Ammonificins C and D, Hydroxyethylamine Chromene Derivatives from a Cultured Marine Hydrothermal Vent Bacterium, Thermovibrio ammonificans
Abstract
:1. Introduction
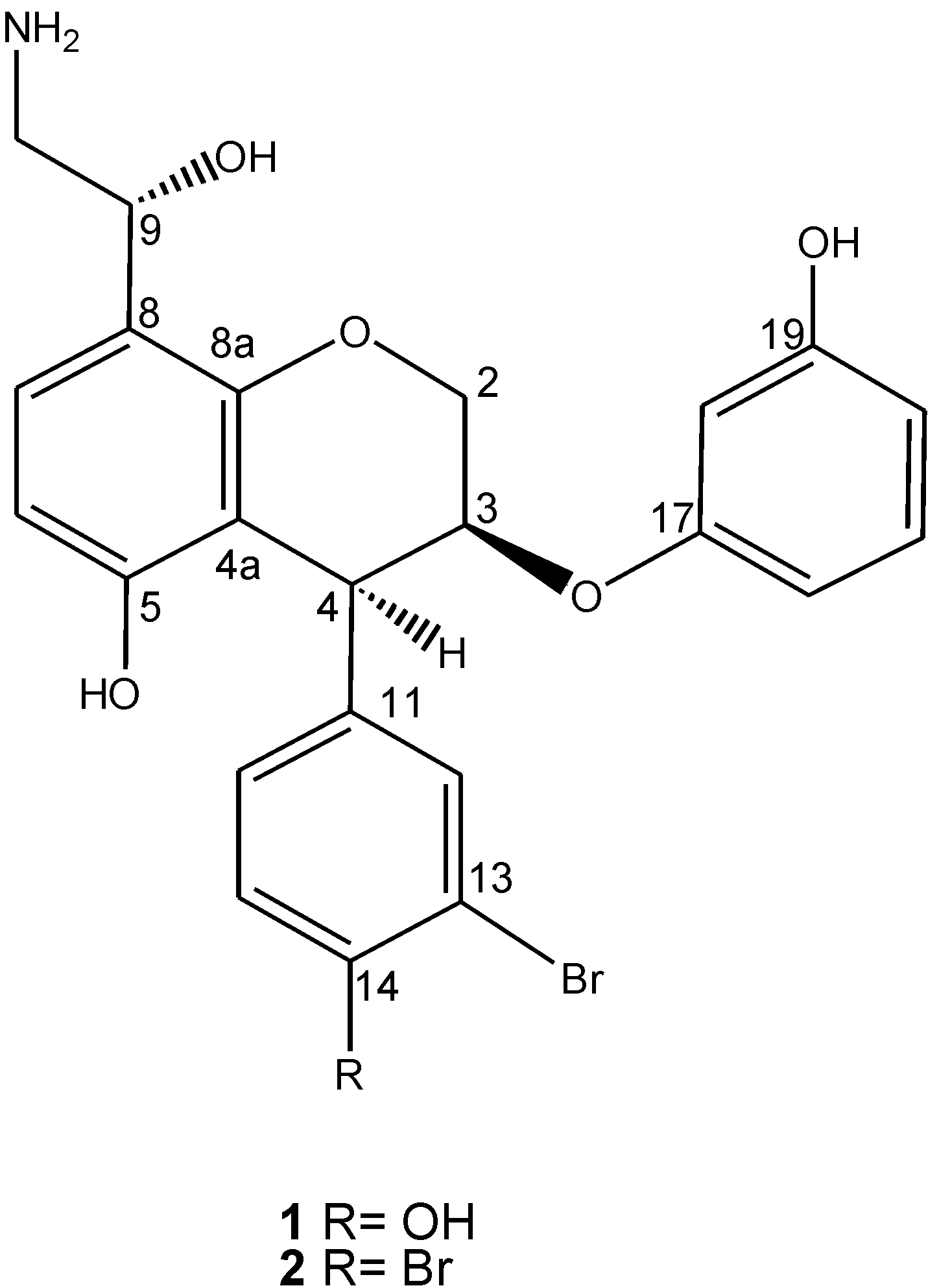
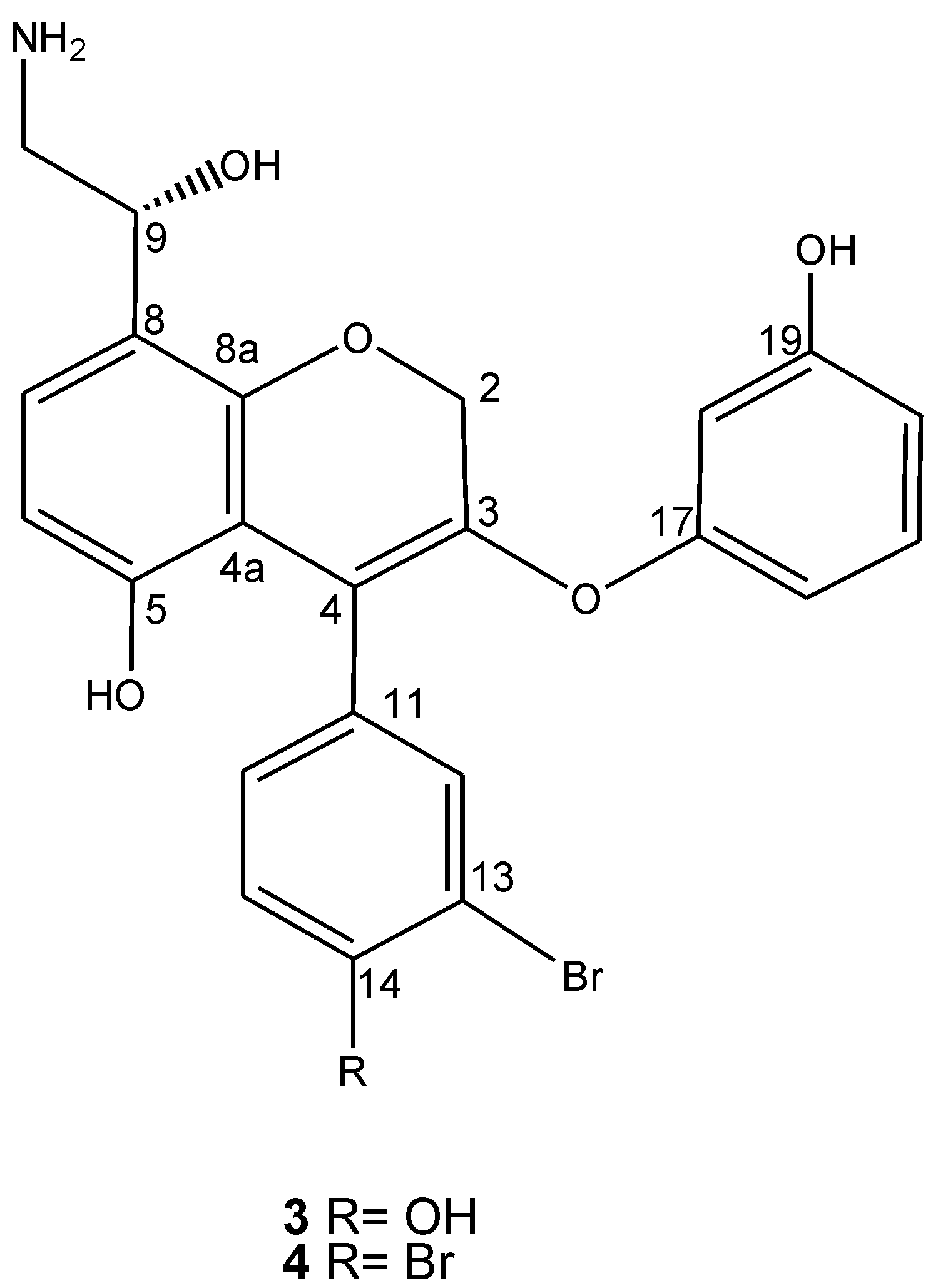
2. Results and Discussion
 ) of 3 with the spectrum calculated (...........) for 9R.
