New Azalomycin F Analogs from Mangrove Streptomyces sp. 211726 with Activity against Microbes and Cancer Cells
Abstract
:1. Introduction
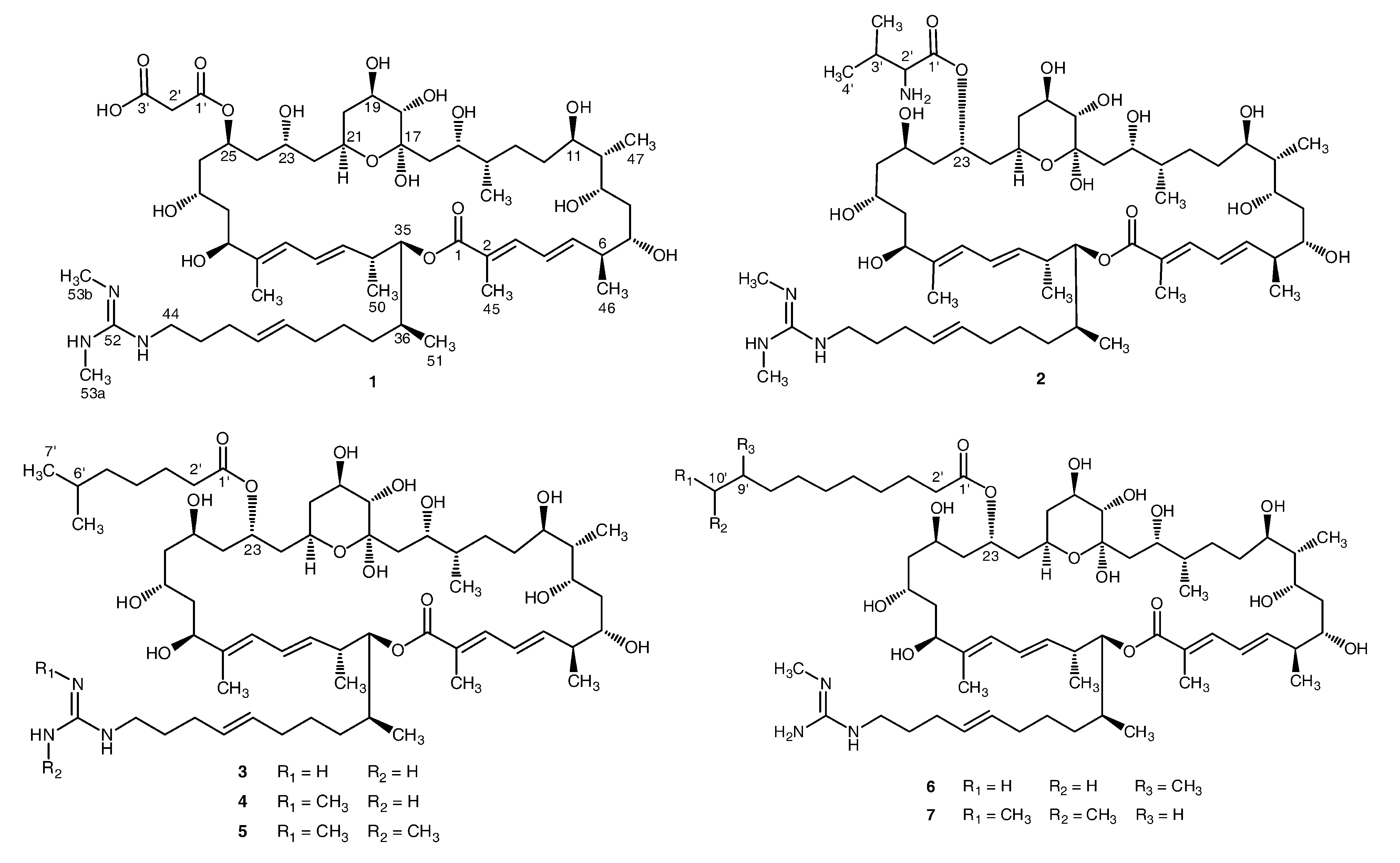
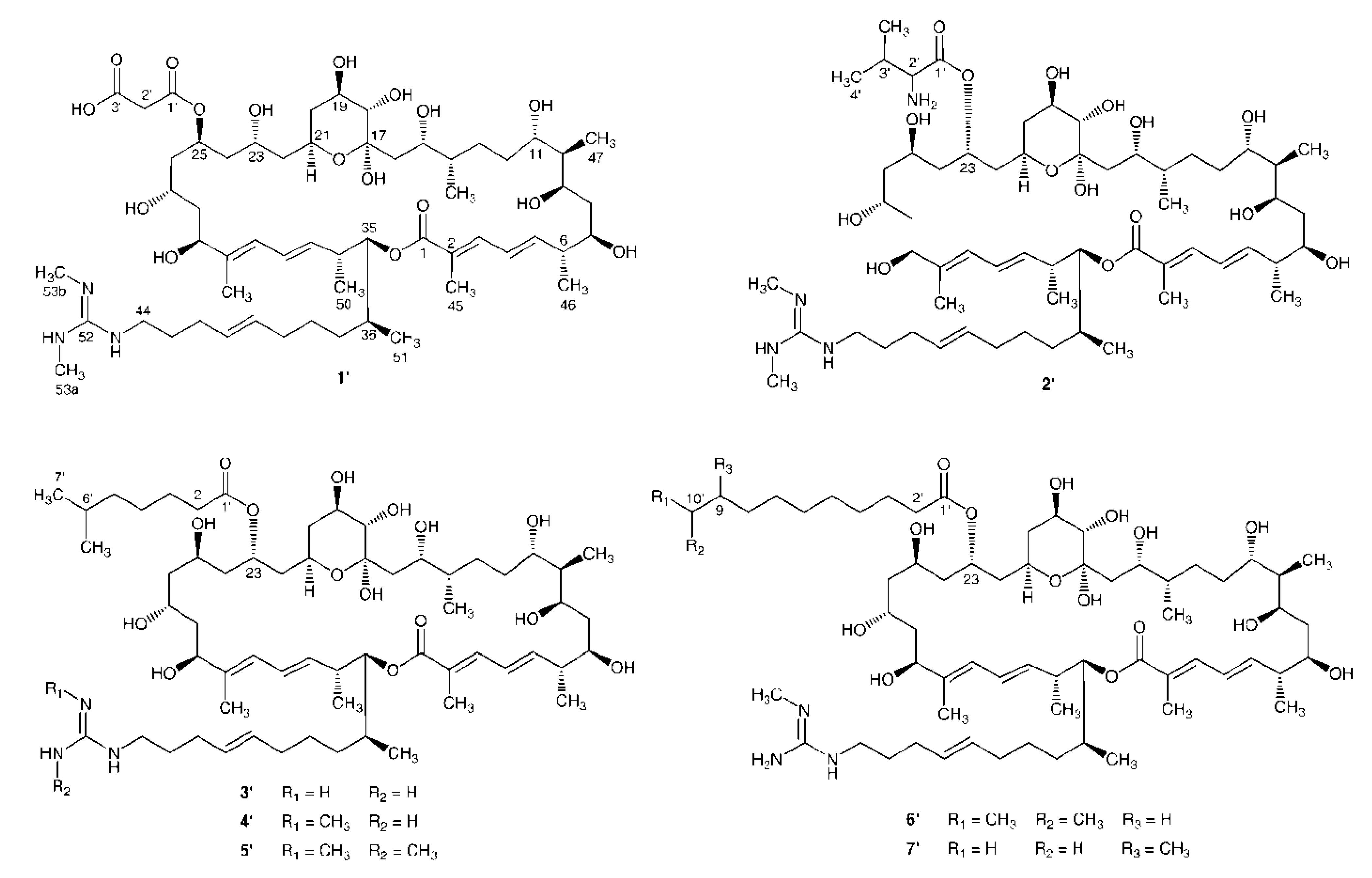
2. Results and Discussion
2.1. Structural Elucidation
| Position | 1 | 2 | 3 | 6 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| δC | δH (J in Hz) | δC | δH (J in Hz) | δC | δH (J in Hz) | δC | δH (J in Hz) | ||||
| C-1 | 170.2 | - | 170.1 | - | 170.1 | - | 170.1 | - | |||
| C-2 | 126.8 | - | 126.7 | - | 126.8 | - | 126.8 | - | |||
| C-3 | 140.3 | 7.09 d (11.2) | 140.3 | 7.10 d (11.0) | 140.2 | 7.10 d (11.2) | 140.3 | 7.10 d (11.0) | |||
| C-4 | 127.6 | 6.43 dd (11.5, 14.9) | 127.6 | 6.43 dd (11.5, 14.7) | 127.6 | 6.43 dd (11.9, 14.3) | 127.6 | 6.43 dd (11.5, 14.8) | |||
| C-5 | 146.2 | 6.07 dd (15.1, 9.0) | 146.1 | 6.08 dd (14.8, 9.0) | 146.1 | 6.08 dd (14.0, 9.0) | 146.1 | 6.08 dd (14.9, 9.0) | |||
| C-6 | 44.8 | 2.43 m | 44.7 | 2.44 m | 44.6 | 2.43 m | 44.7 | 2.44 m | |||
| C-7 | 75.9 | 3.80 m | 75.8 | 3.77 m | 75.8 | 3.77 m | 75.8 | 3.77 m | |||
| C-8 | 39.3 | 1.50 m, 1.78 m | 39.3 | 1.50 m, 1.78 m | 39.3 | 1.50 m, 1.77 m | 39.3 | 1.50 m, 1.78 m | |||
| C-9 | 75.4 | 3.80 m | 75.2 | 3.80 m | 75.2 | 3.80 m | 75.2 | 3.80 m | |||
| C-10 | 44.7 | 1.54 m | 44.6 | 1.53 m | 44.5 | 1.54 m | 44.6 | 1.53 m | |||
| C-11 | 72.2 | 3.91 m | 72.2 | 3.87 m | 72.2 | 3.91 m | 72.2 | 3.87 m | |||
| C-12 | 33.5 | 1.62 m, 1.38 m | 33.5 | 1.60 m, 1.38 m | 33.5 | 1.62 m, 1.37 m | 33.4 | 1.62 m, 1.37 m | |||
