Evidence of Accelerated Evolution and Ectodermal-Specific Expression of Presumptive BDS Toxin cDNAs from Anemonia viridis
Abstract
:1. Introduction
2. Results and Discussion
2.1. A. viridis BDS cDNA Characterisation

| Accession Number | Toxin | ORF length a | Protein length b | Molecular weight c | pI d |
|---|---|---|---|---|---|
| FK728690 | BDS-1 | 231 | 76 | 8342.8 | 9.02 |
| FK744472 | BDS-3 | 231 | 76 | 8444.9 | 8.86 |
| FK722457 | BDS-4 | 231 | 76 | 8489.0 | 8.86 |
| FK720902 | BDS-5 | 231 | 76 | 8376.9 | 9.02 |
| FK754940 | BDS-6 | 243 | 80 | 8671.2 | 8.14 |
| FK736435 | BDS-7 | 231 | 76 | 8356.8 | 8.86 |
| FK723172 | BDS-8 | 234 | 77 | 8427.9 | 8.68 |
| FK725608 | BDS-10 | 249 | 82 | 9041.5 | 8.68 |
| FK740326 | BDS-11 | 243 | 80 | 8765.2 | 7.52 |
| FK736010 | BDS-12 | 231 | 76 | 8470.0 | 9.02 |
| FK752236 | BDS-13 | 231 | 76 | 8394.8 | 8.44 |
| FK745823 | BDS-14 | 231 | 76 | 8272.7 | 8.96 |
| FK725211 | BDS-15 | 231 | 76 | 8317.8 | 8.93 |
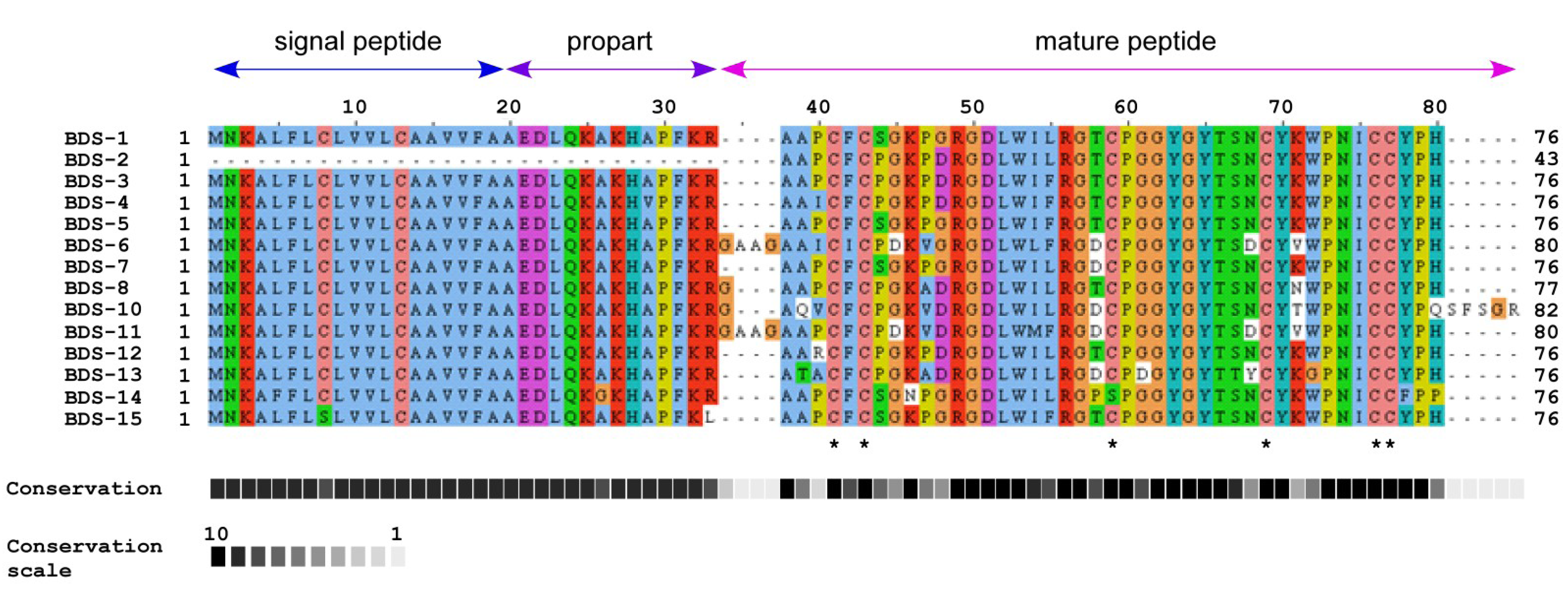
2.2. Comparison of BDS Amino Acid Sequences
2.3. Sequence Similarity with Other Peptide Toxins
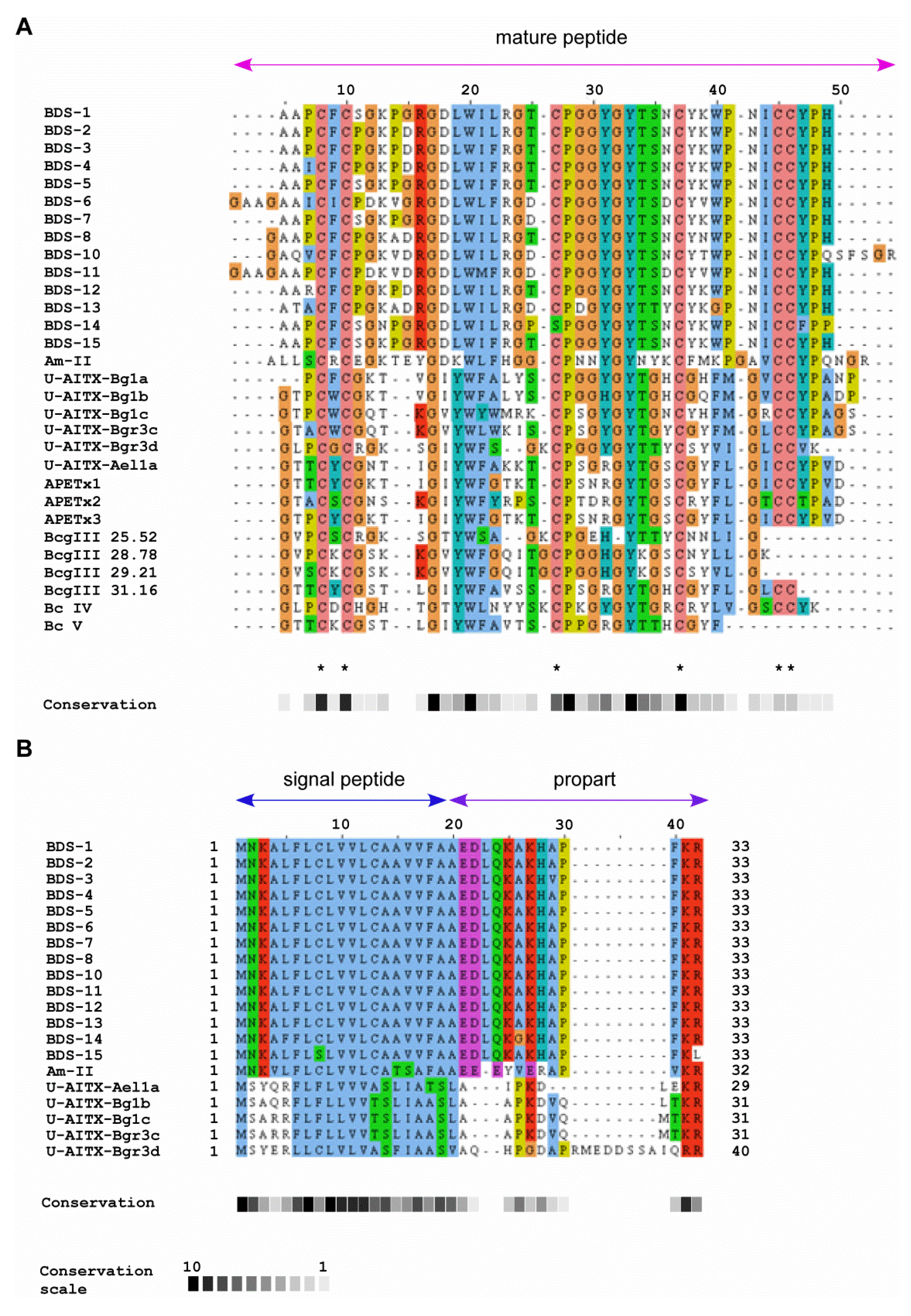
| Species | Toxin | GenBank accession number | References |
|---|---|---|---|
| Bunodosoma cangicum | Bcg III 29.21 | P86464 | Zaharenko et al. 2008 [34] |
| Bcg III 25.52 | P86463 | ||
| Bcg III 28.78 | P86462 | ||
| Bcg III 31.16 | P86461 | ||
| Bunodosoma granuliferum | U-AITX-Bg1a | CCC86602 | Rodriguez et al. 2012 [35] |
| U-AITX-Bg1b | CCC86603 | ||
| U-AITX-Bg1c | CCC86604 | ||
| U-AITX-Bgr3c | CCC86605 | ||
| U-AITX-Bgr3d | CCC86606 | ||
| Bunodosoma caissarum | BcIV | P86470 | Oliveira et al. 2006 [36] |
| BcV | P84919 | Zaharenko, A.Z. [37] | |
| Antheopsis maculata | Am II | P69930 | Honma et al. 2005 [11] |
| Anthopleura elegantissima | APETx1 | P61541 | Diochot et al. 2003 [10] |
| APETx2 | P61542 | ||
| APETx3 | B3EWF9 | Peigneur, S. [38] | |
| U-AITX-Ael1a | FG392547 * | Richier et al. 2008 [39] |
2.4. Phylogenetic Analysis
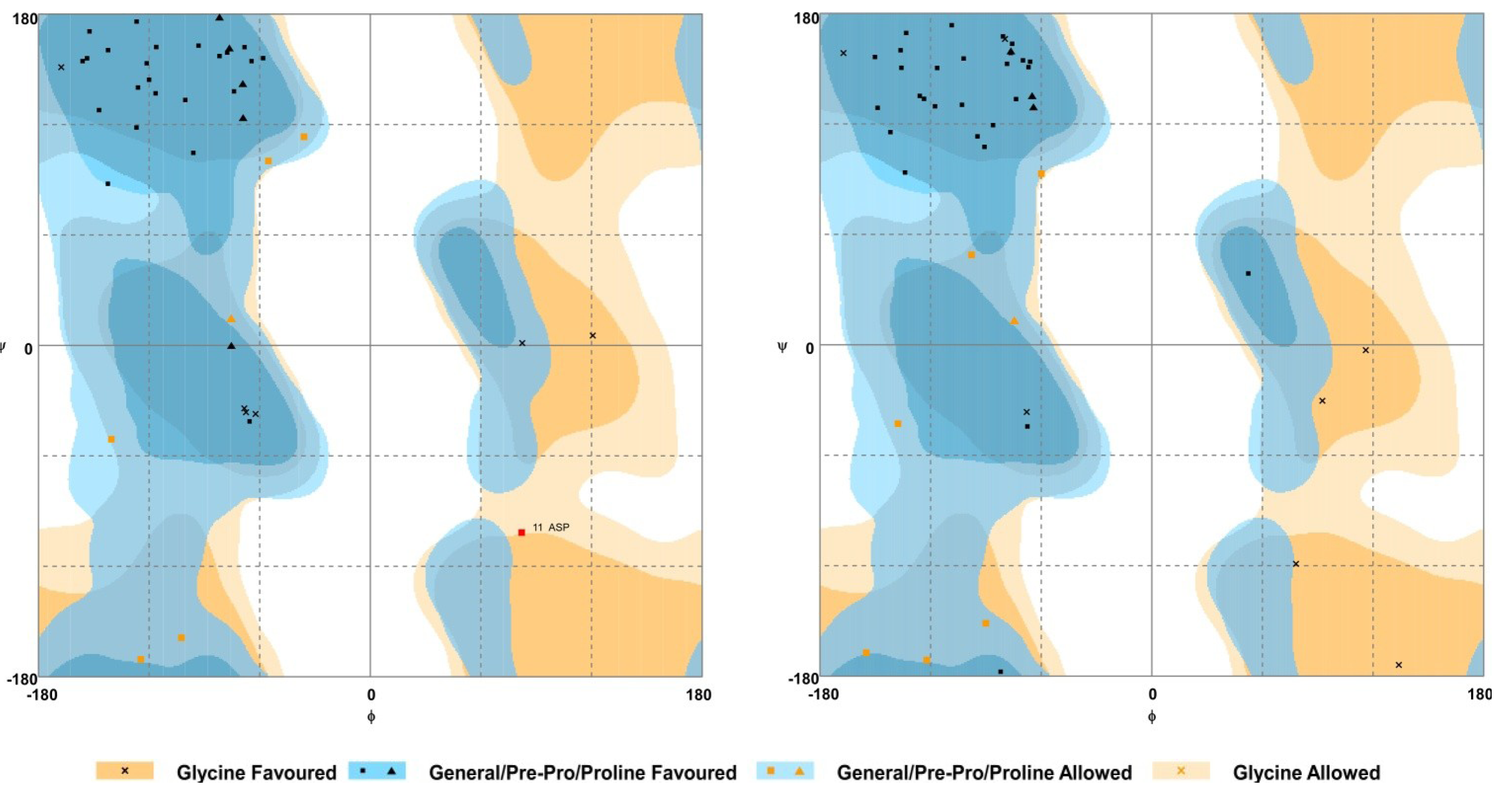
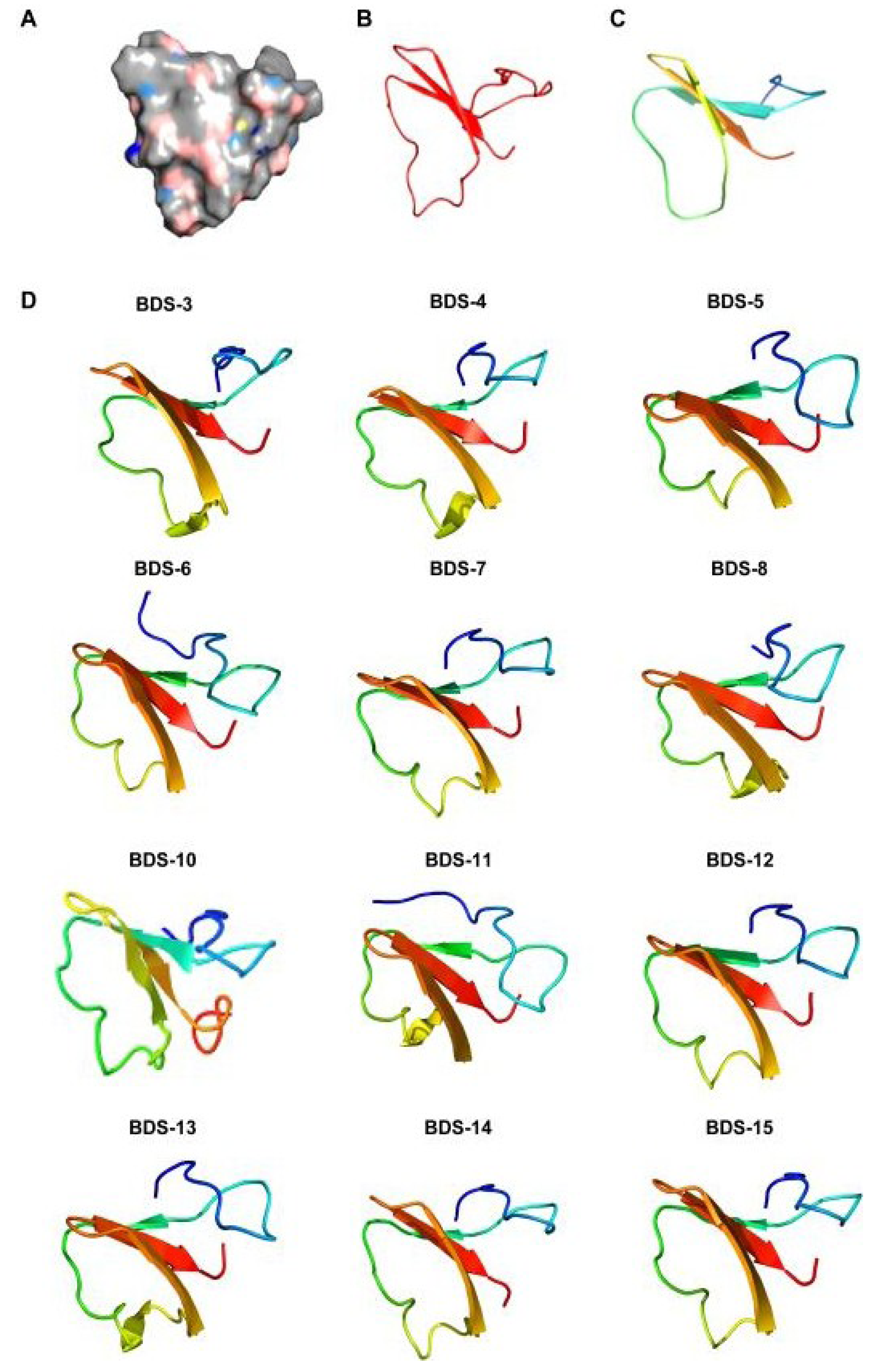
