Improvement of Neutral Lipid and Polyunsaturated Fatty Acid Biosynthesis by Overexpressing a Type 2 Diacylglycerol Acyltransferase in Marine Diatom Phaeodactylum tricornutum
Abstract
:1. Introduction
2. Results
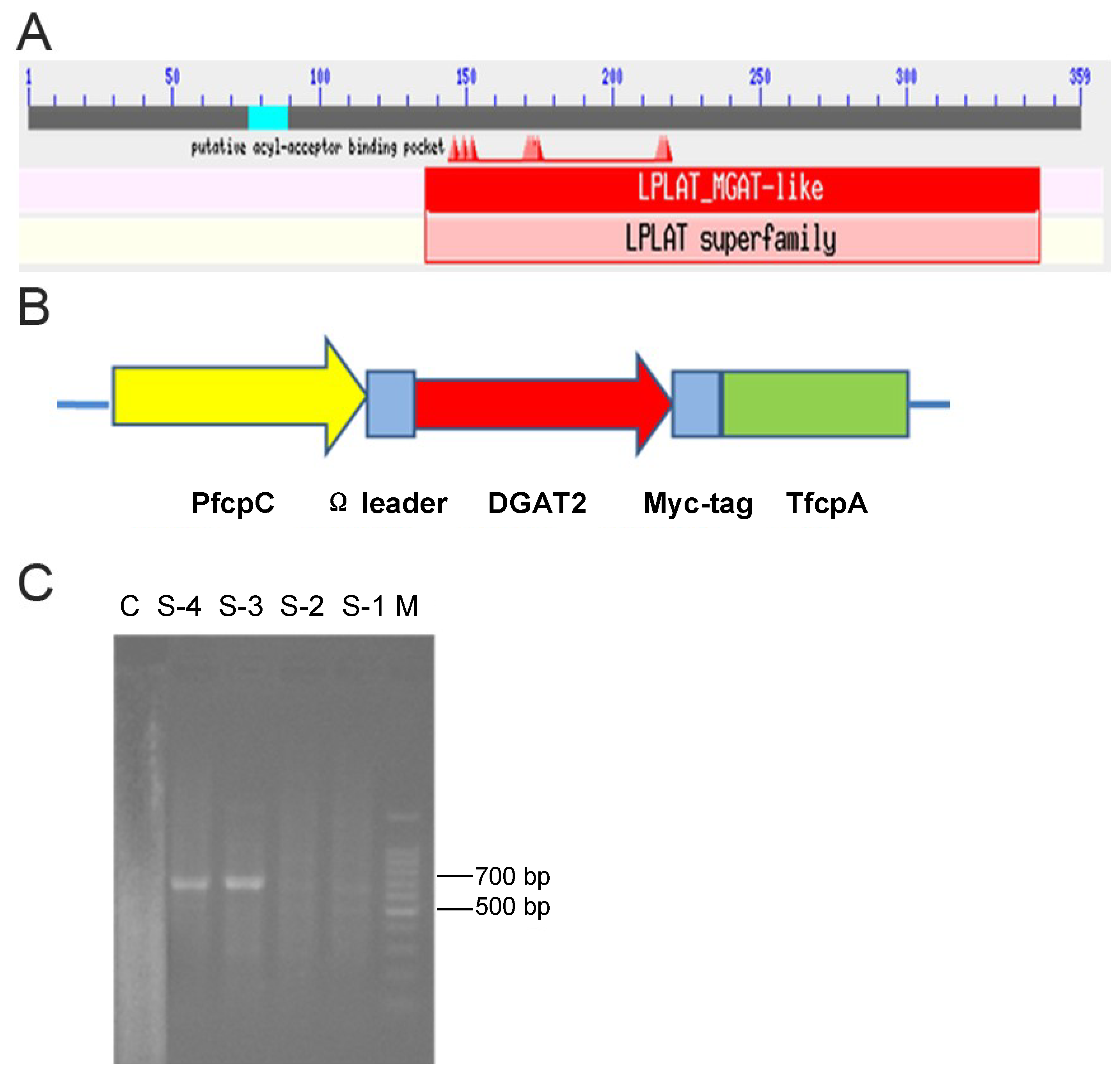
2.1. Sequence-Structure Analysis of DGAT2 Protein
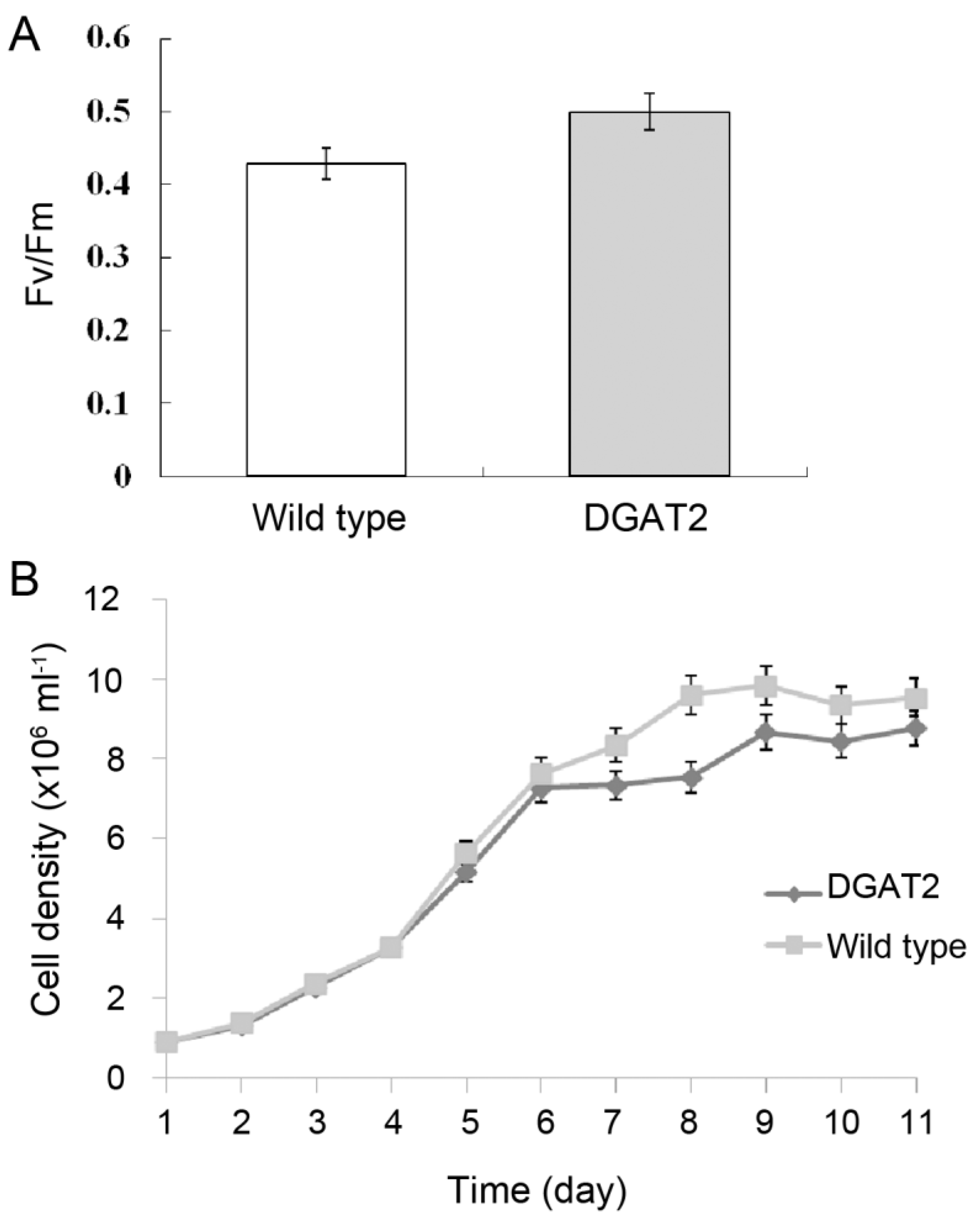
2.2. Photosynthesis Activity and Growth of Transgenic P. tricornutum
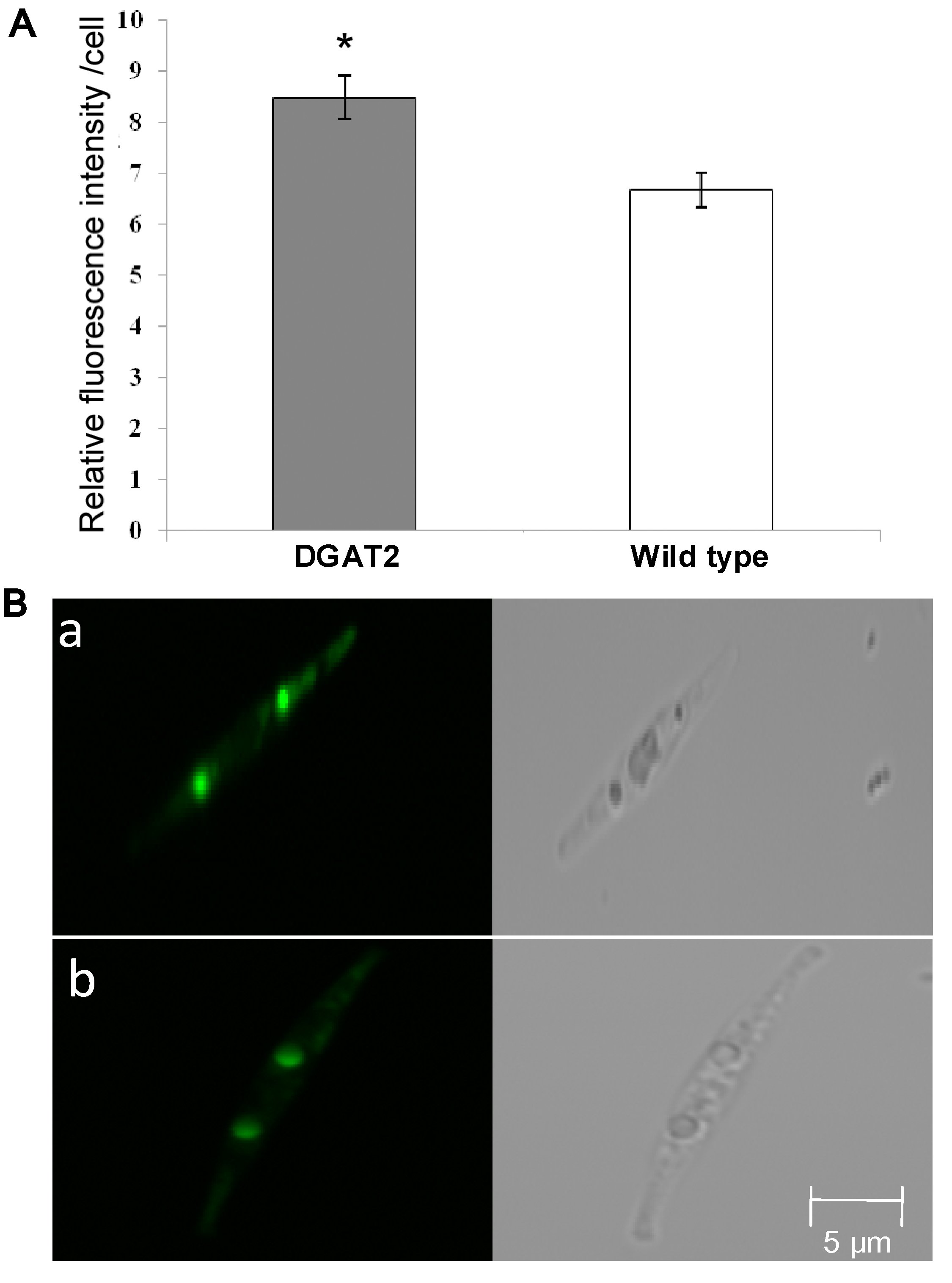
2.3. Analysis on Lipid Content and Fatty Acid Composition
| Fatty Acid | Wild Type | DGAT2 |
|---|---|---|
| C14:0 | 1.62 | 1.36 |
| C15:0 | 0.18 | 0.36 |
| C16:0 | 6.43 | 6.69 |
| C17:0 | 0.1 | 0.05 |
| C18:0 | 3.35 | 5.5 |
| C20:0 | 0.08 | 1.5 |
| C21:0 | 0.49 | 0.48 |
| C22:0 | 0.05 | 0.15 |
| C24:0 | 0.7 | 1.54 |
| SUM SFA | 13 | 17.63 |
| C16:1 | 6.11 | 9.31 |
| C18:1 | 1.03 | 0.29 |
| C22:1 | ND | 0.04 |
| C24:1 | ND | 0.04 |
| SUM MUFA | 7.14 | 9.68 |
| C16:3 | 2.26 | 1.87 |
| C18:2 | 0.78 | 0.61 |
| C18:3 | 0.08 | ND |
| C20:2 | 0.05 | 0.05 |
| C20:5 | 4.21 | 7.42 |
| C22:6 | ND | ND |
| SUM PUFA | 7.38 | 9.95 |
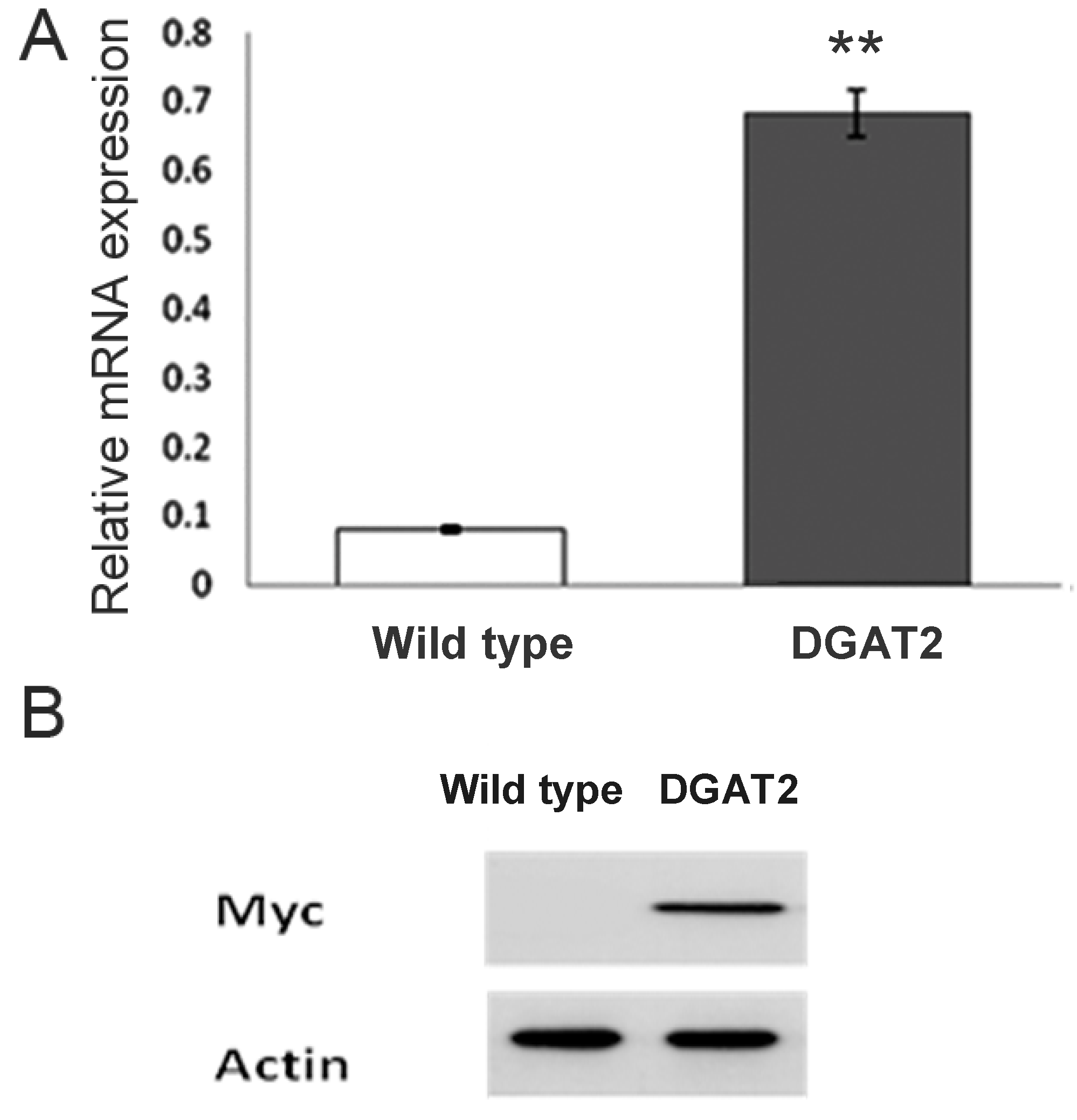
2.4. Expression of DGAT2 Determined by qPCR and Western Blot Analysis
3. Discussion
4. Experimental Section
4.1. Diatom Strain and Culture Conditions
4.2. Cloning of DGAT2 and Generation of Transgenic P. tricornutum by Electroporation
