Penicillinolide A: A New Anti-Inflammatory Metabolite from the Marine Fungus Penicillium sp. SF-5292
Abstract
:1. Introduction
2. Results and Discussion
2.1. Structure Determination of Penicillinolide A (1)
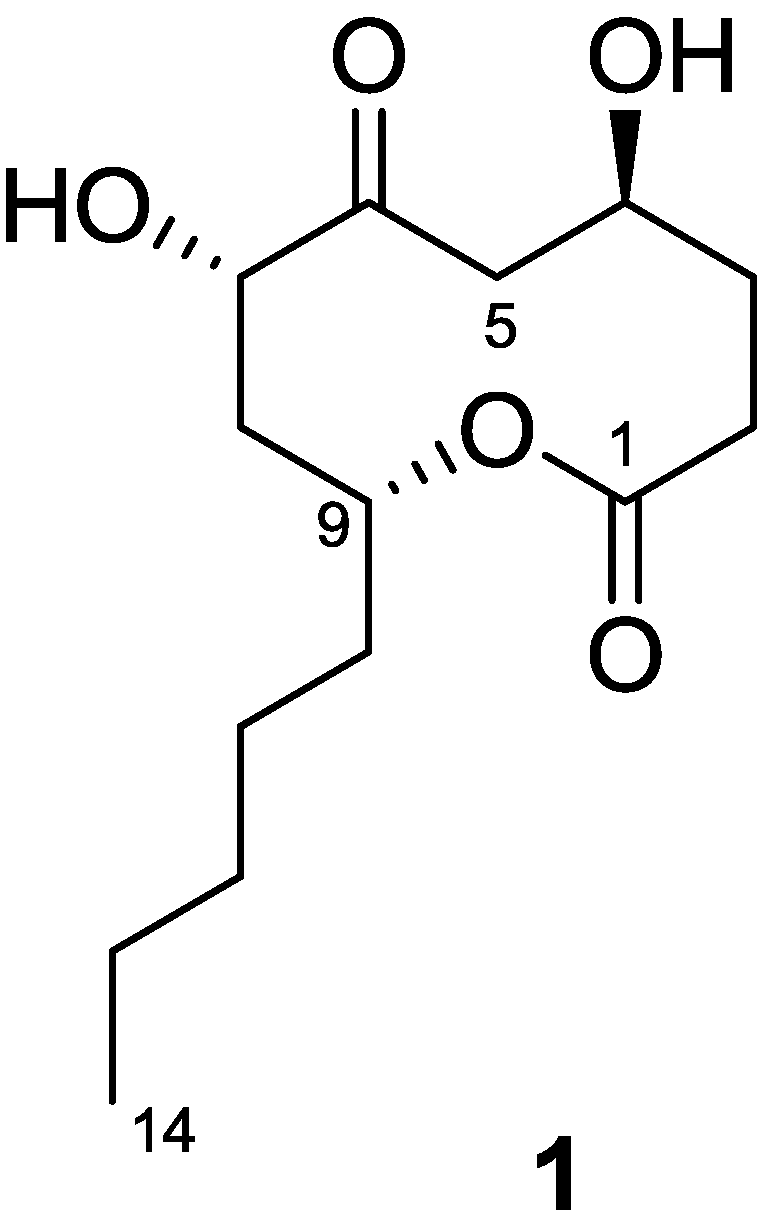
| Position | δC a | δH, mult. (J in Hz) b | Key NOESY | HMBC (H→C#) |
|---|---|---|---|---|
| 1 | 172.9 | - | - | |
| 2 | 28.9 | 3.11, m; 2.56, m | - | 1, 3, 4 |
| 3 | 28.6 | 2.65, m; 2.00, m | - | 1, 2, 4, 5 |
| 4 | 65.2 | 5.00, m | H-7 | - |
| 5 | 46.2 | 3.12, dd (18.1, 11.0); 2.93, dd (18.1, 4.8) | - | 3, 4, 6 |
| 6 | 211.0 | - | - | - |
| 7 | 75.2 | 4.57, dd (7.7, 3.7) | H-4, H-9 | 8, 9 |
| 8 | 39.3 | 2.58, m; 2.41, m | - | 6, 7, 9, 10 |
| 9 | 73.1 | 5.27, m | H-7 | 1, 7 |
| 10 | 34.3 | 1.81, m; 1.70, m | - | 8, 9, 11, 12 |
| 11 | 25.8 | 1.27, m | - | 10, 12, 13 |
| 12 | 31.8 | 1.23–1.09, m | - | - |
| 13 | 22.7 | 1.21–1.11, m | - | - |
| 14 | 14.1 | 0.76, t (6.6) | - | 12, 13 |
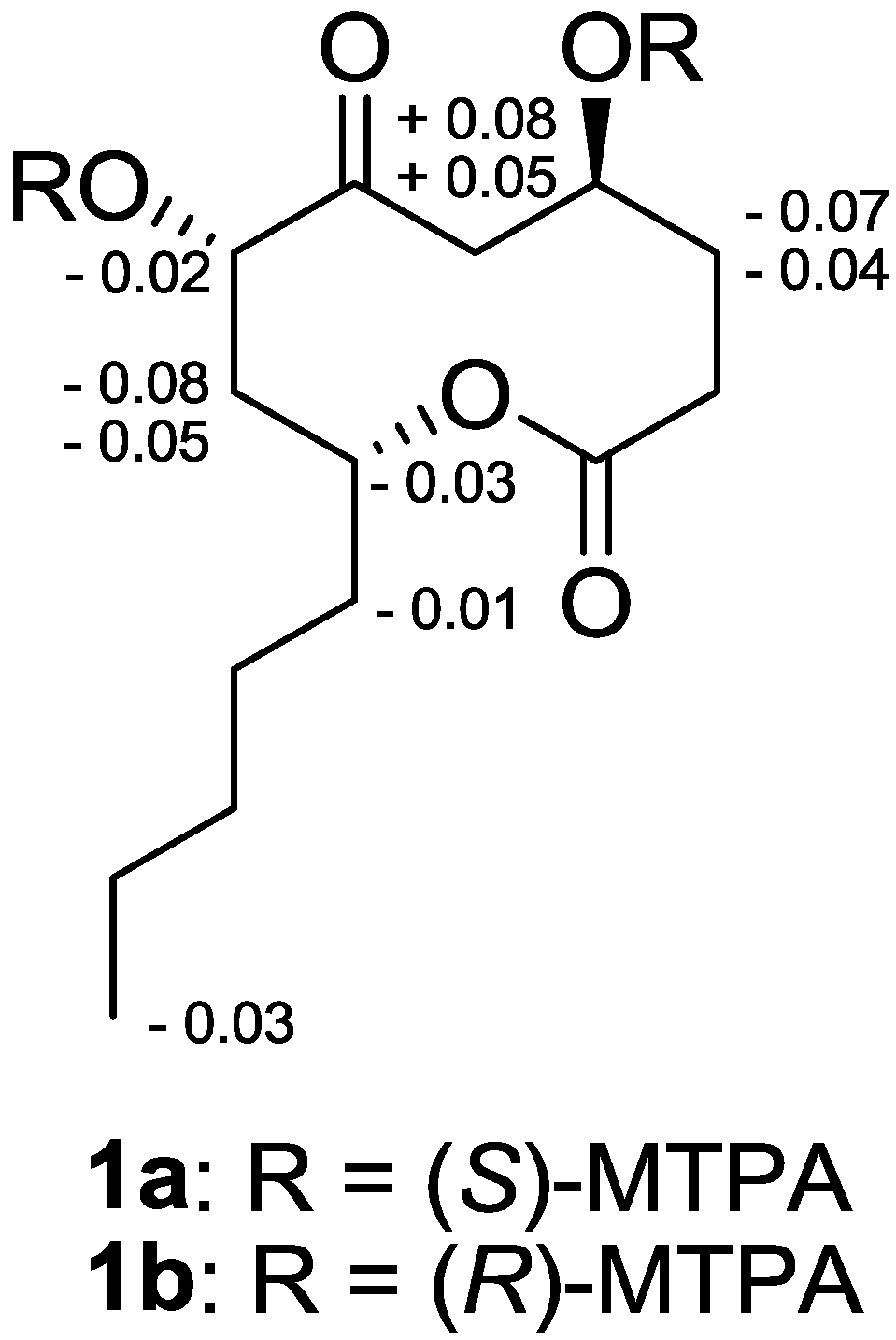
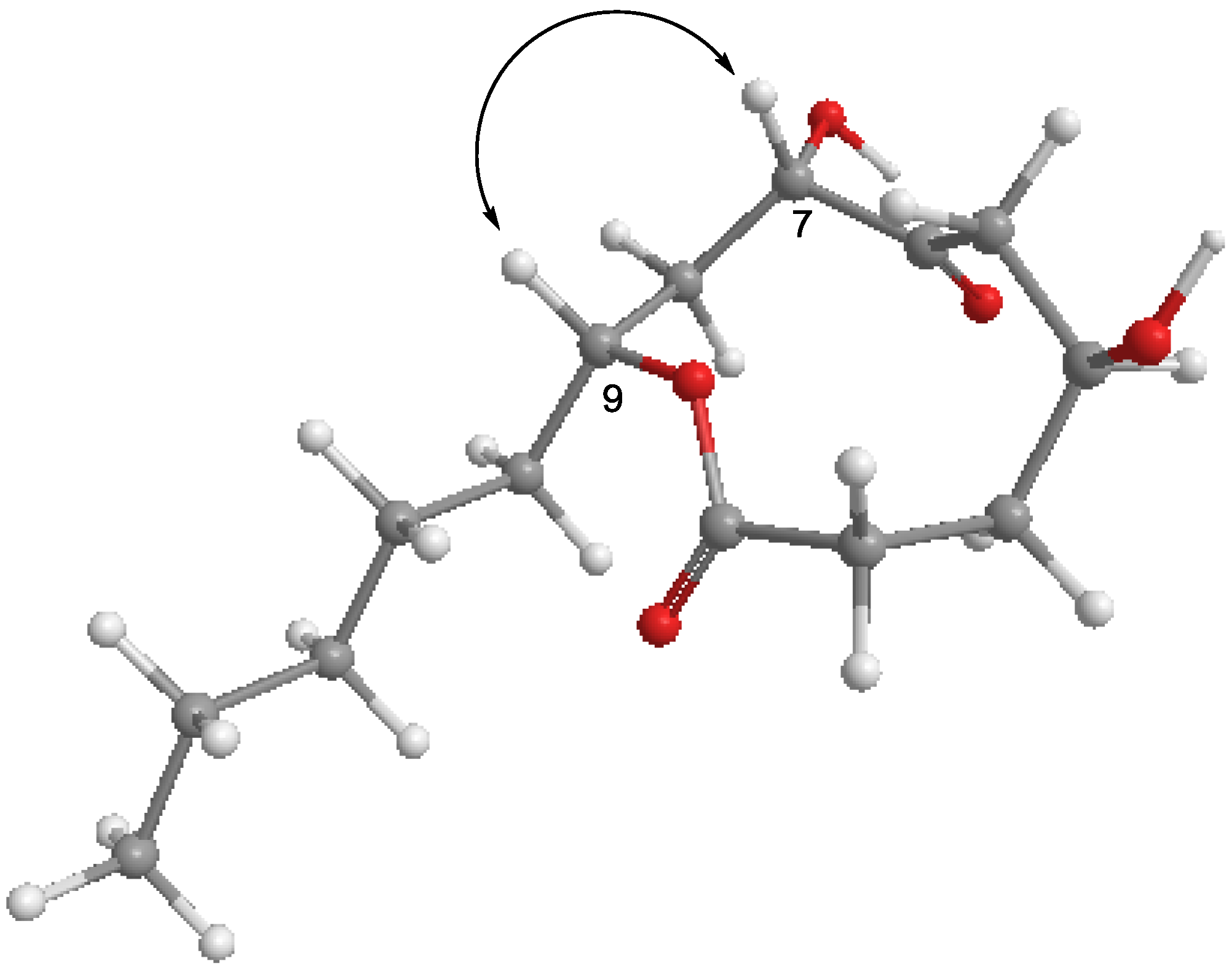
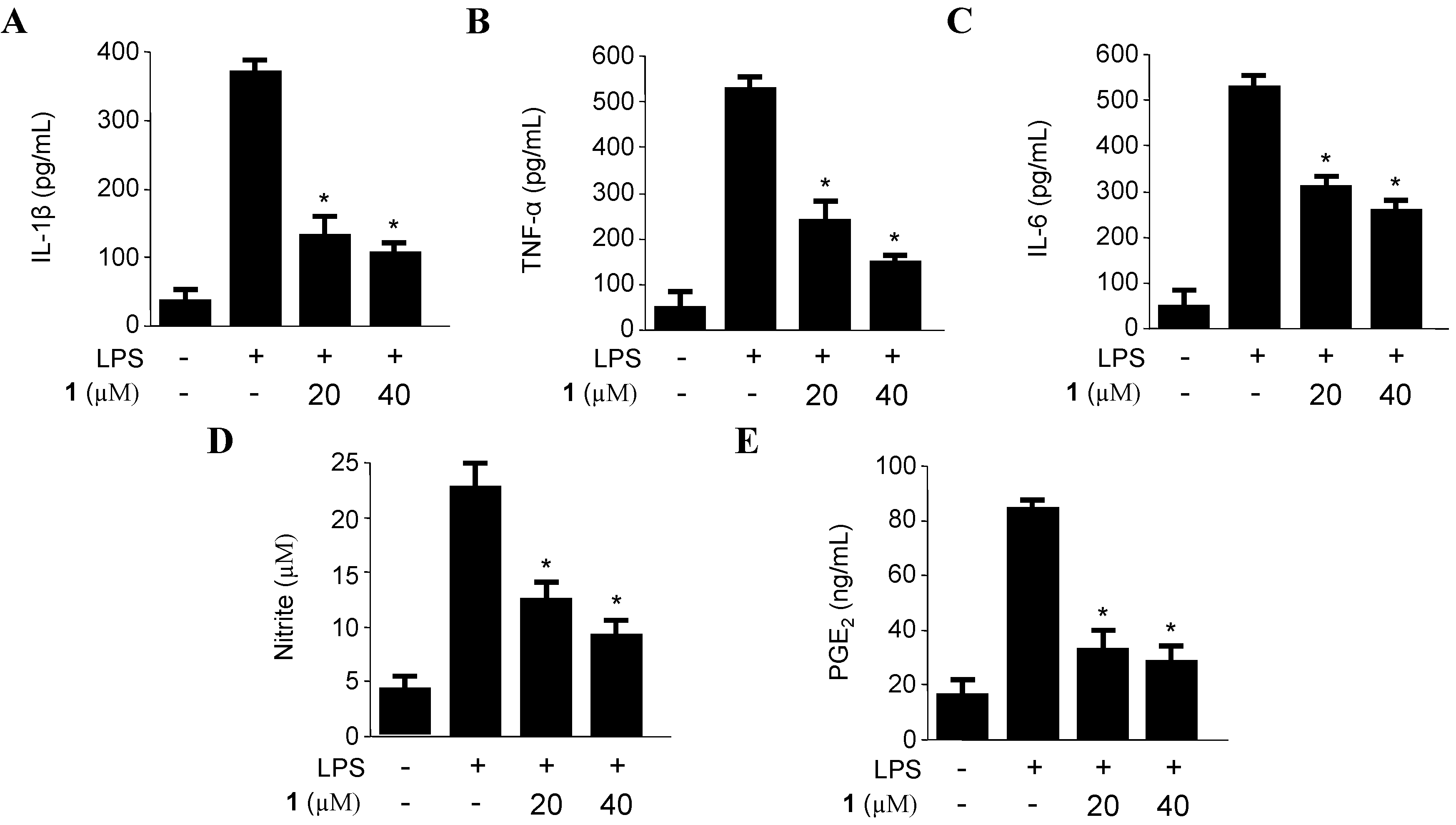
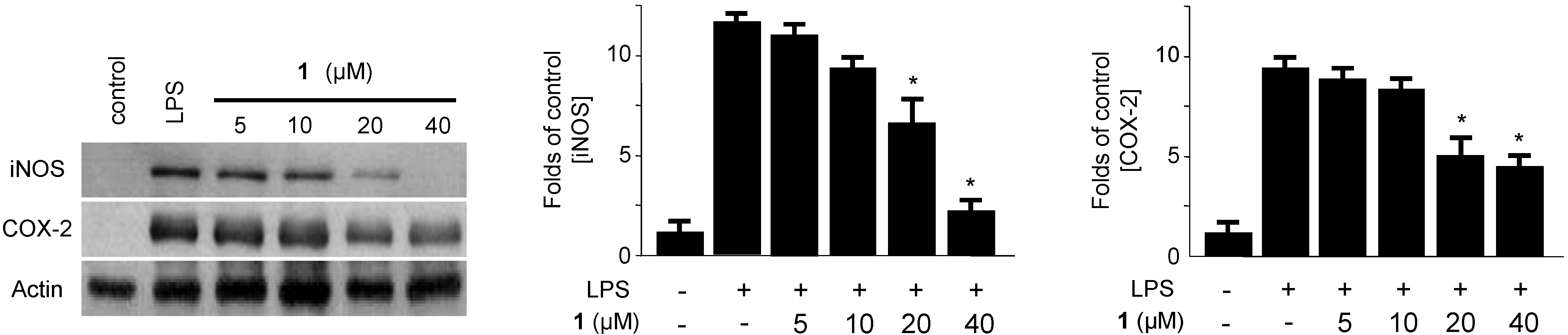
2.2. Effects of Penicillinolide A (1) on the Expression of Pro-Inflammatory Proteins and Production of Pro-Inflammatory Cytokines in Murine Peritoneal Macrophages Stimulated with LPS
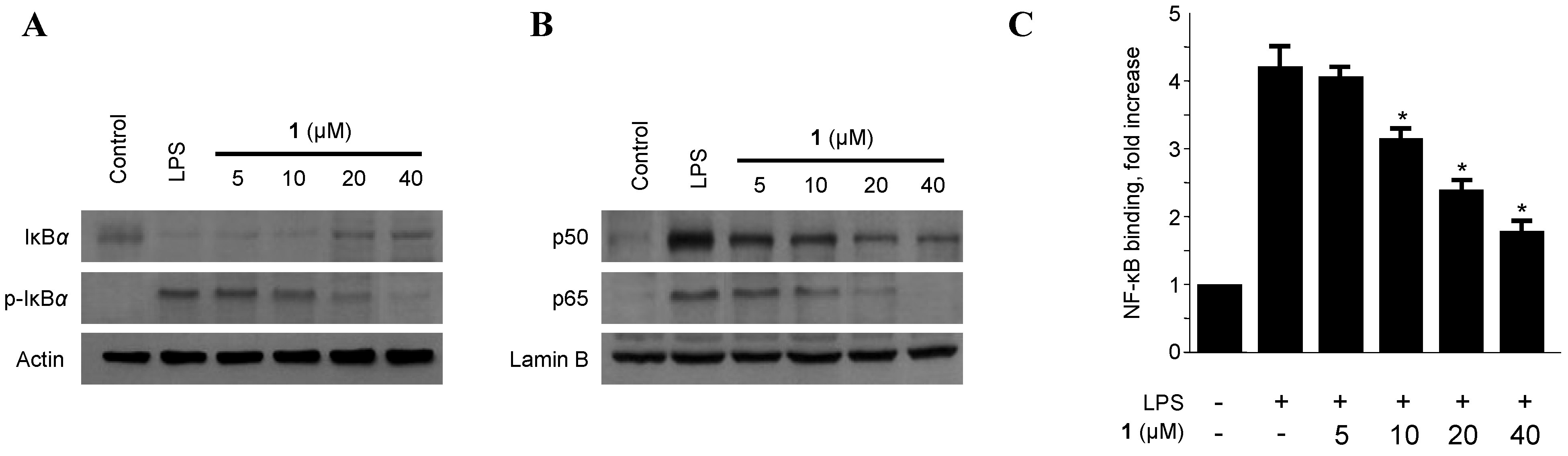
2.3. Effects of Penicillinolide A (1) on the Protein Expression Levels of IκB-α Phosphorylation and Degradation as well as NF-κB Translocation and DNA Binding Activity in Murine Peritoneal Macrophages
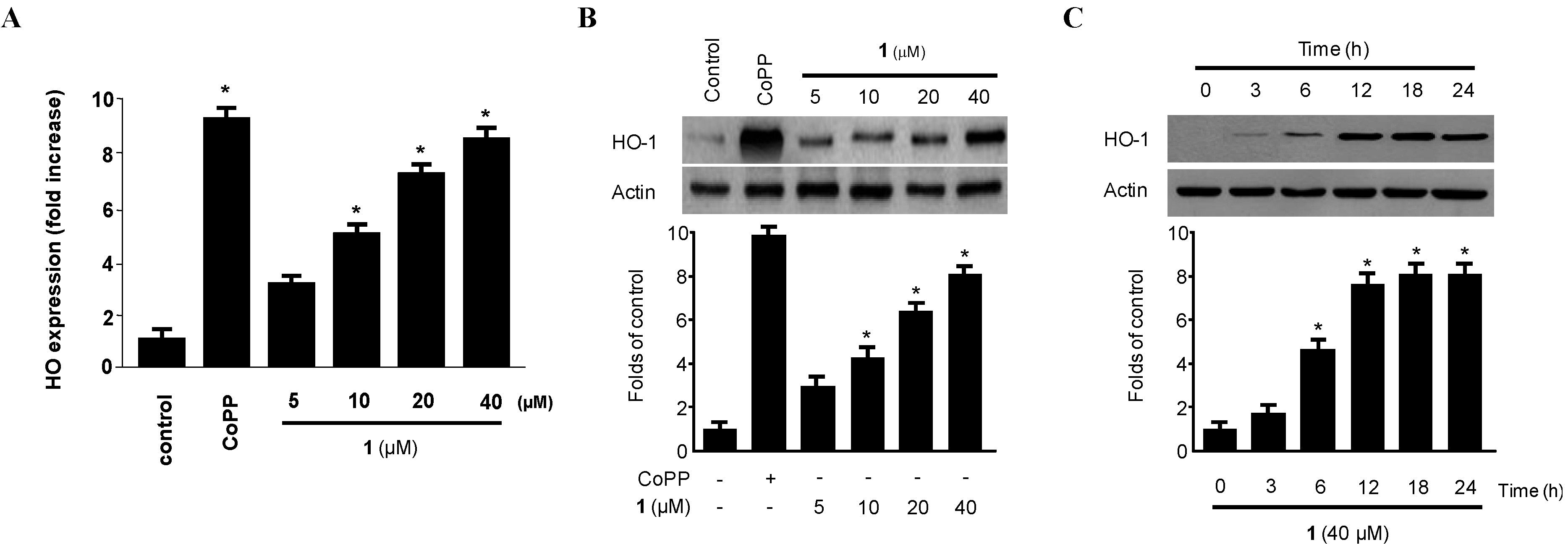
2.4. Effect of Penicillinolide (1) on HO-1 Expression via Nuclear Translocation of Nrf2 in Murine Peritoneal Macrophages
