Detection of Staphylococcus epidermidis by a Quartz Crystal Microbalance Nucleic Acid Biosensor Array Using Au Nanoparticle Signal Amplification
Abstract
:1. Introduction
2. Experimental Section
2.1. Reagents and oligonucleotides
2.2. Apparatus
2.3. Standard bacterial strains and clinical bacterial samples
2.4. DNA extraction and PCR amplification
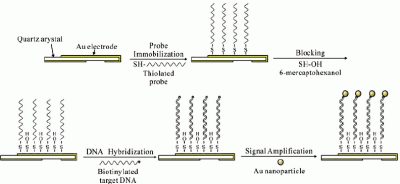
2.5. Immobilization of the oligonucleotide probes
2.6. Detection procedure
2.7. Electrochemical characterization of the QCM electrodes
2.8. Clinical sample detection
2.9. Data analysis
3. Results and Discussion
3.1. Immobilization of synthesized oligonucleotide probes
3.2. Optimization of the experimental conditions for the signal amplifying biosensing system
3.3. Validation of the fabrication and detection of the QCM biosensor by electrochemical impedance spectroscopy
3.4. Detection of PCR-amplified DNAs
3.5. Specificity of the QCM system in detecting S. epidermidis
3.6. Clinical sample detection and comparison
4. Conclusions
Acknowledgments
References and Notes
- Raad, I.; Alrahwan, A.; Rolston, K. Staphylococcus epidermidis: Emerging Resistance and Need for Alternative Agents. Clin. Infect. Dis. 1998, 26, 1182–1187. [Google Scholar]
- Heilmann, C.; Schweitzer, O.; Gerke, C.; Vanittanakom, N.; Mack, D.; Götz, F. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol. Microbiol. 1996, 20, 1083–1091. [Google Scholar]
- Friedman, L.E.; Brown, A.E.; Miller, D.R.; Armstrong, D. Staphylococcus epidermidis septicemia in children with leukemia and lymphoma. Am. J. Dis. Child. 1984, 138, 715–719. [Google Scholar]
- Curtin, J.J.; Donlan, R.M. Using bacteriophages to reduce formation of catheter-associated biofilms by Staphylococcus epidermidis. Antimicrob. Agents Chemother. 2006, 50, 1268–1275. [Google Scholar]
- Yao, C.; Zhu, T.; Tang, J.; Wu, R.; Chen, Q.; Chen, M.; Zhang, B.; Huang, J.; Fu, W. Hybridization assay of hepatitis B virus by QCM peptide nucleic acid biosensor. Biosens. Bioelectron. 2008, 23, 879–885. [Google Scholar]
- Pantalei, S.; Zampetti, E.; Macagnano, A.; Bearzotti, A.; Venditti, I.; Russo, M.V. Enhanced sensory properties of a multichannel quartz crystal microbalance coated with polymeric nanobeads. Sensors 2007, 7, 2920–2928. [Google Scholar]
- Chou, S.F.; Hsu, W.L.; Hwang, J.M.; Chen, C.Y. Determination of alpha-fetoprotein in human serum by a quartz crystal microbalance-based immunosensor. Clin. Chem. 2002, 48, 913–918. [Google Scholar]
- de Abreu, F.C.; de Paula, F.S.; Ferreira, D.C.M.; Nascimento, V.B.; Santos, A.M.C.; Santoro, M.M.; Salas, C.E.; Lopes, J.C.D.; Goulart, M.O.F. The application of DNA-biosensors and differential scanning calorimetry to the study of the DNA-binding agent berenil. Sensors 2008, 8, 1519–1538. [Google Scholar]
- Sauerbrey, G.Z. Use of quartz crystal vibrator for weighting thin films on a microbalance. Z. Phys. 1959, 155, 206–222. [Google Scholar]
- Kanazawa, K.K.; Gordon, J.G. Frequency of a quartz microbalance in contact with liquid. Anal. Chem. 1985, 57, 1770–1771. [Google Scholar]
- Theisen, L.A.; Martin, S.J.; Hillman, A.R. A model for the quartz crystal microbalance frequency response to wetting characteristics of corrugated surfaces. Anal. Chem. 2004, 76, 796–804. [Google Scholar]
- Su, X.; Robelek, R.; Wu, Y.; Wang, G.; Knoll, W. Detection of point mutation and insertion mutations in DNA using a quartz crystal microbalance and MutS, a mismatch binding protein. Anal. Chem. 2004, 76, 489–494. [Google Scholar]