2.5. Homology Modelling

2.6. Tissue-Specific Gene Expression Pattern

3. Experimental Section
3.1. GenBank Accession Numbers
3.2. Sequence and Phylogenetic Analyses
3.3. Homology Modelling
3.4. RNA Extraction and First-Strand cDNA Synthesis
3.5. Tissue-Specific BDS cDNA Library Construction
| Primers | Sequences (5′–3′) | Amplicon size (bp) |
|---|---|---|
| BDS-F | GAAAATGAACAAAGCTCTTTCC | 278–290 |
| BDS-R | GATCGGACTGATGTTACTGG | |
| CA2-m-F | CTTTGGCGGCATTTCACTTG | 129 |
| CA2-m-R | GTGATTGGTTGGAGCCATCG | |
| HMG-F | AGTATGTGAAGCCATAGTGC | 311 |
| HMG-R | TAGTACCACCACCAACAGTC |
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Ruppert, E.E.; Barnes, R.D. The Cnidarians. In Invertebrate Zoology, 6th ed.; Saunders College Publishing: Philadelphia, PA, USA, 1994; pp. 103–154. [Google Scholar]
- Moran, Y.; Weinberger, H.; Sullivan, J.C.; Reitzel, A.M.; Finnerty, J.R.; Gurevitz, M. Concerted evolution of sea anemone neurotoxin genes is revealed through analysis of the Nematostella vectensis genome. Mol. Biol. Evol. 2008, 25, 737–747. [Google Scholar]
- Moran, Y.; Weinberger, H.; Reitzel, A.M.; Sullivan, J.C.; Kahn, R.; Gordon, D.; Finnerty, J.R.; Gurevitz, M. Intron retention as a posttranscriptional regulatory mechanism of neurotoxin expression at early life stages of the starlet anemone Nematostella vectensis. J. Mol. Biol. 2008, 380, 437–443. [Google Scholar]
- Moran, Y.; Weinberger, H.; Lazarus, N.; Gur, M.; Kahn, R.; Gordon, D.; Gurevitz, M. Fusion and retrotransposition events in the evolution of the sea anemone Anemonia viridis neurotoxin genes. J. Mol. Evol. 2009, 69, 115–124. [Google Scholar] [CrossRef]
- Venn, A.A.; Tambutte, E.; Lotto, S.; Zoccola, D.; Allemand, D.; Tambutte, S. Imaging intracellular pH in a reef coral and symbiotic anemone. Proc. Natl. Acad. Sci. USA 2009, 106, 16574–16579. [Google Scholar]
- Castaneda, O.; Harvey, A.L. Discovery and characterization of cnidarian peptide toxins that affect neuronal potassium ion channels. Toxicon 2009, 54, 1119–1124. [Google Scholar] [CrossRef]