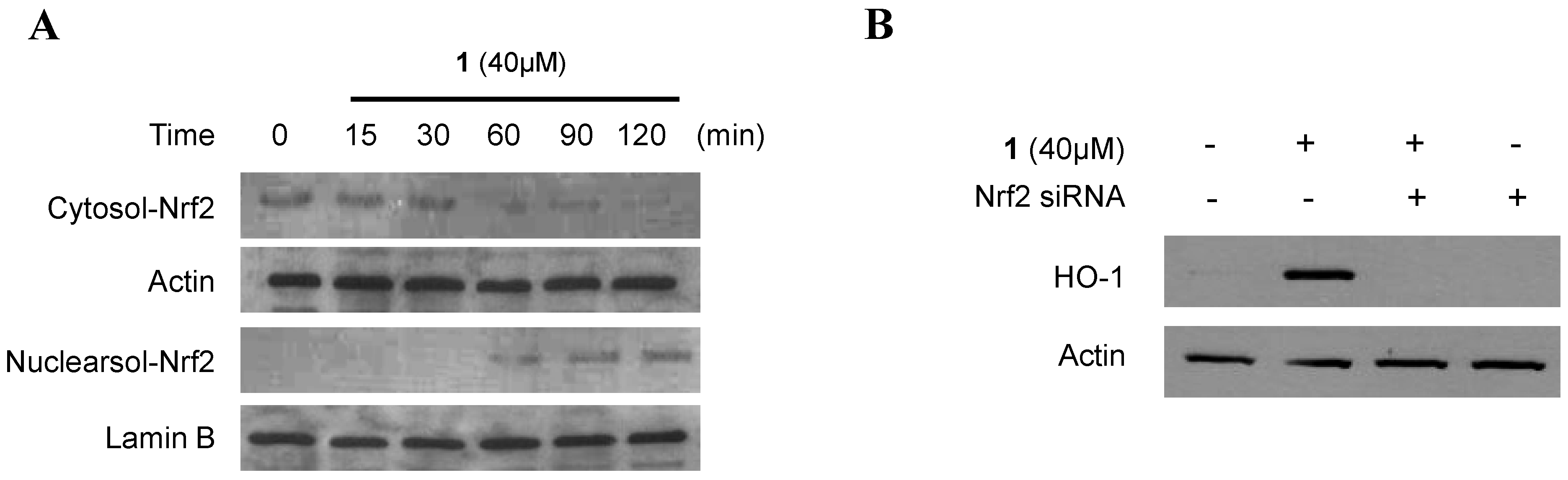
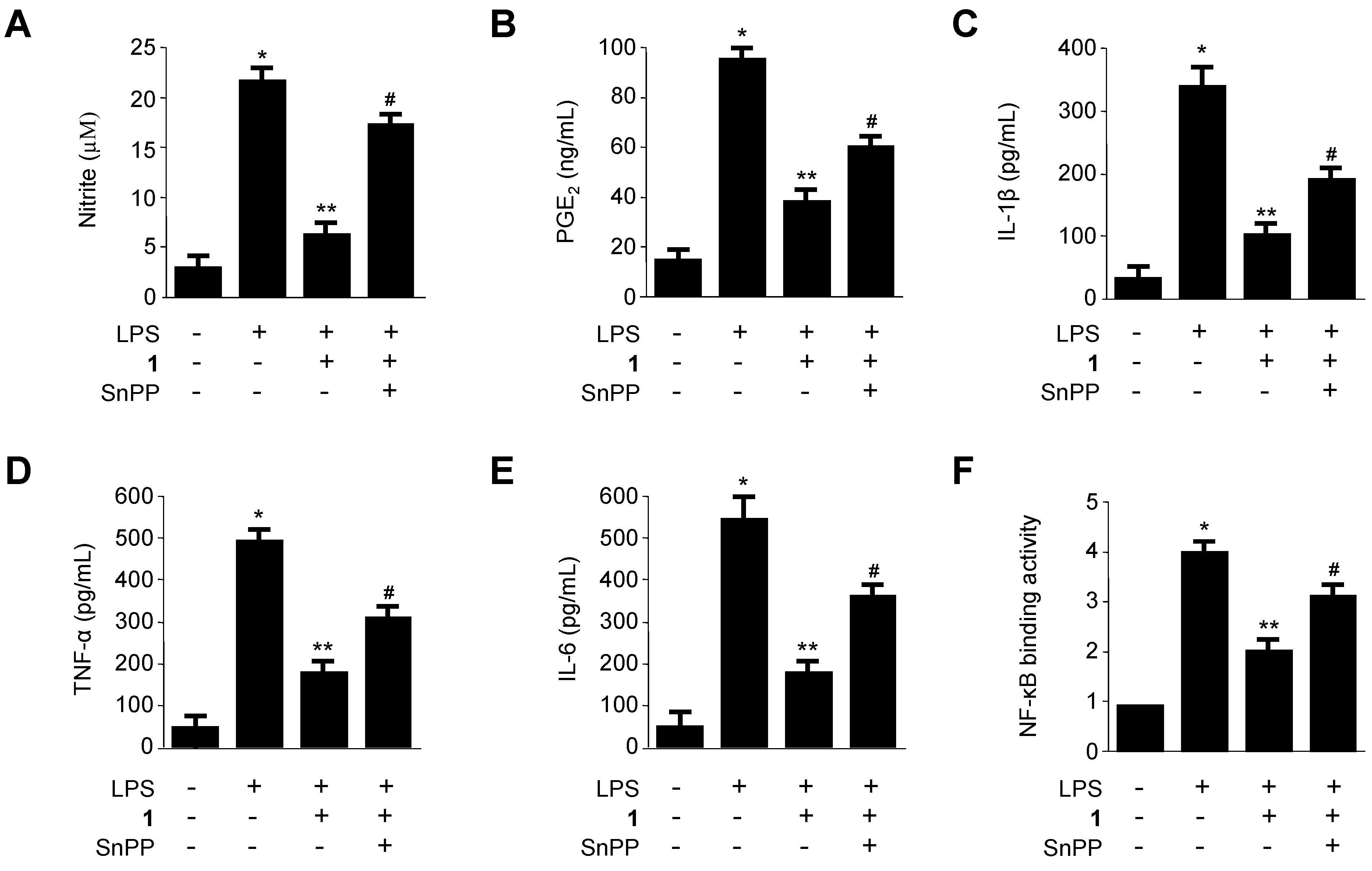
2.5. Effects of SnPP on the Inhibition of Production of Pro-Inflammatory Mediators via Pre-Treatment of Penicillinolide A (1) in LPS-Stimulated Murine Peritoneal Macrophages
3. Experimental Section
3.1.General Experimental Procedures and Materials
3.2. Specimen Collection and Identification of the Marine-Derived Fungus Penicillium sp. SF-5292
3.3. Fermentation, Extraction and Isolation of Penicillinolide A (1) from Penicillium sp. SF-5292
3.4. Preparation of Mosher Esters of Penicillinolide A (1)
3.4.1. Selected 1H NMR (CDCl3, 400 MHz) Data for 1a
3.4.2. Selected 1H NMR (CDCl3, 400 MHz) Data for 1b
3.5. Peritoneal Macrophage Cultures and Cell Viability Assay
3.6. Determination of Nitrite Production and PGE2, TNF-α, IL-1β and IL-6 Assays
3.7. Preparation of Cytosolic and Nuclear Fractions
3.8. Western Blot Analysis
3.9. Real-Time PCR
3.10. DNA-Binding Activity of NF-κB
3.11. Transfection of Nrf2 siRNA
3.12. Statistical Analysis
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Heller, R.A.; Schena, M.; Chai, A.; Shalon, D.; Bedilion, T.; Gilmore, J.; Woolley, D.E.; Davis, R.W. Discovery and analysis of inflammatory disease-related genes using cdna microarrays. Proc. Natl. Acad. Sci. USA 1997, 94, 2150–2155. [Google Scholar] [CrossRef]
- Abraham, N.G.; Kappas, A. Pharmacological and clinical aspects of heme oxygenase. Pharmcol. Rev. 2008, 60, 79–127. [Google Scholar] [CrossRef]
- Otterbein, L.E.; Mantell, L.L.; Choi, A.M. Carbon monoxide provides protection against hyperoxic lung injury. Am. J. Physiol. 1999, 276, L688–L694. [Google Scholar]