| C-13 | 30.7 | 1.30 m, 1.45 m | 30.6 | 1.30 m, 1.42 m | 30.6 | 1.30 m, 1.43 m | 30.6 | 1.30 m, 1.43 m | |||
| C-14 | 40.6 | 1.60 m | 40.5 | 1.61 m | 40.7 | 1.61 m | 40.5 | 1.61 m | |||
| C-15 | 72.7 | 3.86 m | 72.7 | 3.87 m | 72.4 | 3.86 m | 72.7 | 3.87 m | |||
| C-16 | 41.9 | 1.80 m | 41.8 | 1.81 m | 41.9 | 1.82 m | 41.8 | 1.81 m | |||
| C-17 | 99.9 | - | 100.0 | - | 99.8 | - | 99.9 | - | |||
| C-18 | 77.5 | 3.34 d (9.2) | 77.4 | 3.35 d (9.1) | 77.2 | 3.35 d (9.2) | 77.5 | 3.35 d (9.1) | |||
| C-19 | 69.9 | 3.87 m | 69.9 | 3.88 m | 69.7 | 3.87 m | 69.8 | 3.88 m | |||
| C-20 | 41.4 | 1.89 m, 1.30 m | 41.3 | 1.89 m, 1.30 m | 41.2 | 1.90 m, 1.31 m | 41.3 | 1.89 m, 1.31 m | |||
| C-21 | 65.7 | 4.17 m | 66.2 | 4.16 m | 66.3 | 4.16 m | 66.3 | 4.16 m | |||
| C-22 | 44.5 | 1.52 m | 41.9 | 1.82 m | 41.8 | 1.88 m | 41.9 | 1.81 m | |||
| C-23 | 66.3 | 4.03 m | 70.9 | 5.29 m | 70.7 | 5.27 m | 70.9 | 5.29 m | |||
| C-24 | 44.6 | 1.69 m | 44.0 | 1.70 m | 44.0 | 1.72 m | 44.1 | 1.76 m, 1.66 m | |||
| C-25 | 70.8 | 5.28 m | 65.7 | 3.86 m | 65.6 | 3.87 m | 65.7 | 3.86 m | |||
| C-26 | 44.0 | 1.61 m, 1.83 m | 46.3 | 1.51 m | 46.4 | 1.51 m | 46.3 | 1.51 m | |||
| C-27 | 65.7 | 3.88 m | 65.8 | 4.04 m | 65.6 | 4.04 m | 65.8 | 4.04 m | |||
| C-28 | 44.2 | 1.78 m | 44.1 | 1.54 m | 44.1 | 1.53 m | 44.1 | 1.63 m | |||
| C-29 | 74.2 | 4.18 m | 74.2 | 4.18 m | 74.2 | 4.18 m | 74.2 | 4.18 m | |||
| C-30 | 140.2 | - | 140.1 | - | 140.1 | - | 140.1 | - | |||