4.3. Selection and PCR Screening of Transgenic Microalgae
4.4. Measurement of Photosynthesis Activity
4.5. Lipid Analysis
4.6. Transcript Abundance of DGAT2 Analyzed by qPCR
4.7. DGAT2 Protein Expression Analyzed by Western Blot Analysis
5. Conclusions
Abbreviations
| CAT | chloramphenicol acetyltransferase |
| DGAT | diacylglycerol acyltranferase |
| Fcp | fucoxanthin chlorophyll a/c binding protein |
| NCBI | National Center for Biotechnology Information |
| PAGE | polyacrylamide gel electrophoresis |
| PUFA | polyunsaturated fatty acid |
| TAG | triacylglyceride |
Acknowledgements
Conflicts of Interest
References
- Kornberg, A.; Pricer, W.E., Jr. Enzymatic esterification of α-glycerophosphate by long chain fatty acids. J. Biol. Chem. 1953, 204, 329–343. [Google Scholar]
- Kennedy, E.P.; Weiss, S.B. The function of cytidine coenzymes in the biosynthesis of phospholipides. J. Biol. Chem. 1956, 222, 193–214. [Google Scholar]
- Lung, S.-C.; Weselake, R.J. Diacylglycerol acyltransferase: A key mediator of plant triacylglycerol synthesis. Lipids 2006, 41, 1073–1088. [Google Scholar] [CrossRef]
- Jako, C.; Kumar, A.; Wei, Y.; Zou, J.; Barton, D.; Giblin, E.; Covello, P.S.; Taylor, D. Seed-specific over-expression of an arabidopsis cDNA encoding a diacylglycerol acyltransferase enhances seed oil content and seed weight. Plant Physiol. 2001, 126, 861–874. [Google Scholar] [CrossRef]