- Ryter, S.W.; Alam, J.; Choi, A.M. Heme oxygenase-1/carbon monoxide: From basic science to therapeutic applications. Physiol. Rev. 2006, 86, 583–650. [Google Scholar] [CrossRef]
- Otterbein, L.E.; Bach, F.H.; Alam, J.; Soares, M.; Tao Lu, H.; Wysk, M.; Davis, R.J.; Flavell, R.A.; Choi, A.M. Carbon monoxide has anti-inflammatory effects involving the mitogen-activated protein kinase pathway. Nat. Med. 2000, 6, 422–428. [Google Scholar] [CrossRef]
- Lee, T.S.; Tsai, H.L.; Chau, L.Y. Induction of heme oxygenase-1 expression in murine macrophages is essential for the anti-inflammatory effect of low dose 15-deoxy-delta 12,14-prostaglandin J2. J. Biol. Chem. 2003, 278, 19325–19330. [Google Scholar]
- Wiesel, P.; Foster, L.C.; Pellacani, A.; Layne, M.D.; Hsieh, C.M.; Huggins, G.S.; Strauss, P.; Yet, S.F.; Perrella, M.A. Thioredoxin facilitates the induction of heme oxygenase-1 in response to inflammatory mediators. J. Biol. Chem. 2000, 275, 24840–24846. [Google Scholar] [CrossRef]
- Morse, D.; Pischke, S.E.; Zhou, Z.; Davis, R.J.; Flavell, R.A.; Loop, T. Suppression of inflammatory cytokine production by carbon monoxide involves the JNK pathway and AP-1. J. Biol. Chem. 2003, 278, 36993–36998. [Google Scholar]
- Suh, G.Y.; Jin, Y.; Yi, A.K.; Wang, X.M.; Choi, A.M. CCAAT/enhancer-binding protein mediates carbon monoxide-induced suppression of cyclooxygenase-2. Am. J. Respir. Cell Mol. Biol. 2006, 35, 220–226. [Google Scholar] [CrossRef]
