Multiscale Unsupervised Segmentation of SAR Imagery Using the Genetic Algorithm
Abstract
:1. Introduction
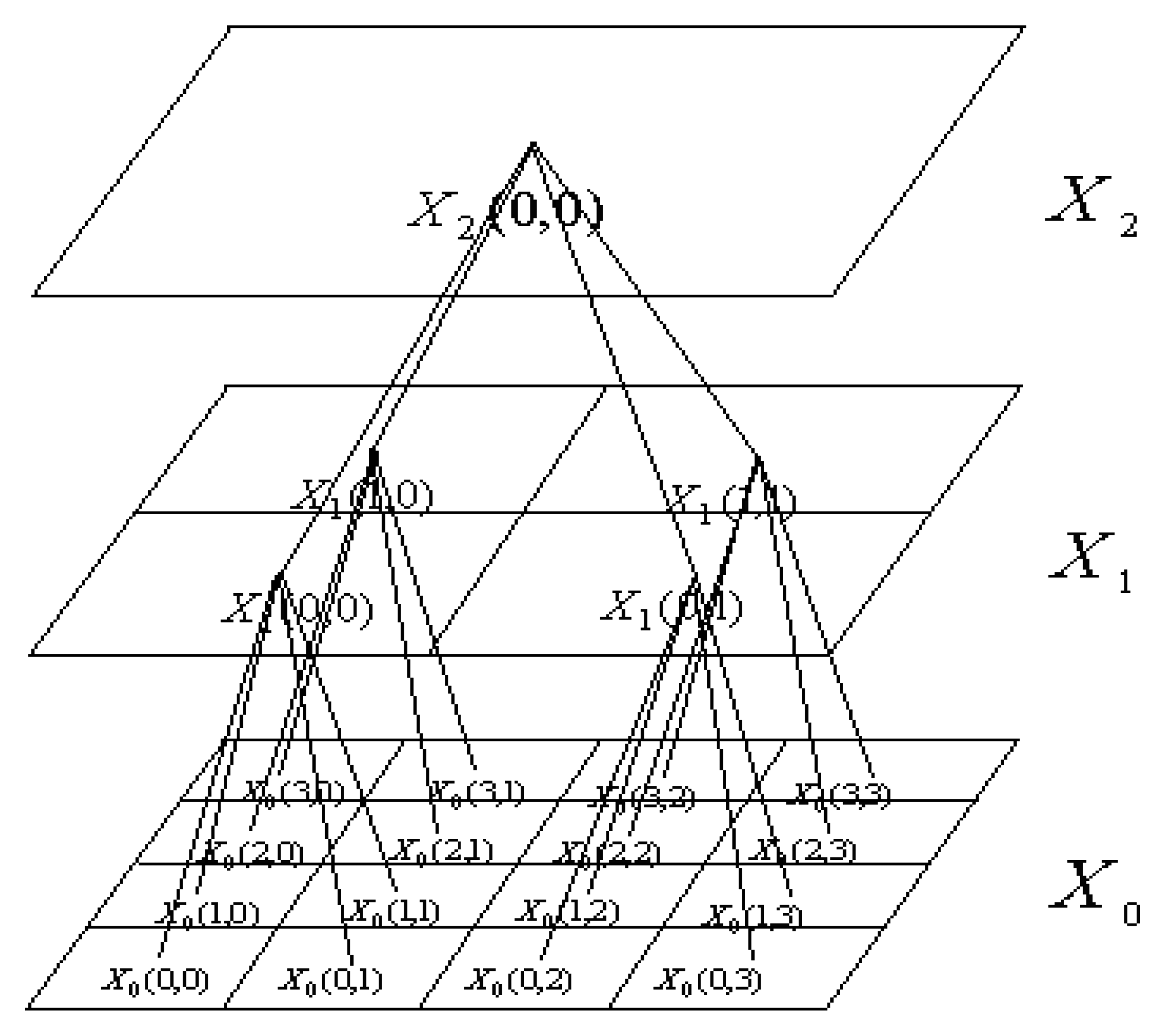
2. Quadtree Interpretation of SAR Imagery and Its MMAR Model
A. Expectation Step
B. Maximization Step
3. Hybrid method of GA and EM Algorithm
| procedure GA-EM |
| begin |
| t ← 0 |
| Oldsize ← 0 |
| cend ← 0 |
| initialize P(t) |
| while (cend ≠ 5) |
| P′(t) ← perform R EM steps on P(t) |
| MDL′ ← evaluate P′(t) |
| P′(t) ← recombine P′(t) |
| P‴(t) ← perform R EM steps on P′(t) |
| MDL′ ← evaluate P‴(t) |
| [P″(t),MDL] ← select{[P‴(t),MDL′ ] ∪ [P′(t),MDL′ ] |
| MDLmin ← min(MDL) |
| amin ← argminMDL (P″(t)) |
| if (|amin| ≠ Oldsize) then |
| cend ← 0 |
| Oldsize ←| amin| |
| else |
| cend ← cend + 1 |
| end |
| P″‴(t) ← enforce mutation P‴(t) |
| P(t +1) ← mutate (P″‴(t)) |
| t ← t + 1 |
| end |
| EM(amin) until convergence of the log likelihood is reached |
| end |
Encoding
Recombination