| C-31 | 125.3 | 5.98 d (10.4) | 125.3 | 5.98 d (10.7) | 125.2 | 5.98 d (10.5) | 125.3 | 5.98 d (10.7) | |||
| C-32 | 128.6 | 6.22 dd (10.9, 14.5) | 128.5 | 6.23 dd (10.9, 14.9) | 128.6 | 6.22 dd (10.9, 14.9) | 128.5 | 6.23 dd (10.9, 14.8) | |||
| C-33 | 136.3 | 5.43 m | 136.3 | 5.44 m | 136.2 | 5.45 m | 136.3 | 5.44 m | |||
| C-34 | 41.0 | 2.57 m | 40.7 | 2.57 m | 41.0 | 2.57 m | 40.9 | 2.57 m | |||
| C-35 | 80.9 | 4.78 dd (7.6, 4.0) | 80.9 | 4.79 dd (7.7, 4.1) | 80.8 | 4.78 dd (7.8, 3.9) | 80.9 | 4.79 dd (7.6, 4.1) | |||
| C-36 | 35.3 | 1.82 m | 35.3 | 1.82 m | 35.2 | 1.81 m | 35.3 | 1.82 m | |||
| C-37 | 34.4 | 1.15 m, 1.35 m | 34.4 | 1.15 m, 1.35 m | 34.5 | 1.15 m, 1.35 m | 34.4 | 1.15 m, 1.35 m | |||
| C-38 | 27.9 | 1.42 m | 27.8 | 1.42 m | 27.9 | 1.41 m | 27.9 | 1.42 m | |||
| C-39 | 33.6 | 1.99 m | 33.6 | 1.99 m | 33.6 | 1.99 m | 33.6 | 1.99 m | |||
| C-40 | 132.6 | 5.44 m | 132.5 | 5.44 m | 132.6 | 5.44 m | 132.6 | 5.44 m | |||
| C-41 | 130.3 | 5.44 m | 130.3 | 5.44 m | 130.1 | 5.50 m | 130.2 | 5.44 m | |||
| C-42 | 30.7 | 2.07 m | 30.4 | 2.07 m | 30.8 | 2.07 m | 30.6 | 2.07 m | |||
| C-43 | 29.9 | 1.67 m | 29.8 | 1.64 m | 29.8 | 1.64 m | 29.8 | 1.64 m | |||
| C-44 | 42.2 | 3.17 t (7.3) | 42.0 | 3.15 t (7.1) | 42.0 | 3.15 t (7.0) | 42.0 | 3.15 t (7.1) | |||
| C-45 | 12.9 | 1.92 s | 12.9 | 1.92 s | 12.9 | 1.92 s | 12.9 | 1.92 s | |||
| C-46 | 17.1 | 1.11 d (6.8) | 17.1 | 1.12 d (6.8) | 17.1 | 1.11 d (6.8) | 17.1 | 1.12 d (6.8) | |||
| C-47 | 10.5 | 0.89 d (6.9) | 10.5 | 0.89 d (6.9) | 10.5 | 0.89 d (6.9) | 10.5 | 0.89 d (6.9) | |||