- Oh, G.S.; Pae, H.O.; Lee, B.S.; Kim, B.N.; Kim, J.M.; Kim, H.R.; Jeon, S.B.; Jeon, W.K.; Chae, H.J.; Chung, H.T. Hydrogen sulfide inhibits nitric oxide production and nuclear factor kappa B via heme oxygenase-1 expression in RAW264.7 macrophages stimulated with lipopolysaccharide. Free Radic. Biol. Med. 2006, 41, 106–119. [Google Scholar] [CrossRef]
- Oh, G.S.; Pae, H.O.; Choi, B.M.; Chae, S.C.; Lee, H.S.; Ryu, D.G. 3-Hydroxyanthranilic acid, one of metabolites of tryptophan via indoleamine 2,3-dioxygenase pathway, suppresses inducible nitric oxide synthase expression by enhancing heme oxygenase-1 expression. Biochem. Biophys. Res. Commun. 2004, 320, 1156–1162. [Google Scholar] [CrossRef]
- Van-Assche, T.; Huygelen, V.; Crabtree, M.J.; Antoniades, C. Gene therapy targeting inflammation in atherosclerosis. Curr. Pharm. Des. 2011, 17, 4210–4223. [Google Scholar] [CrossRef]
- Lee, D.S.; Jeong, G.S.; Li, B.; Park, H.; Kim, Y.C. Anti-inflammatory effects of sulfuretin from Rhus verniciflua stokes via the induction of heme oxygenase-1 expression in murine macrophages. Int. Immunopharmacol. 2010, 10, 850–858. [Google Scholar] [CrossRef]
- Willoughby, D.A.; Moore, A.R.; Colville-Nash, P.R.; Gilroy, D. Resolution of inflammation. Int. J. Immunopharmacol. 2000, 22, 1131–1135. [Google Scholar] [CrossRef]
- Gueler, F.; Park, J.K.; Rong, S.; Kirsch, T.; Lindschau, C.; Zheng, W.; Elger, M.; Fiebeler, A.; Fliser, D.; Luft, F.C.; et al. Statins attenuate ischemia-reperfusion injury by inducing heme oxygenase-1 in infiltrating macrophages. Am. J. Pathol. 2007, 170, 1192–1199. [Google Scholar] [CrossRef]
- Gonçalves, G.M.; Cenedeze, M.A.; Feitoza, C.Q.; Wang, P.M.; Bertocchi, A.P.; Damiao, M.J.; Pinheiro, H.S.; Antunes Teixeira, V.P.; dos Reis, M.A.; Pacheco-Silva, A.; et al. The role of heme oxygenase 1 in rapamycin-induced renal dysfunction after ischemia and reperfusion injury. Kidney Int. 2006, 70, 1742–1749. [Google Scholar] [CrossRef]