- Oakes, J.; Brackenridge, D.; Colletti, R.; Daley, M.; Hawkins, D.J.; Xiong, H.; Mai, J.; Screen, S.E.; Val, D.; Lardizabal, K.; et al. Expression of fungal diacylglycerol acyltransferase2 genes to increase kernel oil in maize. Plant Physiol. 2011, 155, 1146–1157. [Google Scholar] [CrossRef]
- Xu, J.; Francis, T.; Mietkiewska, E.; Giblin, E.; Barton, D.; Zhang, Y.; Zhang, M.; Taylor, D. Cloning and characterization of an acyl-CoA-dependent diacylglycerol acyltransferase 1 (DGAT1) gene from Tropaeolum majus, and a study of the functional motifs of the DGAT protein using site-directed mutagenesis to modify enzyme activity and oil content. Plant Biotechnol. J. 2008, 6, 799–818. [Google Scholar] [CrossRef]
- Monetti, M.; Levin, M.C.; Watt, M.J.; Sajan, M.P.; Marmor, S.; Hubbard, B.K.; Stevens, R.D.; Bain, J.R.; Newgard, C.B.; Farese, R.V., Jr. Dissociation of hepatic steatosis and insulin resistance in mice overexpressing DGAT in the liver. Cell Metab. 2007, 6, 69–78. [Google Scholar] [CrossRef]
- Wagner, M.; Hoppe, K.; Czabany, T.; Heilmann, M.; Daum, G.; Feussner, I.; Fulda, M. Identification and characterization of an acyl-CoA:diacylglycerol acyltransferase 2 (DGAT2) gene from the microalga O. tauri. Plant Physiol. Biochem. 2010, 48, 407–416. [Google Scholar] [CrossRef]