Selection
Enforced Mutation
Mutation
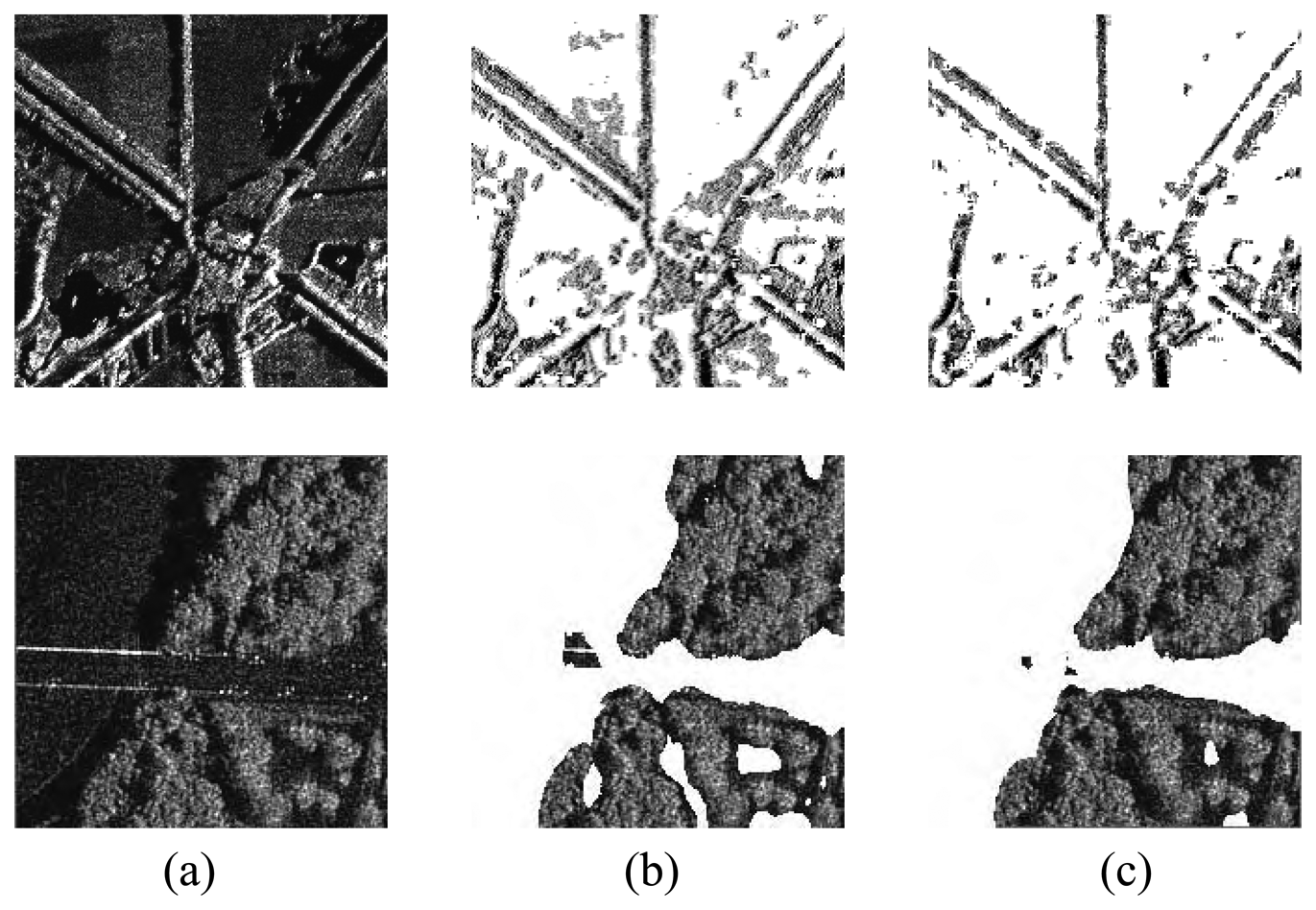
4. Experiments
5. Conclusions
Acknowledgments
References and Notes
- Fosgate, C.; Irving, W.W.; Karl, W.; Willsky, A.S. Multiscale segmentation and anomaly enhancement of SAR imagery. IEEE Trans. Image Process. 1997, 6, 7–20. [Google Scholar]
- Irving, W.W.; Novak, L.M.; Willsky, A.S. A multiresolution approach to discrimination in SAR imagery. IEEE Tran. Aerosp. Electron. Syst. 1997, 33, 1157–1169. [Google Scholar]
- Kim, A.; Kim, H. Hierarchical stochastic modeling of SAR imagery for segmentation/compression. IEEE Trans. Signal Process. 1999, 47, 458–468. [Google Scholar]
- Wen, X.B.; Tian, Z. Mixture multiscale autoregressive modeling of SAR imagery for segmentation. Electron. Lett. 2003, 39, 1272–1274. [Google Scholar]
- Pernkopf, F.; Bouchaffra, D. Genetic-based EM algorithm for learning Gaussian mixture model. IEEE Trans. Pattern Anal. Machine Intel. 2005, 27, 1344–1348. [Google Scholar]
- Back, T. Evolutionary algorithma in theory and practice; Oxford Univ. Press: Oxford, UK, 1996. [Google Scholar]
- Back, T.; Schwefel, H. Evolutionary computation: an overview. Proc. IEEE Conf. Evolut. Comput. 1996, 20–29. [Google Scholar]
© 2008 by MDPI Reproduction is permitted for noncommercial purposes.
Share and Cite
Wen, X.-B.; Zhang, H.; Jiang, Z.-T. Multiscale Unsupervised Segmentation of SAR Imagery Using the Genetic Algorithm. Sensors 2008, 8, 1704-1711. https://doi.org/10.3390/s8031704
Wen X-B, Zhang H, Jiang Z-T. Multiscale Unsupervised Segmentation of SAR Imagery Using the Genetic Algorithm. Sensors. 2008; 8(3):1704-1711. https://doi.org/10.3390/s8031704
Chicago/Turabian StyleWen, Xian-Bin, Hua Zhang, and Ze-Tao Jiang. 2008. "Multiscale Unsupervised Segmentation of SAR Imagery Using the Genetic Algorithm" Sensors 8, no. 3: 1704-1711. https://doi.org/10.3390/s8031704