- Frazão, B.; Vasconcelos, V.; Antunes, A. Sea anemone (Cnidaria, Anthozoa, Actiniaria) toxins: An overview. Mar. Drugs 2012, 10, 1812–1851. [Google Scholar] [CrossRef]
- Honma, T.; Shiomi, K. Peptide toxins in sea anemones: Structural and functional aspects. Mar. Biotechnol. 2006, 8, 1–10. [Google Scholar] [CrossRef]
- Honma, T.; Kawahata, S.; Ishida, M.; Nagai, H.; Nagashima, Y.; Shiomi, K. Novel peptide toxins from the sea anemone Stichodactyla haddoni. Peptides 2008, 29, 536–544. [Google Scholar]
- Diochot, S.; Loret, E.; Bruhn, T.; Beress, L.; Lazdunski, M. APETx1, a new toxin from the sea anemone Anthopleura elegantissima, blocks voltage-gated human ether-a-go-go-related gene potassium channels. Mol. Pharmacol. 2003, 64, 59–69. [Google Scholar] [CrossRef]
- Honma, T.; Hasegawa, Y.; Ishida, M.; Nagai, H.; Nagashima, Y.; Shiomi, K. Isolation and molecular cloning of novel peptide toxins from the sea anemone Antheopsis maculata. Toxicon 2005, 45, 33–41. [Google Scholar] [CrossRef]
- Béress, L.; Doppelfeld, I.S.; Etschenberg, E.; Graf, E.; Henschen, A.; Zwick, J. Polypeptides, Methods of Production and Their Uses as Antihypertensives. German Patent DE3324689A1, 17 January 1985. [Google Scholar]
- Diochot, S.; Schweitz, H.; Beress, L.; Lazdunski, M. Sea anemone peptides with a specific blocking activity against the fast inactivating potassium channel Kv3.4. J. Biol. Chem. 1998, 273, 6744–6749. [Google Scholar]
- Yeung, S.Y.M.; Thompson, D.; Wang, Z.; Fedida, D.; Robertson, B. Modulation of Kv3 subfamily potassium currents by the sea anemone toxin BDS: Significance for CNS and biophysical studies. J. Neurosci. 2005, 38, 8735–8745. [Google Scholar]
- Abbott, G.W.; Butler, M.H.; Bendahhou, S.; Dalakas, M.C.; Ptacek, L.J.; Goldstein, S.A. MiRP2 forms potassium channels in skeletal muscle with Kv3.4 and is associated with periodic paralysis. Cell 2001, 104, 217–231. [Google Scholar] [CrossRef]
- Liu, P.; Jo, S.; Bean, B.P. Modulation of neuronal sodium channels by the sea anemone peptide BDS-I. J. Neurophysiol. 2012, 107, 3155–3167. [Google Scholar] [CrossRef]