- Aburaya, M.; Tanaka, K.; Hoshino, T.; Tsutsumi, S.; Suzuki, K.; Makise, M.; Akagi, R.; Mizushima, T. Heme oxygenase-1 protects gastric mucosal cells against nonsteroidal anti-inflammatory drugs. J. Biol. Chem. 2006, 281, 33422–33432. [Google Scholar]
- Baird, L.; Dinkova-Kostova, A.T. The cytoprotective role of the Keap1–Nrf2 pathway. Arch. Toxicol. 2011, 85, 241–272. [Google Scholar] [CrossRef]
- Jaiswal, A.K. Regulation of genes encoding NAD(P)H: Quinine oxidoreductases. Free Radic. Biol. Med. 2000, 29, 254–262. [Google Scholar] [CrossRef]
- Balogun, E.; Hoque, M.; Gong, P.; Killeen, E.; Green, C.J.; Foresti, R.; Alam, J.; Motterlini, R. Curcumin activates the haem oxygenase-1 gene via regulation of NRF2 and he antioxidant-responsive element. Biochem. J. 2003, 371, 887–895. [Google Scholar] [CrossRef]
- Lee, D.S.; Jeong, G.S.; Li, B.; Lee, S.U.; Oh, H.; Kim, Y.C. Asperlin from the marine-derived fungus Aspergillus sp. SF-5044 exerts anti-inflammatory effects through heme oxygenase-1 expression in murine macrophages. J. Pharmacol. Sci. 2011, 116, 283–295. [Google Scholar] [CrossRef]
- Lee, D.S.; Jang, J.H.; Ko, W.; Kim, K.S.; Sohn, J.H.; Kang, M.S.; Ahn, J.S.; Kim, Y.C.; Oh, H. PTP1B inhibitory and anti-inflammatory effects of secondary metabolites isolated from the marine-derived fungus Penicillium sp. JF-55. Mar. Drugs 2013, 11, 1409–1426. [Google Scholar]
- Fenical, W.; Jensen, P.R. Developing a new resource for drug discovery: Marine actinomycete bacteria. Nat. Chem. Biol. 2006, 2, 666–673. [Google Scholar] [CrossRef]