| C-48 | 15.2 | 0.91 d (6.7) | 15.3 | 0.92 d (6.7) | 15.3 | 0.91 d (6.7) | 15.3 | 0.92 d (6.7) | |||
| C-49 | 13.1 | 1.65 s | 13.1 | 1.65 s | 13.3 | 1.65 s | 13.1 | 1.65 s | |||
| C-50 | 17.8 | 1.01 d (6.7) | 17.9 | 1.00 d (6.7) | 17.9 | 1.00 d (6.6) | 17.9 | 1.00 d (6.8) | |||
| C-51 | 14.4 | 0.94 d (6.7) | 14.5 | 0.95 d (6.7) | 14.4 | 0.94 d (6.7) | 14.5 | 0.94 d (6.7) | |||
| C-52 | 157.4 | - | 157.4 | - | 158.7 | - | 158.3 | - | |||
| C-53a | 28.4 | 2.85 s | 28.4 | 2.85 s | 28.4 | 2.84 s | |||||
| C-53b | 28.4 | 2.85 s | 28.4 | 2.85 s | |||||||
| C-1′ | 171.9 | - | 174.1 | - | 175.5 | - | 175.4 | - | |||
| C-2′ | 46.0 | 3.22 m | 61.7 | 3.44 d (4.0) | 35.0 | 2.36 t (7.4) | 35.0 | 2.36 t (7.5) | |||
| C-3′ | 173.9 | - | 30.8 | 2.28 m | 26.0 | 1.62 m | 26.0 | 1.61 m | |||
| 3′-CH3 | 17.9 | 1.02 d (6.8) | |||||||||
| C-4′ | 19.2 | 1.07 d (6.8) | 30.4 | 1.42 m | 30.3 | 1.35 m | |||||
| C-5′ | 40.3 | 1.18 m | 30.4 | 1.31 m | |||||||
| C-6′ | 29.2 | 1.29 m | 30.8 | 1.30 m | |||||||
| 6′-CH3 | 23.8 | 0.88 d (6.6) | |||||||||
| C-7′ | 23.7 | 0.88 d (6.6) | 28.5 | 1.29 m | |||||||
| C-8′ | 40.3 | 1.17 m | |||||||||
| C-9′ | 29.2 | 1.31 m | |||||||||
| 9′-CH3 | 23.1 | 0.89 d (6.8) | |||||||||
| C-10′ | 23.1 | 0.89 d (6.8) | |||||||||
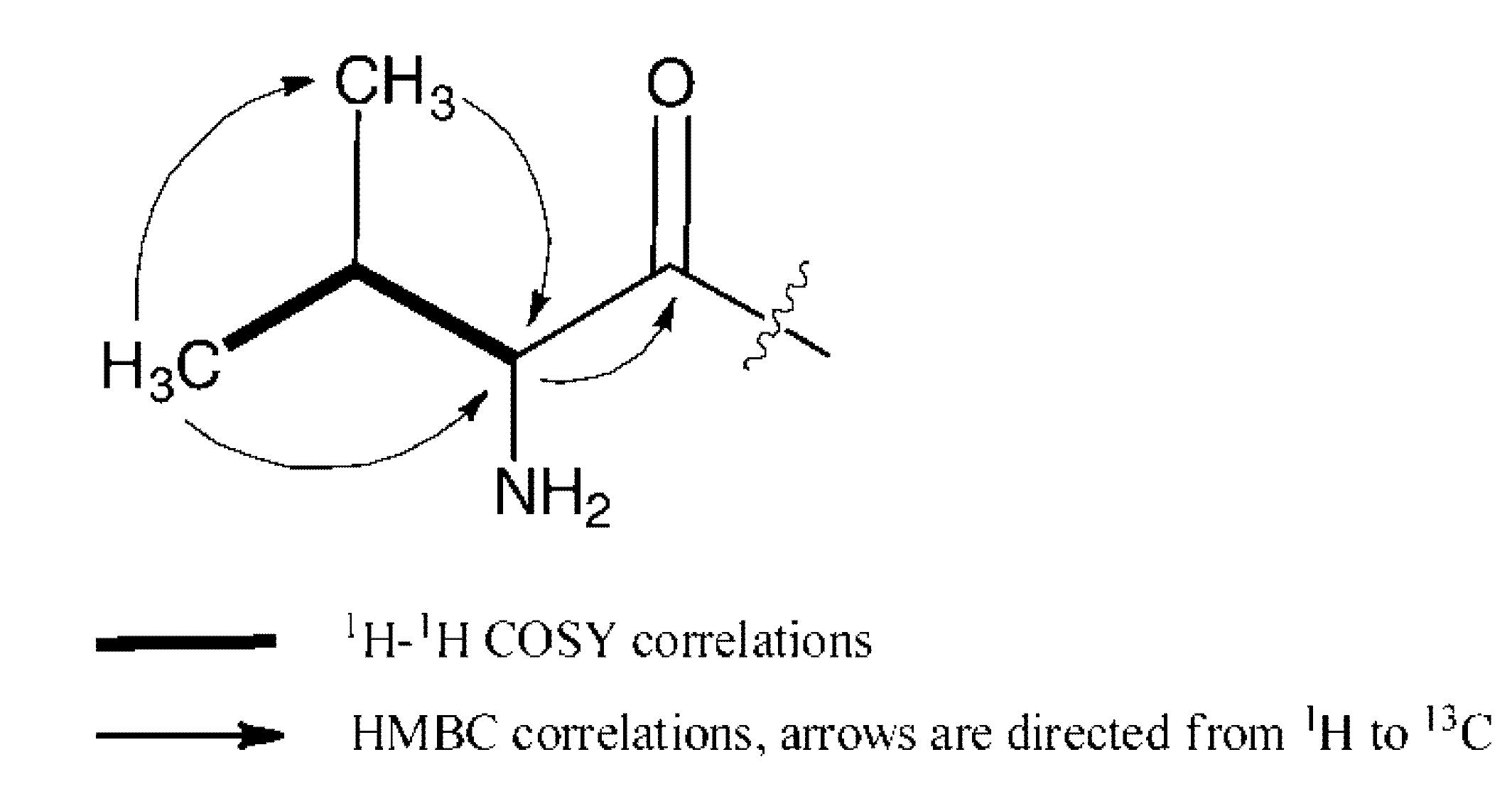
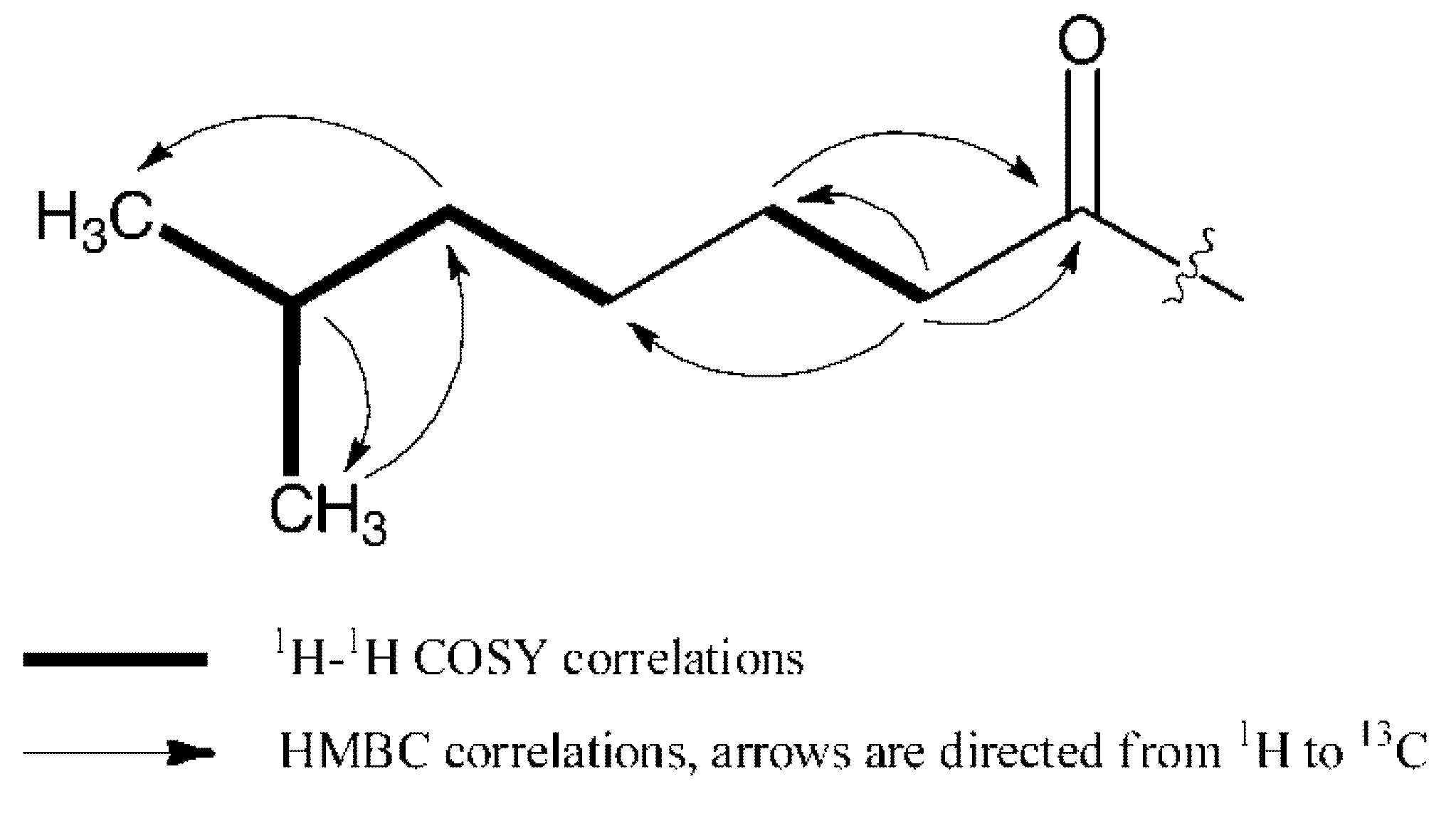
2.2. Biological Assays
| Compounds | MICs (μg/mL) | IC50 (μg/mL) | |||
|---|---|---|---|---|---|
