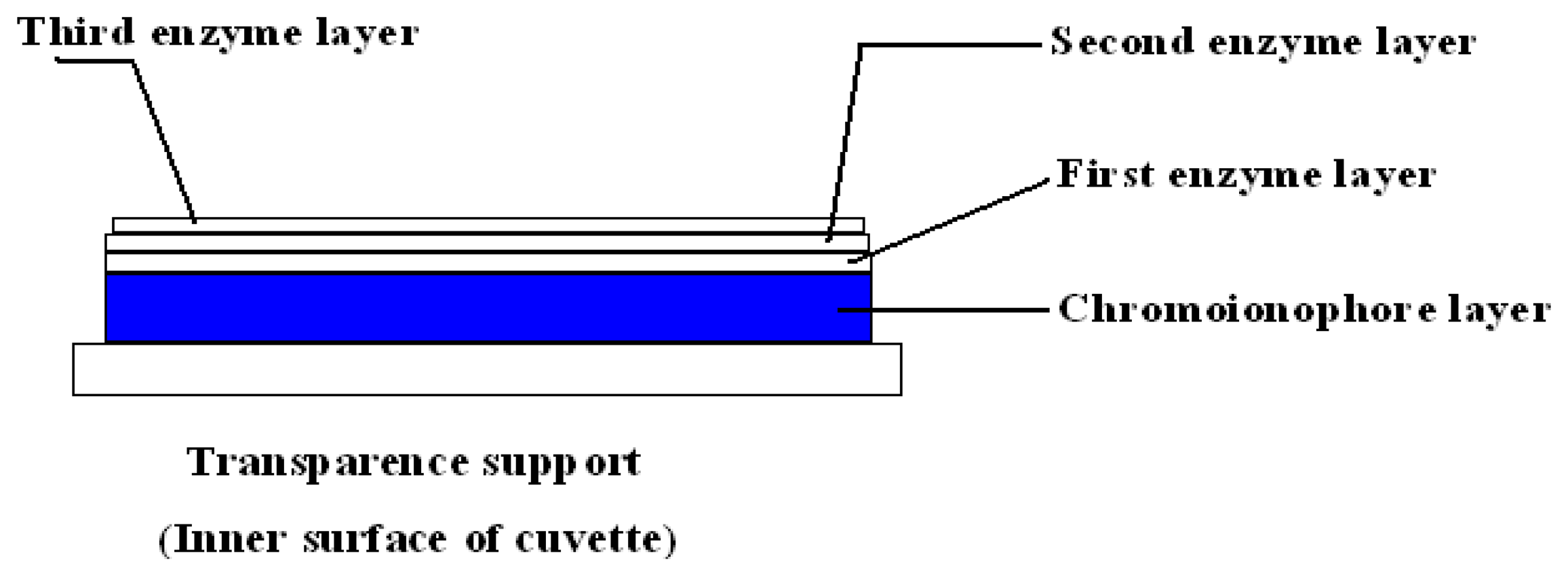
3.3 Preparation urea biosensor and evaluation of biosensor response
3.4 Assessment of the reproducibility and repeatability of the optical urea biosensor
3.5 Validation and recovery studies using urine samples
3.6 Procedure for long term stability evaluation of the urea biosensor
Acknowledgments
References
- Gupta, R.; Chaudhury, N.K. Entrapment of biomolecules in sol-gel matrix for applications in biosensors: Problems and future prospects. Biosensors and Bioelectronics 2007, 22, 2387–2399. [Google Scholar]
- Jeronimo, P.C.A.; Araujo, A.N.; Montenegro, M.; Conceicao, B.S.M. Optical sensors and biosensors based on sol–gel films. Talanta 2007. In press. [Google Scholar]
- Tsai, H.C.; Doong, R.A.; Chiang, H.C.; Chen, K.T. Sol–gel derived urease-based optical biosensor for the rapid determination of heavy metals. Analytica Chimica Acta. 2003, 481, 75–84. [Google Scholar]
- Gulcev, M.D.; Goring, L.G.; Gillian, R.M.; Brennan, J.D. Reagentless pH-based biosensing using a fluorescently-labelled dextran co-entrapped with a hydrolytic enzyme in sol–gel derived nanocomposite films. Analytica Chimica Acta. 2002, 457, 47–59. [Google Scholar]
- Wofbeis, O.S.; Li, H. Fluorescence optical urea biosensor with an ammonium optrode as transducer. Biosensor Bioelectronics 1993, 8, 161–166. [Google Scholar]
- Preininger, C.; Wolfbeis, O.S. Disposable cuvette test with integrated sensor layer for enzymatic determination of heavy metals. Biosensor Bioelectronics 1996, 11, 981–990. [Google Scholar]
- Kovács, B.; Nagy, G.; Dombi, R.; Tóth, K. Optical biosensor for urea with improved response time. Biosensors and Bioelectronics 2003, 18, 111–118. [Google Scholar]
- Wong, F.C.M.; Ahmad, M.; Heng, L.Y.; Peng, L.B. An optical biosensor for dichlovos using stacked sol-gel containing acetylcholinesterase and a lipophilic chromoionophore. Talanta 2006, 69, 888–893. [Google Scholar]
- Williams, S. Official Methods of Analysis of the Association of Official Analytical Chemists; Association of Official Analytical Chemists: Colombia, Maryland, 1984. [Google Scholar]
- Heng, L.Y.; Fang, T.H.; Chern, L.H.; Ahmad, M. Influence of methacrylic-acrylic copolymer composition on plasticizer-free optode films for pH sensors. Sensors 2003, 3, 83–90. [Google Scholar]
- Rajesh, B.V.; Takashima, W.; Kaneto, K. An amperometric urea biosensor based on covalent immobilization of urease onto an electrochemically prepared copolymer poly (N-3-aminopropyl pyrrole-co-pyrrole) film. Biomaterials 2005, 26, 3683–3690. [Google Scholar]
- Conn, E.E.; Stumpf, P.K. Outlines of Biochemistry, 3rd Edition ed; John Wiley & Sons: New York, 1972. [Google Scholar]