- La Russa, M.; Bogen, C.; Uhmeyer, A.; Doebbe, A.; Filippone, E.; Kruse, O.; Mussgnug, J.H. Functional analysis of three type-2 DGAT homologue genes for triacylglycerol production in the green microalga Chlamydomonas reinhardtii. J. Biotechnol. 2012, 162, 13–20. [Google Scholar] [CrossRef]
- Nguyen, H.M.; Baudet, M.; Cuiné, S.; Adriano, J.-M.; Barthe, D.; Billon, E.; Bruley, C.; Beisson, F.; Peltier, G.; Ferro, M.; et al. Proteomic profiling of oil bodies isolated from the unicellular green microalga Chlamydomonas reinhardtii: With focus on proteins involved in lipid metabolism. Proteomics 2011, 11, 4266–4273. [Google Scholar] [CrossRef]
- Chen, J.E.; Smith, A.G. A look at diacylglycerol acyltransferases (DGATs) in algae. J. Biotechnol. 2012, 162, 28–39. [Google Scholar] [CrossRef]
- Xu, J.; Kazachkov, M.; Jia, Y.; Zheng, Z.; Zou, J. Expression of a type 2 diacylglycerol acyltransferase from Thalassiosira pseudonana in yeast leads to incorporation of docosahexaenoic acid β-oxidation intermediates into triacylglycerol. FEBS J. 2013, in press. [Google Scholar]
- Guihéneuf, F.; Leu, S.; Zarka, A.; Khozin-Goldberg, I.; Khalilov, I.; Boussiba, S. Cloning and molecular characterization of a novel acyl-CoA: Diacylglycerol acyltransferase 1-like gene (PtDGAT1) from the diatom Phaeodactylum tricornutum. FEBS J. 2011, 278, 3651–3666. [Google Scholar] [CrossRef]