- Pannaccione, A.; Boscia, F.; Scorziello, A.; Adornetto, A.; Castaldo, P.; Sirabella, R.; Taglialatela, M.; di Renzo, G.F.; Annunziato, L. Upregulation and increased activity of KV3.4 channels and their accessory subunit MinK-related peptide 2 induced by amyloid peptide are involved in apoptotic neuronal death. Mol. Pharmacol. 2007, 272, 665–673. [Google Scholar]
- Urbarova, I.; Karlsen, B.O.; Okkenhaug, S.; Seternes, O.M.; Johansen, S.D.; Emblem, A. Digital marine bioprospecting: Mining new neurotoxin drug candidates from the transcriptomes of cold-water sea anemones. Mar. Drugs 2012, 10, 2265–2279. [Google Scholar] [CrossRef] [Green Version]
- Sabourault, C.; Ganot, P.; Deleury, E.; Allemand, D.; Furla, P. Comprehensive EST analysis of the symbiotic sea anemone, Anemonia viridis. BMC Genomics 2009, 10, 333. [Google Scholar] [CrossRef]
- Kozlov, S.; Grishin, E. The mining of toxin-like polypeptides from EST database by single residue distribution analysis. BMC Genomics 2011, 12, 88. [Google Scholar] [CrossRef]
- Kimura, M. The Neutral Theory of Molecular Evolution; Cambridge University Press: Cambridge, UK, 1983. [Google Scholar]
- Ogawa, T.; Oda, N.; Nakashima, K.; Sasaki, H.; Hattori, M.; Sakaki, Y.; Kihara, H.; Ohno, M. Unusually high conservation of untranslated sequences in cDNAs for Trimeresurus flavoviridis phospholipase A2 isozymes. Proc. Natl. Acad. Sci. USA 1992, 89, 8557–8561. [Google Scholar]
- Deshimaru, M.; Ogawa, T.; Nakashima, K.; Nobuhisa, I.; Chijiwa, T.; Shimohigashi, Y.; Fukumaki, Y.; Niwa, M.; Yamashina, I.; Hattori, S.; et al. Accelerated evolution of crotalinae snake venom gland serine proteases. FEBS Lett. 1996, 397, 83–88. [Google Scholar]
- Zhu, S.; Bosmans, F.; Tytgat, J. Adaptive evolution of scorpion sodium channel toxins. J. Mol. Evol. 2004, 58, 145–153. [Google Scholar]
- Anderluh, G.; Podlesek, Z.; Macek, P. A common motif in proparts of Cnidarian toxins and nematocyst collagens and its putative role. Biochim. Biophys. Acta 2000, 1476, 372–376. [Google Scholar] [CrossRef]