| Candida albicans ATCC 10231 | Staphylococcus aureus S014 | Bacillus subtilis S028 | Eschzerichia coli S002 | HCT-116 | |
| 1 | 3.13 | 0.39 | 0.20 | 3.13 | 5.00 |
| 2 | 6.25 | 1.56 | 0.39 | 6.25 | 1.95 |
| 3 | 3.13 | 0.78 | 0.39 | 3.13 | 2.46 |
| 4 | 1.56 | 1.56 | 0.20 | 6.25 | 2.45 |
| 5 | 1.56 | 0.78 | 0.78 | 12.5 | 1.81 |
| 6 | 3.13 | 0.39 | 0.39 | 25.00 | 1.54 |
| 7 | 3.13 | 0.39 | 0.39 | 3.13 | 2.46 |
| Positive controls * | 2.0 | 0.50 | 0.20 | 2.0 | 0.18 |
2.3. Discussion
3. Experimental Section
3.1. General Experimental Procedures
3.2. Actinomycetes Material and Fermentation
3.3. Extract and Isolation
3.4. Biological Assays
4. Conclusions
Acknowledgments
References and Notes
- Costanza, R.; d’Arge, R.; Groot, R.; Farberk, S.; Grasso, M.; Hannon, B.; Limburg, K.; Naeem, S.; O’Neill, R.V.; Paruelo, J.; et al. The value of the world’s ecosystem services and natural capital. Nature 1997, 387, 253–260. [Google Scholar]
- Wang, B.; Liang, S.; Zhang, W.; Zan, Q. Mangrove flora of the world. Acta. Bot. Sin. 2003, 45, 644–653. [Google Scholar]
- Li, J.; Li, M.Y.; Feng, G.; Zhang, J.; Karonen, M.; Sinkkonen, J.; Satyanandamurty, T.; Wu, J.; Moluccensins, R.-Y. Limonoids from the seeds of a mangrove, Xylocarpus moluccensis. J. Nat. Prod. 2012, 75, 1277–1283. [Google Scholar] [CrossRef]
- Wu, J.; Xiao, Q.; Xu, J.; Li, M.Y.; Pan, J.Y.; Yang, M.H. Natural products from true mangrove flora: Source, chemistry and bioactivities. Nat. Prod. Rep. 2008, 25, 955–981. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, W.; Li, Z.; Xiang, W.; Guo, Y.; Krohn, K. Elucidation of excogallochaols A-D, four unusual diterpenoids from the Chinese mangrove Excoecaria agallocha. Phytochemistry 2007, 68, 2426–2431. [Google Scholar] [CrossRef]