- Gong, Y.; Zhang, J.; Guo, X.; Wan, X.; Liang, Z.; Hu, C.J.; Jiang, M. Identification and characterization of PtDGAT2B, an acyltransferase of the DGAT2 acyl-Coenzyme A: Diacylglycerol acyltransferase family in the diatom Phaeodactylum tricornutum. FEBS Lett. 2013, 587, 481–487. [Google Scholar] [CrossRef]
- Williams, N. New biofuel questions. Curr. Biol. 2010, 20, R219–R220. [Google Scholar] [CrossRef]
- Radakovits, R.; Jinkerson, R.E.; Darzins, A.; Posewitz, M.C. Genetic engineering of algae for enhanced biofuel production. Eukaryot. Cell 2010, 9, 486–501. [Google Scholar] [CrossRef]
- Andrianov, V.; Borisjuk, N.; Pogrebnyak, N.; Brinker, A.; Dixon, J.; Spitsin, S.; Flynn, J.; Matyszczuk, P.; Andryszak, K.; Laurelli, M.; et al. Tobacco as a production platform for biofuel: Overexpression of Arabidopsis DGAT and LEC2 genes increases accumulation and shifts the composition of lipids in green biomass. Plant Biotechnol. J. 2010, 8, 277–287. [Google Scholar] [CrossRef]
- Park, J.; Rho, H.K.; Kim, K.H.; Choe, S.S.; Lee, Y.S.; Kim, J.B. Overexpression of glucose-6-phosphate dehydrogenase is associated with lipid dysregulation and insulin resistance in obesity. Mol. Cell Biol. 2005, 25, 5146–5157. [Google Scholar] [CrossRef]
- Radakovits, R.; Eduafo, P.M.; Posewitz, M.C. Genetic engineering of fatty acid chain length in Phaeodactylum tricornutum. Metab. Eng. 2011, 13, 89–95. [Google Scholar] [CrossRef]