) of 3 with the spectrum calculated (...........) for 9R.
 ) of 3 with the spectrum calculated (...........) for 9R.
) of 3 with the spectrum calculated (...........) for 9R.
 ) of 4 with the spectrum calculated (...........) for 9R.
) of 4 with the spectrum calculated (...........) for 9R.
 ) of 4 with the spectrum calculated (...........) for 9R.
) of 4 with the spectrum calculated (...........) for 9R.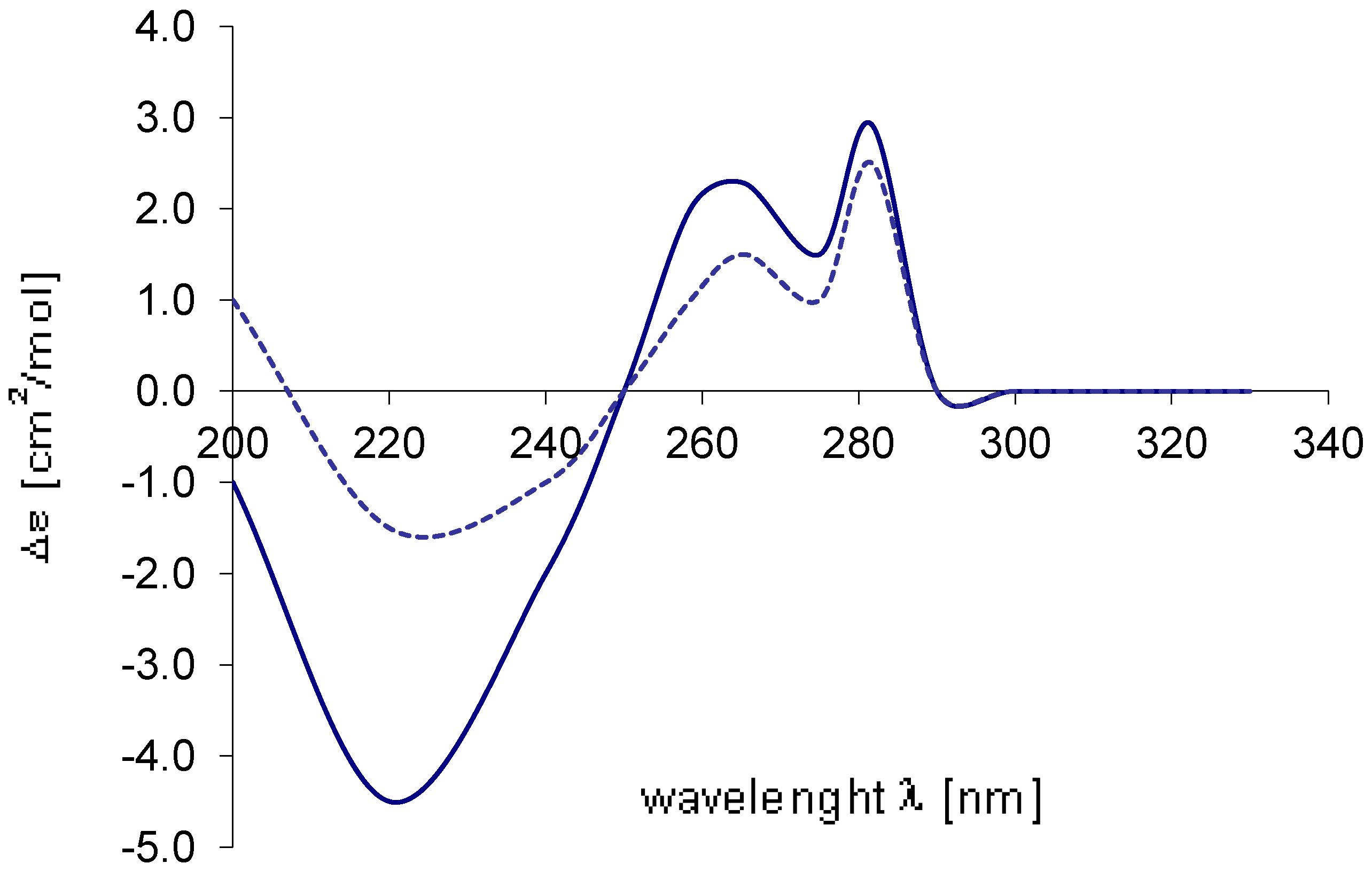
3. Experimental Section
3.1. General Experimental Procedures
3.2. Extraction and Isolation Procedures
3.2.1. Collection of Organism [4]
3.2.2. Culture [4]
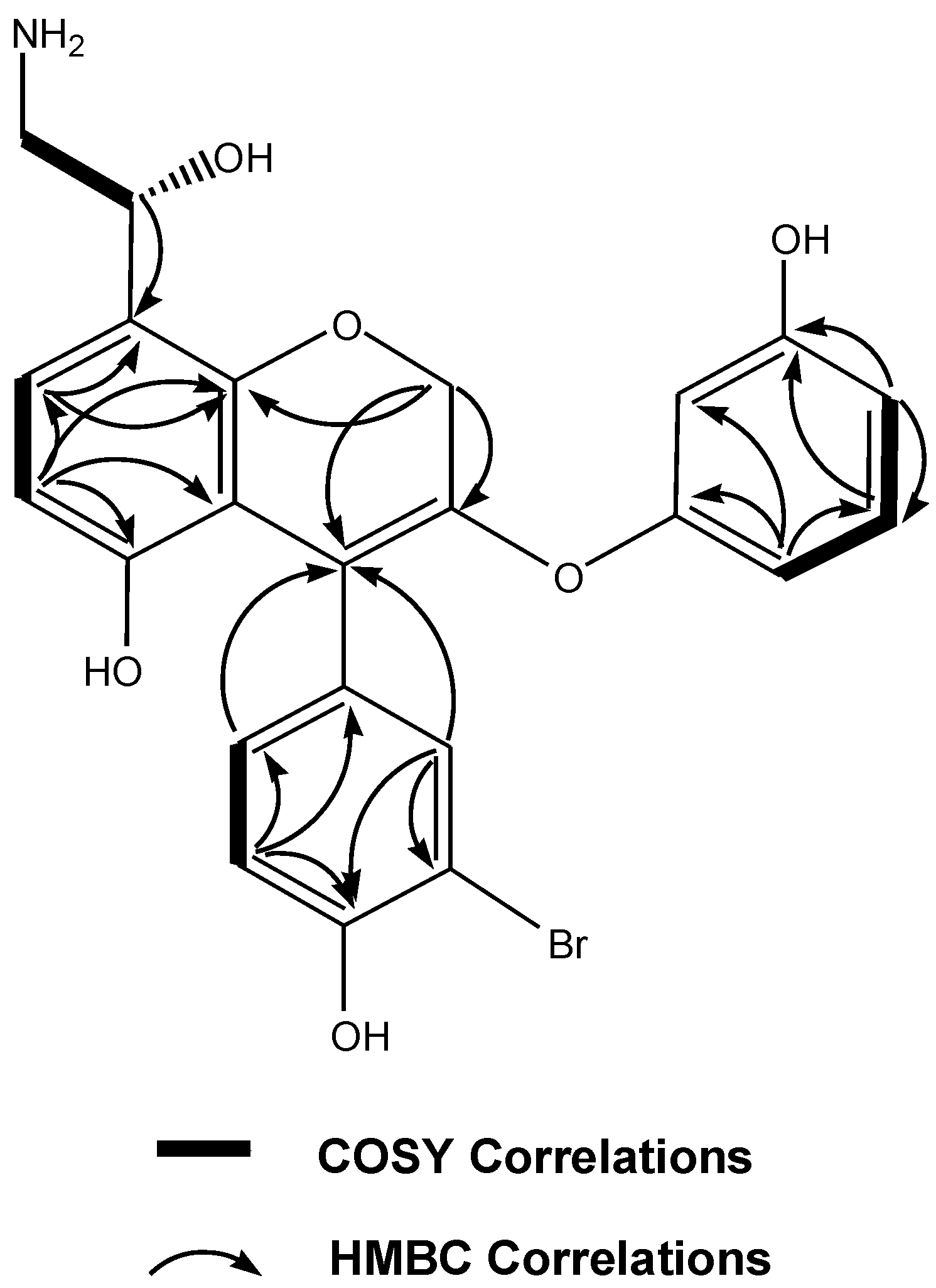
| Position | δC type | δH mult. ( J in Hz) | HMBC a |
|---|---|---|---|
| 2a | 76.2, CH2 | 4.41, d (6.1) | 8 a, 4 |
| 2b | 4.10, d (6.1) | 3 | |
| 3 | 141.5, qC | ||
| 4 | 111.7, qC | ||
| 4a | 107.9, qC | ||
| 5 | 155.7, qC | ||
| 6 | 108.0, CH | 6.67, d (7.9) | 4 a, 5, 7, 8 a |
| 7 | 126.8, CH | 7.09, d (7.9) | 5, 8, 8 a |
| 8 | 119.7, qC | ||
| 8a | 157.9, qC | ||
| 9 | 69.6, CH | 4.70, m | 7, 8 |
| 10 | 49.2, CH2 | 3.45, m | 8, 9 |
| 11 | 129.7, qC | ||
| 12 | 133.1, CH | 7.30, s | 4,11, 13, 14, 16 |
| 13 | 110.8, qC | ||
| 14 | 157.1, qC | ||
| 15 | 118.4, CH | 6.77, d (7.2) | 11, 13, 14, 16 |
| 16 | 128.8, CH | 7.26, d (7.2) | 4,11, 12, 14, 15 |
| 17 | 158.9, qC | ||
| 18 | 101.4, CH | 6.65, s | 17, 19, 20, 22 |
| 19 | 156.9, qC | ||
| 20 | 107.3, CH | 6.74, d (7.8) | 18, 19, 21, 22 |
| 21 | 130.5, CH | 7.01, dd (7.6,7.8) | 17, 19, 20, 22 |
| 22 | 106.8, CH | 6.81, d (7.6) | 17, 18, 20, 21 |
| OH on C-5 | 9.26, br s | ||
| OH on C-14 | 8.48, br s | ||
| OH on C-19 | 9.47, br s |
| Position | δC type | δH mult. ( J in Hz) | HMBC a |
|---|---|---|---|