- Bugni, T.S.; Ireland, C.M. Marine-derived fungi: A chemically and biologically diverse group of microorganisms. Nat. Prod. Rep. 2004, 21, 143–163. [Google Scholar] [CrossRef]
- Saleem, M.; Ali, M.S.; Hussain, S.; Jabbar, A.; Ashraf, M.; Lee, Y.S. Marine natural products of fungal origin. Nat. Prod. Rep. 2007, 24, 1142–1152. [Google Scholar] [CrossRef]
- Lee, S.U.; Asami, Y.; Lee, D.; Jang, J.-H.; Ahn, J.S.; Oh, H. Protuboxepins A and B and protubonines A and B from the marine-derived fungus Aspergillus sp. SF-5044. J. Nat. Prod. 2011, 74, 1284–1287. [Google Scholar] [CrossRef]
- Kim, M.-Y.; Sohn, J.H.; Ahn, J.S.; Oh, H. Alternaramide, a cyclic depsipeptide from the marine-derived fungus Alternaria sp. SF-5016. J. Nat. Prod. 2005, 72, 77–98. [Google Scholar]
- Palmer, R.M.; Ashton, D.S.; Moncada, S. Vascular endothelial cells synthesize nitric oxide from l-Arginine. Nature 1988, 333, 664–666. [Google Scholar] [CrossRef]
- Block, M.L.; Hong, J.S. Microglia and inflammation-mediated neurodegeneration: Multiple triggers with a common mechanism. Prog. Neurobiol. 2005, 76, 77–98. [Google Scholar] [CrossRef]
- Surh, Y.J.; Chun, K.S.; Cha, H.H.; Han, S.S.; Keum, Y.S.; Park, K.K.; Lee, S.S. Molecular mechanisms underlying chemopreventive activities of anti-inflammatory phytochemicals: Down-regulation of COX-2 and iNOS through suppression of NF-kappa B activation. Mutat. Res. 2001, 480–481, 243–268. [Google Scholar]
- Verma, I.M.; Stevenson, J.K.; Schwarz, E.M.; van Antwerp, D.; Miyamoto, S. Rel/NF-kappaB/I kappaB family: Intimate tales of association and dissociation. Genes Dev. 1995, 9, 2723–2735. [Google Scholar] [CrossRef]