- Anderluh, G.; Macek, P. Cytolytic peptide and protein toxins from sea anemones (Anthozoa: Actiniaria). Toxicon 2002, 40, 111–124. [Google Scholar] [CrossRef]
- Nagai, H.; Oshiro, N.; Takuwa-Kuroda, K.; Iwanaga, S.; Nozaki, M.; Nakajima, T. A new polypeptide toxin from the nematocyst venom of an Okinawan sea anemone Phyllodiscus semoni (Japanese name “unbachi-isoginchaku”). Biosci. Biotechnol. Biochem. 2002, 66, 2621–2625. [Google Scholar]
- Spagnuolo, A.; Zanetti, L.; Cariello, L.; Piccoli, R. Isolation and characterization of two genes encoding calitoxins, neurotoxic peptides from Calliactis parasittica (Cnidaria). Gene 1994, 138, 187–191. [Google Scholar] [CrossRef]
- Ghadessy, F.J.; Chen, D.; Kini, R.M.; Chung, M.C.; Jeyaseelan, K.; Khoo, H.E.; Yuen, R. Stonustoxin is a novel lethal factor from stonefish (Synanceja horrida) venom. cDNA cloning and characterization. J. Biol. Chem. 1996, 271, 25575–25581. [Google Scholar]
- Menon, R.P.; Hughes, R.C. Determinants in the N-terminal domains of galectin-3 for secretion by a novel pathway circumventing the endoplasmic reticulum-Golgi complex. Eur. J. Biochem. 1999, 264, 569–576. [Google Scholar] [CrossRef]
- Liu, W.H.; Wang, L.; Wang, Y.L.; Peng, L.S.; Wu, W.Y.; Peng, W.L.; Jiang, X.Y.; Tu, H.B.; Chen, H.P.; Ou-Yang, P.; et al. Cloning and characterization of a novel neurotoxin from the sea anemone Anthopleura sp. Toxicon 2003, 41, 793–801. [Google Scholar] [CrossRef]
- Muesch, A.; Hartmann, E.; Rohde, K.; Rubartelli, A.; Sitia, R.; Rapoport, T.A. A novel pathway for secretory proteins? Trends Biochem. Sci. 1990, 15, 86–88. [Google Scholar] [CrossRef]
- Carbonetti, N.H.; Mays, R.M.; Artamonova, G.V.; Plaut, R.D.; Worthington, Z.E. Proteolytic cleavage of pertussis toxin S1 subunit is not essential for its activity in mammalian cells. BMC Microbiol. 2005, 5, 7. [Google Scholar] [CrossRef] [Green Version]
- Zaharenko, A.J.; Ferreira, W.A., Jr.; Oliveira, J.S.; Richardson, M.; Pimenta, D.C.; Konno, K.; Portaro, F.C.V.; Freitas, J.C. Proteomics of the neurotoxic fraction from the sea anemone Bunodosoma cangicum venom: Novel peptides belonging to new classes of toxins. Comp. Biochem. Physiol. Part D 2008, 3, 219–225. [Google Scholar]