| 2a | 76.2, CH2 | 4.41, d (6.1) | 8 a, 4 |
| 2b | 4.10, d (6.1) | 3 | |
| 3 | 141.5, qC | ||
| 4 | 111.7, qC | ||
| 4a | 107.91, qC | ||
| 5 | 155.7, qC | ||
| 6 | 108.0, CH | 6.67, d (7.9) | 4 a, 5, 7, 8 a |
| 7 | 126.8, CH | 7.09, d (7.9) | 5, 8, 8 a |
| 8 | 119.7, qC | ||
| 8a | 157.9, qC | ||
| 9 | 69.6, CH | 4.65, t (4.3) | 7, 8 |
| 10 | 49.2, CH2 | 3.45, m | 8, 9 |
| 11 | 136.1, qC | ||
| 12 | 133.9, CH | 7.26, s | 4, 11, 13, 14, 16 |
| 13 | 126.9, qC | ||
| 14 | 123.9, qC | ||
| 15 | 132.5, CH | 7.28, d (7.2) | 11, 13, 14, 16 |
| 16 | 129.6, CH | 7.24, d (7.2) | 4, 11, 12, 14, 15 |
| 17 | 158.9, qC | ||
| 18 | 101.4, CH | 6.65, s | 17, 19, 20, 22 |
| 19 | 156.9, qC | ||
| 20 | 107.3, CH | 6.74, d (7.8) | 18, 19, 21, 22 |
| 21 | 130.5, CH | 7.01, dd (7.6,7.8) | 17, 19, 20, 22 |
| 22 | 106.8, CH | 6.81, d (7.6) | 17, 18, 20, 21 |
| OH on C-5 | 9.26, br s | ||
| OH on C-19 | 9.47, br s |
3.3. Computational Methods
3.4. Biological Evaluation—Apoptosis Induction
4. Conclusions
Acknowledgments
References
- Gärtner, A.; Wiese, J.; Imhoff, J.F. Amphritea atlantica gen. nov., sp. nov., a gammaproteobacterium from the Logatchev hydrothermal vent field. Int. J. Syst. Evol. Microbiol. 2008, 58, 34–39. [Google Scholar] [CrossRef] [Green Version]
- Thornburg, C.C.; Zabriskie, T.M.; McPhail, K.L. Deep-sea hydrothermal vents: Potential hot spots for natural products discovery? J. Nat. Prod. 2010, 73, 489–499. [Google Scholar] [CrossRef]
- Kicklighter, C.E.; Fisher, C.R.; Hay, M.E. Chemical defense of hydrothermal vent and hydrocarbon seep organisms: A preliminary assessment using shallow-water consumers. Mar. Ecol. Prog. Ser. 2004, 275, 11–19. [Google Scholar] [CrossRef]