- Tombelli, S.; Mascini, M.; Braccini, L.; Anichini, M.; Turner, A.P.F. Coupling of a DNA piezoelectric biosensor and polymerase chain reaction to detect apolipoprotein E polymorphisms. Biosens. Bioelectron. 2000, 15, 363–370. [Google Scholar]
- Mannelli, I.; Minunni, M.; Tombelli, S.; Mascini, M. Quartz crystal microbalance (QCM) affinity biosensor for genetically modified organisms (GMOs) detection. Biosens. Bioelectron. 2003, 18, 129–140. [Google Scholar]
- Ryu, S.; Jung, S.; Kim, N.; Kim, W. Chemisorption of thiolated Listeria monocytogenes-specific DNA onto the gold surface of piezoelectric quartz crystal. Agri. Chem. Biotechnol. 2001, 44, 163–166. [Google Scholar]
- Mo, X.; Zhou, Y.; Lei, H.; Deng, L. Microbalance-DNA probe method for the detection of specific bacteria in water. Enzyme Microb. Technol. 2002, 30, 583–589. [Google Scholar]
- Wu, V.C.H.; Chen, S.H.; Lin, C.S. Real-time detection of Escherichia coli O157:H7 sequences using a circulating-flow system of quartz crystal microbalance. Biosens. Bioelectron. 2007, 22, 2967–2975. [Google Scholar]
- Bardea, A.; Dagan, A.; Willner, I. Amplified electronic transduction of oligonucleotide interactions novel routes for Tay-Sachs biosensors. Anal. Chim. Acta 1999, 385, 33–43. [Google Scholar]
- Patolsky, F.; Lichtenstein, A.; Willner, I. Amplified microgravimetric quartz-crystal-microbalance assay of DNA using oligonucleotide-functionalized liposomes or biotinylated liposomes. J. Am. Chem. Soc. 2000, 122, 418–419. [Google Scholar]
- Patolsky, F.; Lichtenstin, A.; Willner, I. Detection of single-base DNA mutations by enzyme-amplified electronic transduction. Nat. Biotechnol. 2001, 19, 253–257. [Google Scholar]
- Liu, T.; Tang, J.; Han, M.; Jiang, L. A novel microgravimetric DNA sensor with high sensitivity. Biochem. Biophys. Res. Commun. 2003, 304, 98–100. [Google Scholar]
- Mao, X.; Yang, L.; Su, X.L.; Li, Y. A nanoparticle amplification based quartz crystal microbalance DNA sensor for detection of Escherichia coli O157:H7. Biosens. Bioelectron. 2006, 21, 1178–1185. [Google Scholar]
- Chen, S.H.; Wu, V.C.; Chuang, Y.C.; Lin, C.S. Using oligonucleotide-functionalized Au nanoparticles to rapidly detect foodborne pathogens on a piezoelectric biosensor. J. Microbiol. Methods 2008, 73, 7–17. [Google Scholar]
- Weizmann, Y.; Patolsky, F.; Willner, I. Amplified detection of DNA and analysis of single-base mismatches by the catalyzed deposition of gold on Au-nanoparticles. Analyst 2001, 126, 1502–1504. [Google Scholar]
- Katz, E.; Willner, I. Integrated nanoparticle-biomolecule hybrid systems: Synthesis, properties and applications. Angew. Chem. Int. Ed. 2004, 43, 6042–6108. [Google Scholar]
- Tan, W.B.; Zhang, Y. Surface modification of gold and quantum dot nanoparticles with chitosan for bioapplications. Biomed. Mater. Res. A. 2005, 75, 56–62. [Google Scholar]
- Liu, T; Tang, J; Jiang, L. The enhancement effect of gold nanoparticles as a surface modifier on DNA sensor sensitivity. Biochem. Biophys. Res. Commun. 2004, 313, 3–7. [Google Scholar]
- Millar, B.C.; Jiru, X.; Moore, J.E.; Earle, J.A. A simple and sensitive method to extract bacterial, yeast and fungal DNA from blood culture material. J. Microbiol. Methods 2000, 42, 139–147. [Google Scholar]
- Caruso, F.; Rodda, E.; Furlong, D.; Niikura, K.; Okahata, Y. Quartz crystal microbalance study of DNA immobilization and hybridization for nucleic acid sensor development. Anal. Chem. 1997, 69, 2043–2049. [Google Scholar]