- Xu, M.; Liu, X.; Zhao, Y.; Liu, D.; Liu, D.; Xu, Z.; Ao, P.; Lin, W.; Yang, S.; Zhang, Z.; Xu, J. Identification and characterization of an anti-fibrotic benzopyran compound isolated from mangrove-derived Streptomyces xiamenensis. Mar. Drugs 2012, 10, 639–654. [Google Scholar]
- Hong, K.; Gao, A.; Xie, Q.; Gao, H.; Zhuang, L.; Lin, H.; Yu, H.; Li, J.; Yao, X.; Goodfellow, M.; Ruan, J. Actinomycetes for marine drug discovery isolated from mangrove soils and plants in China. Mar. Drugs 2009, 7, 24–44. [Google Scholar] [CrossRef]
- Fu, P.; Yang, C.; Wang, Y.; Liu, P.; Ma, Y.; Xu, L.; Su, M.; Hong, K.; Zhu, W. Streptocarbazoles A and B, two novel 2 indolocarbazoles from the marine-derived actinomycete strain Streptomyces sp. FMA. Org. Lett. 2012, 14, 2422–2425. [Google Scholar]
- Chen, G.D.; Gao, H.; Tang, J.S.; Huang, Y.F.; Chen, Y.; Wang, Y.; Zhao, H.N.; Lin, H.P.; Xie, Q.Y.; Hong, K.; et al. Benzamides and quinazolines from a mangrove actinomycetes Streptomyces sp. (No. 061316) and their inhibiting caspase-3 catalytic activity in vitro. Chem. Pharm. Bull. 2011, 59, 447–451. [Google Scholar]
- Lin, W.; Li, L.; Fu, H.; Sattler, I.; Huang, X.; Grabley, S. New cyclopentenone derivatives from an endophytic Streptomyces sp. isolated from the mangrove plant Aegiceras comiculatum. J. Antibiot. 2005, 58, 594–598. [Google Scholar] [CrossRef]