- Vetriani, C.; Speck, M.D.; Ellor, S.V.; Lutz, R.A.; Starovoytov, V. Thermovibrio ammonificans sp. nov., a thermophilic, chemolithotrophic, nitrate ammonifying bacterium from deep-sea hydrothermal vent. Int. J. Syst. Evol. Microbiol. 2004, 54, 175–181. [Google Scholar] [CrossRef]
- Andrianasolo, E.H.; Haramaty, L.; Degenhardt, K.; Mathew, R.; White, E.; Lutz, R.; Falkowski, P. Induction of apoptosis by diterpenes from the soft coral Xenia elongata. J. Nat. Prod. 2007, 70, 1551–1557. [Google Scholar] [CrossRef]
- Mathew, R.; Degenhart, K.; Haramaty, L.; Karp, C.M.; White, E. Immortalized mouse epithelial cell models to study the role of apoptosis in cancer. Methods Enzymol. 2008, 446, 77–106. [Google Scholar]
- Karantza-Wadsworth, V.; White, E. Programmed Cell Death. In Cancer: Principles and Practice of Oncology; DeVita, V.T., Lawrence, T.S., Rosenberg, S.A., Eds.; Lippincott, Williams, and Wilkins: Philadelphia, PA, USA, 2008; Chapter 7, pp. 93–101. [Google Scholar]
- Andrianasolo, E.H.; Haramaty, L.; Rosario-Passapera, R.; Bidle, K.; White, E.; Vetriani, C.; Falkowski, P.; Lutz, R. Ammonificins A and B, hydroxyethylamine chroman derivatives from a cultured marine hydrothermal vent bacterium, Thermovibrio ammonifican. J. Nat. Prod. 2009, 72, 1216–1219. [Google Scholar] [CrossRef]
- Fahy, E.; Potts, B.C.M.; Faulkner, J. 6-bromotryptamine derivatives from the Gulf of California tunicate Didemnum candidum. J. Nat. Prod. 1991, 54, 564–569. [Google Scholar] [CrossRef]
- Ahlrichs, R.; Furche, F.; Hättig, C.; Klopper, W.; Sierka, M.; Weigend, F. TURBOMOLE Software, version 5.10; COSMOlogic GmbH & Co. KG: Leverkusen, Germany, 2008. [Google Scholar]