- Paine, A.; Eiz-Vesper, B.; Blasczyk, R.; Immenschuh, S. Signaling to heme oxygenase-1 and its anti-inflammatory therapeutic potential. Biochem. Pharmacol. 2010, 80, 1895–1903. [Google Scholar] [CrossRef]
- Copple, I.M.; Goldring, C.E.; Kitteringham, N.R.; Park, B.K. The Nrf2-Keap1 defence pathway: Role in protection against drug-induced toxicity. Toxicology 2008, 246, 24–33. [Google Scholar] [CrossRef]
- Li, W.; Khor, T.O.; Xu, C.; Shen, G.; Jeong, W.S.; Yu, S.; Kong, A.N. Activation of Nrf2-antioxidant signaling attenuates NFkappaB-inflammatory response and elicits apoptosis. Biochem. Pharmacol. 2008, 76, 1485–1489. [Google Scholar] [CrossRef]
- Narumi, S.; Finke, J.H.; Hamilton, T.A. Interferon gamma and interleukin 2 synergize to induce selective monokine expression in murine peritoneal macrophages. J. Biol. Chem. 1990, 265, 7036–7041. [Google Scholar]
- Berridge, M.V.; Tan, A.S. Characterization of the cellular reduction of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT): Subcellular localization, substrate dependence, and involvement of mitochondrial electron transport in MTT reduction. Arch. Biochem. Biophys. 1993, 303, 474–482. [Google Scholar] [CrossRef]
- Titheradge, M.A. The Enzymatic Measurement of Nitrate and Nitrite. Meth. Mol. Biol. 1998, 101, 83–91. [Google Scholar]
- Sawle, P.; Foresti, R.; Mann, B.E.; Johnson, T.R.; Green, C.J.; Motterlini, R. Carbon monoxide-releasing molecules (CO-RMs) attenuate the inflammatory response elicited by lipopolysaccharide in RAW264.7 murine macrophages. Br. J. Pharmacol. 2005, 145, 800–810. [Google Scholar] [CrossRef]