- Yuan, G.; Lin, H.; Wang, C.; Hong, K.; Liu, Y.; Li, J. 1H and 13C assignments of two new macrocyclic lactones isolated from Streptomyces sp. 211726 and revised assignments of azalomycins F3a, F4a and F5a. Magn. Reson. Chem. 2011, 49, 30–37. [Google Scholar]
- Yuan, G.; Li, P.; Pan, W.; Pang, H.; Chen, S. The relative configurations of azalomycins F5a, F4a and F3a. J. Mol. Struct. 2013, 1035, 31–37. [Google Scholar] [CrossRef]
- .
- Iwasaki, S.; Namikoshi, M.; Sasaki, K.; Fukushima, K.; Okuda, S. Studies on marcrocyclic lactone antibiotics. V. (1) The structures of azalomycins F3a and F5a. Chem. Pharm. Bull. 1982, 30, 4006–4014. [Google Scholar]
- 13C NMR (MeOH-d4, 100 MHz) data of azalomycin F3a: 170.1 (C-1), 126.8 (C-2), 140.3 (C-3), 127.6 (C-4), 146.2 (C-5), 44.6 (C-6), 75.7 (C-7), 39.3 (C-8), 75.0 (C-9), 44.5 (C-10), 72.2 (C-11), 33.6 (C-12), 30.7 (C-13), 40.5 (C-14), 72.3 (C-15), 41.7 (C-16), 99.9 (C-17), 77.2 (C-18), 69.8 (C-19), 41.2 (C-20), 65.4 (C-21), 41.9 (C-22), 70.8 (C-23), 44.6 (C-24), 65.5 (C-25), 46.2 (C-26), 66.2 (C-27), 44.1 (C-28), 73.9 (C-29), 140.1 (C-30), 125.2 (C-31), 128.5 (C-32), 136.2 (C-33), 40.7 (C-34), 81.2 (C-35), 35.2 (C-36), 34.6 (C-37), 27.9 (C-38), 33.6 (C-39), 132.6 (C-40), 130.3 (C-41), 30.7 (C-42), 29.9 (C-43), 42.1 (C-44), 12.8 (C-45), 17.0 (C-46), 10.5 (C-47), 14.8 (C-48), 13.3 (C-49), 17.7 (C-50), 14.4 (C-51), 158.7 (C-52), 171.8 (C-1′), 45.8 (C-2′), 174.0 (C-3′).