- Stetter, K.O.; König, H.; Stackebrandt, E. Pyrodictium gen. nov., a new genus of submarine disc-shaped sulfur reducing archaebacteria growing optimally at 105 °C. Syst. Appl. Microbiol. 1983, 4, 535–551. [Google Scholar] [CrossRef]
- Balch, W.E.; Fox, G.E.; Magrum, L.J.; Woese, C.R.; Wolfe, R.S. Methanogens: Reevaluation of a unique biological group. Microbiol. Rev. 1979, 43, 260–296. [Google Scholar]
- Holscher, D.; Reichert, M.; Gorls, H.; Ohlenschlager, O.; Bringmann, G.; Schneider, B. Monolaterol, the first configurationally assigned phenylphenalenone derivative with a stereogenic center at C-9, from Monochoria elata. J. Nat. Prod. 2006, 69, 1614–1617. [Google Scholar] [CrossRef]
- Pecul, M.; Ruud, K.; Helgaker, T. Density functional theory calculation of electronic circular dichroism using London orbitals. Chem. Phys. Lett. 2004, 388, 110–119. [Google Scholar] [CrossRef]
- Diedrich, C.; Grimme, S. Systematic investigation of modern quantum chemical methods to predict electronic circular dichroism spectra. J. Phys. Chem. 2003, 107, 2524–2539. [Google Scholar] [CrossRef]
- Antus, S.; Kurtan, T.; Juhász, L.; Kiss, L.; Hollósi, M.; Májer, Z.S. Chiroptical properties of 2,3-dihydrobenzo[b]furan and chromane chromophores in naturally occurring O-heterocycles. Chirality 2001, 13, 493–506. [Google Scholar] [CrossRef]
- Danial, N.; Korsmeyer, S. Cell death: Critical control points. Cell 2004, 116, 205–219. [Google Scholar] [CrossRef]
- Gelinas, C.; White, E. BH3-only proteins in control: Specificity regulates MCL-1 and BAK-mediated apoptosis. Genes Dev. 2005, 19, 1263–1268. [Google Scholar] [CrossRef]
- Degenhardt, K.; White, E. A new generation of mouse models of cancer for translational research. Clin. Cancer Res. 2006, 12, 5274–5276. [Google Scholar] [CrossRef]