- Sun, P.; Lu, S.; Ree, T.V.; Krohn, K.; Li, L.; Zhang, W. Nonanolides of natural origin: Structure, synthesis, and biological activity. Curr. Med. Chem. 2012, 19, 3417–3455. [Google Scholar] [CrossRef]
- Riatto, V.B.; Pilli, R.A.; Victor, M.M. Fifteen years of biological and synthetic studies of decarestrictine family. Tetrahedron 2008, 64, 2279–2300. [Google Scholar] [CrossRef]
- Evidente, A.; Capasso, R.; Andolfi, A.; Vurro, M.; Zonno, M.C. Putaminoxins D and E from Phoma putaminum. Phytochemistry 1998, 48, 941–945. [Google Scholar] [CrossRef]
- Li, Y.Y.; Wang, M.Z.; Huang, Y.J.; Shen, Y.M. Secondary metabolites from Phomopsis sp. A123. Mycology 2010, 1, 254–261. [Google Scholar] [CrossRef]
Supplementary Files
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Lee, D.-S.; Ko, W.; Quang, T.H.; Kim, K.-S.; Sohn, J.H.; Jang, J.-H.; Ahn, J.S.; Kim, Y.-C.; Oh, H. Penicillinolide A: A New Anti-Inflammatory Metabolite from the Marine Fungus Penicillium sp. SF-5292. Mar. Drugs 2013, 11, 4510-4526. https://doi.org/10.3390/md11114510
Lee D-S, Ko W, Quang TH, Kim K-S, Sohn JH, Jang J-H, Ahn JS, Kim Y-C, Oh H. Penicillinolide A: A New Anti-Inflammatory Metabolite from the Marine Fungus Penicillium sp. SF-5292. Marine Drugs. 2013; 11(11):4510-4526. https://doi.org/10.3390/md11114510
Chicago/Turabian StyleLee, Dong-Sung, Wonmin Ko, Tran Hong Quang, Kyoung-Su Kim, Jae Hak Sohn, Jae-Hyuk Jang, Jong Seog Ahn, Youn-Chul Kim, and Hyuncheol Oh. 2013. "Penicillinolide A: A New Anti-Inflammatory Metabolite from the Marine Fungus Penicillium sp. SF-5292" Marine Drugs 11, no. 11: 4510-4526. https://doi.org/10.3390/md11114510