- Moellering, E.R.; Benning, C. RNA Interference Silencing of a Major Lipid Droplet Protein Affects Lipid Droplet Size in Chlamydomonas reinhardtii. Eukaryot. Cell 2010, 9, 97–106. [Google Scholar] [CrossRef]
- Armbrust, E.V.; Berges, J.A.; Bowler, C.; Green, B.R.; Martinez, D.; Putnam, N.H.; Zhou, S.; Allen, A.E.; Apt, K.E.; Bechner, M. The genome of the diatom Thalassiosira pseudonana: Ecology, evolution, and metabolism. Science 2004, 306, 79–86. [Google Scholar] [CrossRef]
- Siaut, M.; Heijde, M.; Mangogna, M.; Montsant, A.; Coesel, S.; Allen, A.; Manfredonia, A.; Falciatore, A.; Bowler, C. Molecular toolbox for studying diatom biology in Phaeodactylum tricornutum. Gene 2007, 406, 23–35. [Google Scholar] [CrossRef]
- Niu, Y.F.; Yang, Z.K.; Zhang, M.H.; Zhu, C.C.; Yang, W.D.; Liu, J.S.; Li, H.Y. Transformation of diatom Phaeodactylum tricornutum by electroporation and establishment of inducible selection marker. Biotechniques 2012. [Google Scholar] [CrossRef]
- Macedo, R.S.; Lombardi, A.T.; Omachi, C.Y.; Rorig, L.R. Effects of herbicide bentazon on growth and photosystem II maximum quantum yield of the marine diatom Skeletonema costatum. Toxicol. in Vitro 2008, 22, 716–722. [Google Scholar] [CrossRef]