- Fesik, S.W. Promoting apoptosis as a strategy for cancer drug discovery. Nat. Rev. Cancer 2005, 5, 876–885. [Google Scholar] [CrossRef]
- Degenhardt, K.S.R.; Chen, G.; Lindsten, T.; Thomson, C.; White, E. BAX and BAK mediate p53-independent suppression of tumorigenesis. J. Bio. Chem. 2002, 277, 14127–14134. [Google Scholar]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Batista, J.M., Jr.; Lopes, A.A.; Ambrόsio, D.L.; Regasini, L.O.; Kato, M.J.; da Silva Bolzani, V.; Cicarelli, R.M.B.; Furlan, M. Natural chromenes and chromene derivatives as potential anti-trypanosomal agents. Biol. Pharm. Bull. 2008, 31, 538–540. [Google Scholar] [CrossRef]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Andrianasolo, E.H.; Haramaty, L.; Rosario-Passapera, R.; Vetriani, C.; Falkowski, P.; White, E.; Lutz, R. Ammonificins C and D, Hydroxyethylamine Chromene Derivatives from a Cultured Marine Hydrothermal Vent Bacterium, Thermovibrio ammonificans. Mar. Drugs 2012, 10, 2300-2311. https://doi.org/10.3390/md10102300
Andrianasolo EH, Haramaty L, Rosario-Passapera R, Vetriani C, Falkowski P, White E, Lutz R. Ammonificins C and D, Hydroxyethylamine Chromene Derivatives from a Cultured Marine Hydrothermal Vent Bacterium, Thermovibrio ammonificans. Marine Drugs. 2012; 10(10):2300-2311. https://doi.org/10.3390/md10102300
Chicago/Turabian StyleAndrianasolo, Eric H., Liti Haramaty, Richard Rosario-Passapera, Costantino Vetriani, Paul Falkowski, Eileen White, and Richard Lutz. 2012. "Ammonificins C and D, Hydroxyethylamine Chromene Derivatives from a Cultured Marine Hydrothermal Vent Bacterium, Thermovibrio ammonificans" Marine Drugs 10, no. 10: 2300-2311. https://doi.org/10.3390/md10102300