- Rodríguez, A.A.; Cassoli, J.S.; Sa, F.; Dong, Z.Q.; de Freitas, J.C.; Pimenta, A.M.; de Lima, M.E.; Konno, K.; Lee, S.M.; Garateix, A.; et al. Peptide fingerprinting of the neurotoxic fractions isolated from the secretions of sea anemones Stichodactyla helianthus and Bunodosoma granulifera. New members of the APETx-like family identified by a 454 pyrosequencing approach. Peptides 2012, 34, 26–38. [Google Scholar] [CrossRef]
- Oliveira, J.S.; Zaharenko, A.J.; Ferreira, W.A., Jr.; Konno, K.; Shida, C.S.; Richardson, M.; Lúcio, A.D.; Beirão, P.S.; de Freitas, J.C. BcIV, a new paralyzing peptide obtained from the venom of the sea anemone Bunodosoma caissarum. A comparison with the Na+ channel toxin BcIII. Biochim. Biophys. Acta 2006, 1764, 1592–1600. [Google Scholar]
- Zaharenko, A.Z. University of São Paulo: São Paulo, SP, Brazil, Unpublished work. 2010.
- Peigneur, S. University of Leuven: Leuven, Belgium, Unpublished work. 2012.
- Richier, S.; Rodriguez-Lanetty, M.; Schnitzler, C.E.; Weis, V.M. Response of the symbiotic cnidarian Anthopleura elegantissima transcriptome to temperature and UV increase. Comp. Biochem. Physiol. Part D 2008, 3, 283–289. [Google Scholar]
- Driscoll, P.C.; Clore, G.M.; Beress, L.; Gronenborn, A.M. A proton nuclear magnetic resonance study of the antihypertensive and antiviral protein BDS-I from the sea anemone Anemonia sulcata: Sequential and stereospecific resonance assignment and secondary structure. Biochemistry 1989, 28, 2178–2187. [Google Scholar]
- Driscoll, P.C.; Gronenborn, A.M.; Beress, L.; Clore, G.M. Determination of the three-dimensional solution structure of the antihypertensive and antiviral protein BDS-I from the sea anemone Anemonia sulcata: A study using nuclear magnetic resonance and hybrid distance geometry-dynamical simulated annealing. Biochemistry 1989, 28, 2188–2198. [Google Scholar]
- Venter, J.C.; Levy, S.; Stockwell, T.; Remington, K.; Halpern, A. Massive parallelism, randomness and genomic advances. Nat. Genet. 2003, 33, 219–227. [Google Scholar]