- Yang, Z.; Niu, Y.; Ma, Y.; Xue, J.; Zhang, M.; Yang, W.; Liu, J.; Lu, S.; Guan, Y.; Li, H. Molecular and cellular mechanisms of neutral lipid accumulation in diatom following nitrogen deprivation. Biotechnol. Biofuels 2013, 6, 67. [Google Scholar] [CrossRef]
- Chomczynski, P.; Sacchi, N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987, 162, 156–159. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Niu, Y.-F.; Zhang, M.-H.; Li, D.-W.; Yang, W.-D.; Liu, J.-S.; Bai, W.-B.; Li, H.-Y. Improvement of Neutral Lipid and Polyunsaturated Fatty Acid Biosynthesis by Overexpressing a Type 2 Diacylglycerol Acyltransferase in Marine Diatom Phaeodactylum tricornutum. Mar. Drugs 2013, 11, 4558-4569. https://doi.org/10.3390/md11114558
Niu Y-F, Zhang M-H, Li D-W, Yang W-D, Liu J-S, Bai W-B, Li H-Y. Improvement of Neutral Lipid and Polyunsaturated Fatty Acid Biosynthesis by Overexpressing a Type 2 Diacylglycerol Acyltransferase in Marine Diatom Phaeodactylum tricornutum. Marine Drugs. 2013; 11(11):4558-4569. https://doi.org/10.3390/md11114558
Chicago/Turabian StyleNiu, Ying-Fang, Meng-Han Zhang, Da-Wei Li, Wei-Dong Yang, Jie-Sheng Liu, Wei-Bin Bai, and Hong-Ye Li. 2013. "Improvement of Neutral Lipid and Polyunsaturated Fatty Acid Biosynthesis by Overexpressing a Type 2 Diacylglycerol Acyltransferase in Marine Diatom Phaeodactylum tricornutum" Marine Drugs 11, no. 11: 4558-4569. https://doi.org/10.3390/md11114558