- Zhou, X.C.; Huang, L.Q.; Li, S.F. Microgravimetric DNA sensor based on quartz crystal microbalance: comparison of oligonucleotide immobilization methods and the application in genetic diagnosis. Biosens. Bioelectron. 2001, 16, 85–95. [Google Scholar]
- Ha, T.H.; Kim, S.; Lim, G.; Kim, K. Influence of liquid medium and surface morphology on the response of QCM during immobilization and hybridization of short oligonucleotides. Biosens. Bioelectron. 2004, 15, 378–389. [Google Scholar]
- Wink, T.; van Zuilen, S.J.; Bult, A.; van Bennekom, W.P. Self-assembled monolayers for biosensors. Analyst 1997, 122, 43R–50R. [Google Scholar]
- Mo, Z.; Wang, H.; Liang, Y.; Liu, F.; Xue, Y. Highly reproducible hybridization assay of zeptomole DNA based on adsorption of nanoparticle-bioconjugate. Analyst 2005, 130, 1589–1594. [Google Scholar]
- Tawa, K.; Knoll, W. Mismatching base-pair dependence of the kinetics of DNA-DNA hybridization studied by surface plasmon fluorescence spectroscopy. Nucleic Acids Res. 2004, 32, 2372–2377. [Google Scholar]
- Okahata., Y.; Kawase, M.; Niikura, K.; Ohtake, F.; Furusawa, H.; Ebara, Y. Kinetic measurements of DNA hybridization on an oligonucleotide-immobilized 27-MHz quartz crystal microbalance. Anal. Chem. 1998, 70, 1288–1296. [Google Scholar]
- Sun, H.; Zhang, Y.; Fung, Y. Flow analysis coupled with PQC/DNA biosensor for assay of E. coli based on detecting DNA products from PCR amplification. Biosens. Bioelectron. 2006, 22, 506–512. [Google Scholar]
- Ren, X.; Pickup, P.G. An impedance study of electron transport and electron transfer in composite polypyrrole + polystyrenesulphonate films. J. Electroanal. Chem. 1997, 420, 251–257. [Google Scholar]
- Anthony, R.M.; Brown, T.J.; French, G.L. Rapid diagnosis of bacteremia by universal amplification of 23S ribosomal DNA followed by hybridization to an oligonucleotide array. J. Clin. Microbiol. 2000, 38, 781–788. [Google Scholar]








| Oligonucleotide | Sequence |
|---|---|
| Thiolated probe | 5′-HS-(CH2)6-CGAGCGAACAGATGAGGAGC-3′ |
| Forward primer | 5′-TGGCGGCGTGCCTAATACATG-3′ |
| Reverse primer | 5′-biotin-CCCGTAGGAGTCTGGACCGTGTC-3′ |
| Positive control oligo | 5′-biotin-GCTCCTCATCTGTTCGCTCG-3′ |
| Negative control oligo | 5′-biotin-CGAGCGAACAGATGAGGAGC-3′ |
| Method | No. of blood cultures | ||
|---|---|---|---|
| No. positive | No. negative | n | |
| conventional microbiological method | 34 | 21 | 55 |
| QCM biosensor | 35 | 20 | 55 |
© 2008 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Xia, H.; Wang, F.; Huang, Q.; Huang, J.; Chen, M.; Wang, J.; Yao, C.; Chen, Q.; Cai, G.; Fu, W. Detection of Staphylococcus epidermidis by a Quartz Crystal Microbalance Nucleic Acid Biosensor Array Using Au Nanoparticle Signal Amplification. Sensors 2008, 8, 6453-6470. https://doi.org/10.3390/s8106453
Xia H, Wang F, Huang Q, Huang J, Chen M, Wang J, Yao C, Chen Q, Cai G, Fu W. Detection of Staphylococcus epidermidis by a Quartz Crystal Microbalance Nucleic Acid Biosensor Array Using Au Nanoparticle Signal Amplification. Sensors. 2008; 8(10):6453-6470. https://doi.org/10.3390/s8106453
Chicago/Turabian StyleXia, Han, Feng Wang, Qing Huang, Junfu Huang, Ming Chen, Jue Wang, Chunyan Yao, Qinghai Chen, Guoru Cai, and Weiling Fu. 2008. "Detection of Staphylococcus epidermidis by a Quartz Crystal Microbalance Nucleic Acid Biosensor Array Using Au Nanoparticle Signal Amplification" Sensors 8, no. 10: 6453-6470. https://doi.org/10.3390/s8106453