- Ganot, P.; Moya, A.; Magnone, V.; Allemand, D.; Furla, P.; Sabourault, C. Adaptations to endosymbiosis in a cnidarian-dinoflagellate association: Differential gene expression and specific gene duplications. PLoS Genet. 2011, 7, e1002187. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DNAsp v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA 5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony method. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Nei, M.; Gojobori, T. Simple methods for estimating the numbers of synonymous and non synonymous nucleotide substitutions. Mol. Biol. Evol. 1986, 3, 418–426. [Google Scholar]
- Kelley, L.A.; Sternberg, M.J.E. Protein structure prediction on the web: A case study using the Phyre server. Nat. Protoc. 2009, 4, 363–371. [Google Scholar] [CrossRef]
- Yamaguchi, Y.; Hasegawa, Y.; Honma, T.; Nagashima, Y.; Shiomi, K. Screening and cDNA cloning of Kv1 potassium channel toxins in sea anemones. Mar. Drugs 2010, 8, 2893–2905. [Google Scholar] [CrossRef]
- Chintiroglou, C.; Koukouras, A. The feeding habits of three Mediterranean sea anemone species, Anemonia viridis (Forskal), Actinia equina (Linnaeus) and Cereus pedunculatus (Pennant). Helgol. Meeresunters. 1992, 46, 53–68. [Google Scholar] [CrossRef]
- Moran, Y.; Gordon, D.; Gurevitz, M. Sea anemone toxins affecting voltage-gated sodium channels: Molecular and evolutionary features. Toxicon 2009, 54, 1089–1101. [Google Scholar] [CrossRef]
- Barlow, A.; Pook, C.E.; Harrison, R.A.; Wuster, W. Coevolution of diet and prey-specific venom activity supports the role of selection in snake venom evolution. Proc. R. Soc. B 2009, 276, 2443–2449. [Google Scholar]
- Fry, B.G.; Vidal, N.; van der Weerd, L.; Kochva, E.; Renjifo, C. Evolution and diversification of the toxicofera reptile venom system. J. Proteomics 2009, 72, 127–136. [Google Scholar]
- Moran, Y.; Genikhovich, G.; Gordon, D.; Wienkoop, S.; Zenkert, C.; Ozbek, S.; Technau, U.; Gurevitz, M. Neurotoxin localization to ectodermal gland cells uncovers an alternative mechanism of venom delivery in sea anemones. Proc. Biol. Sci. B 2012, 279, 1351–1358. [Google Scholar]