- Naz, S. Enzymes And Food; Oxford University Press: Oxford, 2002. [Google Scholar]
- Voet, D.; Voet, J.G. Biochemistry, 3rd Edition ed; John Wiley & Sons: Hoboken, NJ, 2004. [Google Scholar]
- Lee, W.Y.; Kim, S.R.; Kim, T.H.; Lee, K.S.; Shin, M.C.; Park, J.K. Sol-gel derived thick-film conductometric biosensor for urea determination in serum. Anal Chim. Acta. 2000, 404, 195–203. [Google Scholar]
- Sahney, R.; Puri, B.K.; Anand, S. Enzyme coated glass pH-electrode: Its fabrication and applications in the determination of urea in blood samples. Anal. Chim. Acta. 2005, 542, 157–161. [Google Scholar]
- Koncki, R.; Mohr, G.J.; Wolfbeis, O.S. Enzyme biosensor for urea based on a novel pH bulk optode membrane. Biosensors Bioelectronics 1995, 10, 653–659. [Google Scholar]
| TrisHCl (mM) | Mean absorbance change (ΔA) (n = 3) | Relative standard deviation (%) |
|---|---|---|
| 1 | 0.238 ± 0.066 | 27.7 |
| 5 | 0.271 ± 0.075 | 27.6 |
| 10 | 0.294 ± 0.066 | 22.4 |
| pH value | Linear correlation coefficient (R2) (n = 4) | Sensitivity (ΔA/mM) | Urea concentration (mM) |
|---|---|---|---|
| 5 | 0.9854 | 0.0043 | 5-20 |
| 7.5 | 0.9968 | 0.0060 | 5-20 |
| 9 | 0.7136 | 0.0026 | 5-20 |
| Urea spiked (mM) | Urea determined by biosensor (mM) | % of recovery |
|---|---|---|
| 12.76 | 12.06±0.49 | 94.51 |
| 18.36 | 17.89±0.33 | 97.44 |
| 23.10 | 25.31±1.56 | 109.6 |
| 27.17 | 30.58±2.41 | 112.6 |
| Principle of detection | **Sensing matrix | Urea concentration range | Response time | Reference |
|---|---|---|---|---|
| Absorption | Sol-gel/Nile blue indicator | 5.0 – 100 mM | 10-15 min | This work |
| Absorption | Nonactin+Nile blue indicator/Plasticicized PVC | 0.1-1 mM | 16-20s | [17] |
| Fluorescence | Sol-gel(TMOS) FITC-dextran | 1.25μM-50mM | 10 min | [3] |
| Fluorescence | TEOS+ MTES+ DMDMS+ SNARF-1 | 1-30 mM | <2 min | [4] |
| Potentiometric | Sol-gel(TMOS) | 0.03-30 mM | > 5 min | [16] |
| Amperometric | PAPCP\ITO | 0.16-5.02 mM | 40s | [11] |
© 2007 by MDPI ( http://www.mdpi.org). Reproduction is permitted for noncommercial purposes.
Share and Cite
Alqasaimeh, M.S.; Heng, L.Y.; Ahmad, M. A Urea Biosensor from Stacked Sol-Gel Films with Immobilized Nile Blue Chromoionophore and Urease Enzyme. Sensors 2007, 7, 2251-2262. https://doi.org/10.3390/s7102251
Alqasaimeh MS, Heng LY, Ahmad M. A Urea Biosensor from Stacked Sol-Gel Films with Immobilized Nile Blue Chromoionophore and Urease Enzyme. Sensors. 2007; 7(10):2251-2262. https://doi.org/10.3390/s7102251
Chicago/Turabian StyleAlqasaimeh, Muawia Salameh, Lee Yook Heng, and Musa Ahmad. 2007. "A Urea Biosensor from Stacked Sol-Gel Films with Immobilized Nile Blue Chromoionophore and Urease Enzyme" Sensors 7, no. 10: 2251-2262. https://doi.org/10.3390/s7102251