- 13C NMR (MeOH-d4, 100 MHz) data of azalomycin F4a: 170.1 (C-1), 126.8 (C-2), 140.2 (C-3), 127.6 (C-4), 146.1 (C-5), 44.6 (C-6), 75.7 (C-7), 39.2 (C-8), 75.0 (C-9), 44.5 (C-10), 72.2 (C-11), 33.6 (C-12), 30.8 (C-13), 40.5 (C-14), 72.3 (C-15), 41.7 (C-16), 99.8 (C-17), 77.1 (C-18), 69.7 (C-19), 41.2 (C-20), 65.4 (C-21), 42.0 (C-22), 70.7 (C-23), 44.6 (C-24), 65.5 (C-25), 46.5 (C-26), 66.1 (C-27), 44.1 (C-28), 74.2 (C-29), 140.1 (C-30), 125.2 (C-31), 128.6 (C-32), 136.2 (C-33), 41.0 (C-34), 80.7 (C-35), 35.1 (C-36), 34.6 (C-37), 27.9 (C-38), 33.6 (C-39), 132.5 (C-40), 130.3 (C-41), 30.8 (C-42), 29.8 (C-43), 42.1 (C-44), 12.9 (C-45), 17.1 (C-46), 10.5 (C-47), 14.8 (C-48), 13.3 (C-49), 17.6 (C-50), 14.3 (C-51), 158.3 (C-52), 28.4 (C-53a), 171.6 (C-1′), 46.1 (C-2′), 173.9 (C-3′).
- Kobayashi, Y.; Tan, C.H.; Kishi, Y. Toward creation of a universal NMR database for stereochemical assignment: The case of 1,3,5-trisubstituted acyclic systems. Helv. Chim. Acta 2000, 83, 2562–2571. [Google Scholar] [CrossRef]
- Arai, M. Azalomycin F, an antibiotic against fungi and Trichomonas. Arzneim. Forsch. 1968, 18, 1396–1399. [Google Scholar]
- Arai, M. Azalomycins B and F, two new antibiotics. II. Properties of azalomycins B and F. J. Antibiot. Ser. A 1960, 13, 51–56. [Google Scholar]
- Namikoshi, M.; Iwasaki, S.; Sasaki, K.; Yano, M.; Fukushima, K.; Nozoe, S.; Okuda, S. Studies on macrocyclic lactone antibiotics. II. (1) Partial structures of azalomycin F4a. Chem. Pharm. Bull. 1982, 30, 1658–1668. [Google Scholar]
- Iwasaki, S.; Namikoshi, M.; Sasaki, K.; Yano, M.; Fukushima, K.; Nozoe, S.; Okuda, S. Studies on macrocyclic lactone antibiotics. III. (1) Skeletal structure of azalomycin F4a. Chem. Pharm. Bull. 1982, 30, 1669–1673. [Google Scholar]
- Chandra, A.; Nair, M.G. Azalomycin F complex from Streptomyces hygroscopicus, MSU/MN-4-75B. J. Antibiot. 1995, 48, 896–898. [Google Scholar] [CrossRef]
- Berlinck, R.G.S. Natural guanidine derivatives. Nat. Prod. Rep. 1999, 16, 339–365. [Google Scholar] [CrossRef]
- Ubukata, M.; Morita, T.I.; Osada, H. RS-22A, B and C: New macrolide antibiotics from Streptomyces violaceusniger II. Physico-Chemical properties and structure elucidation. J. Antibiot. 1995, 48, 293–299. [Google Scholar]
- Ivanova, V.; Gesheva, V.; Kolarova, M. Dihydroniphimycin: New polyol macrolide antibiotic produced by Streptomyces hygroscopicus 15 isolation and structure elucidation. J. Antibiot. 2000, 53, 627–632. [Google Scholar] [CrossRef]
- Ivanova, V.; Kolarova, M.; Aleksieva, K. Malonyl-4,5-dihydroniphimycin: A new polyol macrolide antibiotic, produced by Streptomyces hygroscopicus. Z. Naturforsch. 2007, 62, 1187–1192. [Google Scholar]
- Stefanelli, S.; Corti, E.; Montanini, N.; Denaro1, M.; Sarubbill, E. Inhibitors of type-I interleukin-1 receptor from microbial metabolites. J. Antibiot. 1997, 50, 484–489. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Yuan, G.; Hong, K.; Lin, H.; She, Z.; Li, J. New Azalomycin F Analogs from Mangrove Streptomyces sp. 211726 with Activity against Microbes and Cancer Cells. Mar. Drugs 2013, 11, 817-829. https://doi.org/10.3390/md11030817
Yuan G, Hong K, Lin H, She Z, Li J. New Azalomycin F Analogs from Mangrove Streptomyces sp. 211726 with Activity against Microbes and Cancer Cells. Marine Drugs. 2013; 11(3):817-829. https://doi.org/10.3390/md11030817
Chicago/Turabian StyleYuan, Ganjun, Kui Hong, Haipeng Lin, Zhigang She, and Jia Li. 2013. "New Azalomycin F Analogs from Mangrove Streptomyces sp. 211726 with Activity against Microbes and Cancer Cells" Marine Drugs 11, no. 3: 817-829. https://doi.org/10.3390/md11030817