- Chabbert, C.; Chambard, J.M.; Sans, A.; Desmadryl, G. Three types of depolarization-activated potassium currents in acutely isolated mouse vestibular neurons. J. Neurophysiol. 2001, 85, 1017–1026. [Google Scholar]
- Baranauskas, G.; Tkatch, T.; Nagata, K.; Yeh, J.Z.; Surmeier, D.J. Kv3.4 subunits enhance the repolarizing efficiency of Kv3.1 channels in fast-spiking neurons. Nat. Neurosci. 2003, 6, 258–266. [Google Scholar]
- Shevchenko, T.; Teruyama, R.; Armstrong, W.E. High-threshold, Kv3-like potassium currents in magnocellular neurosecretory neurons and their role in spike repolarization. J. Neurophysiol. 2004, 92, 3043–3055. [Google Scholar] [CrossRef]
- Kaab, S.; Miguel-Velado, E.; Lopez-Lopez, J.R.; Perez-Garcia, M.T. Down regulation of Kv3.4 channels by chronic hypoxia increases acute oxygen sensitivity in rabbit carotid body. J. Physiol. (Lond.) 2005, 566, 395–408. [Google Scholar]
- Angulo, E.; Noe, V.; Casado, V.; Mallol, J.; Gomez-Isla, T.; Lluis, C.; Ferrer, I.; Ciudad, C.J.; Franco, R. Up-regulation of the Kv3.4 potassium channel subunit in early stages of Alzheimer’s disease. J. Neurochem. 2004, 91, 547–557. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Nicosia, A.; Maggio, T.; Mazzola, S.; Cuttitta, A. Evidence of Accelerated Evolution and Ectodermal-Specific Expression of Presumptive BDS Toxin cDNAs from Anemonia viridis. Mar. Drugs 2013, 11, 4213-4231. https://doi.org/10.3390/md11114213
Nicosia A, Maggio T, Mazzola S, Cuttitta A. Evidence of Accelerated Evolution and Ectodermal-Specific Expression of Presumptive BDS Toxin cDNAs from Anemonia viridis. Marine Drugs. 2013; 11(11):4213-4231. https://doi.org/10.3390/md11114213
Chicago/Turabian StyleNicosia, Aldo, Teresa Maggio, Salvatore Mazzola, and Angela Cuttitta. 2013. "Evidence of Accelerated Evolution and Ectodermal-Specific Expression of Presumptive BDS Toxin cDNAs from Anemonia viridis" Marine Drugs 11, no. 11: 4213-4231. https://doi.org/10.3390/md11114213





