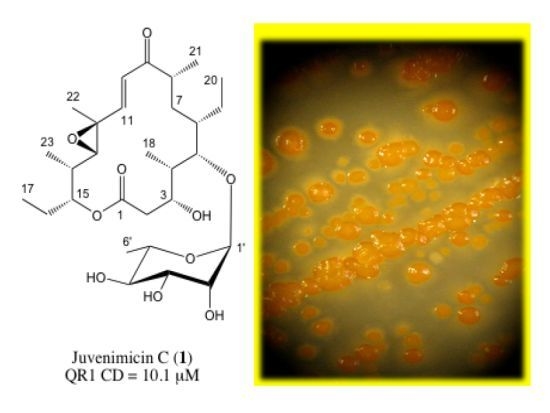
Potential Chemopreventive Activity of a New Macrolide Antibiotic from a Marine-Derived Micromonospora sp.
Abstract
:1. Introduction
2. Results and Discussion
2.1. Structure Elucidation
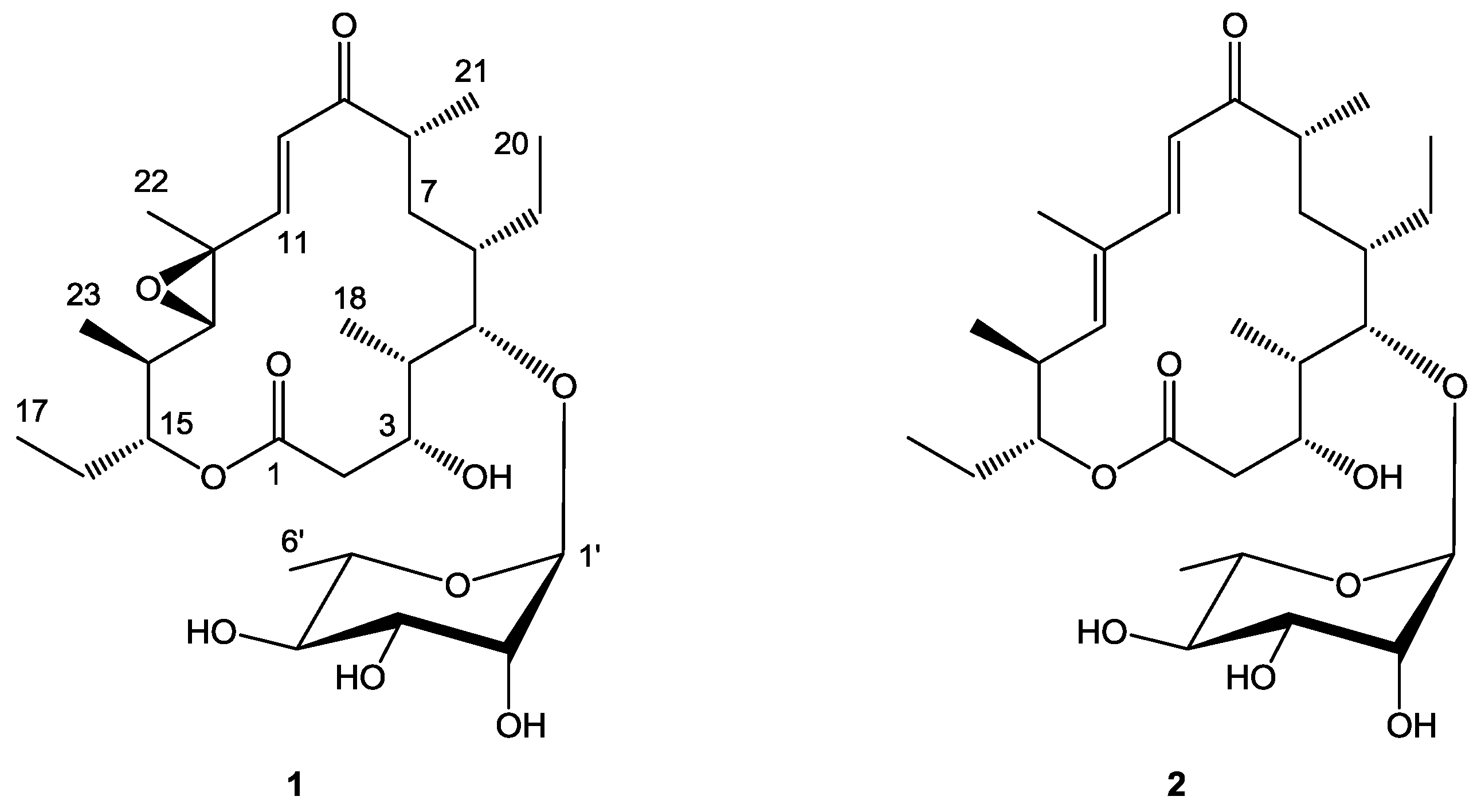
| 1 | ||
|---|---|---|
| Pos | 13C | 1H mult. ( J, Hz) |
| 1 | 174.8 | |
| 2 | 40.4 | 2.59 dd (9.0, 10.2) |
| 2.19 d (9.0) | ||
| 3 Eq | 67.4 | 3.69 d (10.2) |
| 4 | 41.6 | 1.75 m a |
| 5 | 84.7 | 3.58 d (9.6) |
| 6 | 39.6 | 1.14 m |
| 7 | 33.3 | 1.76 m |
| 1.38 m | ||
| 8 | 46.0 | 2.62 m |
| 9 | 202.4 | |
| 10 | 124.8 | 6.65 d (15.9) |
| 11 | 150.5 | 6.34 d (15.9) |
| 12 | 60.7 | |
| 13 | 68.8 | 2.80 d (9.6) |
| 14 | 38.4 | 1.75 m a |
| 15 | 77.7 | 4.83 dt (10.0, 2.4) |
| 16 | 25.1 | 1.80 m |
| 1.49 m | ||
| 17 | 9.3 | 0.88 d (7.2) |
| 18 | 10.1 | 0.95 d (6.6) |
| 19 | 22.5 | 1.50 m |
| 1.36 m | ||
| 20 | 12.5 | 0.87 d (7.2) |
| 21 | 17.5 | 1.16 d (6.6) |
| 22 | 15.3 | 1.43 s |
| 23 | 14.5 | 1.08 d (6.6) |
| 1′ | 104.0 | 4.62 br s |
| 2′ | 72.0 | 3.89 br s |
| 3′ | 72.3 | 3.47 d (7.2) |
| 4′ | 73.3 | 3.29 m |
| 5′ | 69.8 | 3.63 m |
| 6′ | 17.5 | 1.18 d (6.0) |
| 3-OH | 3.22 br s | |
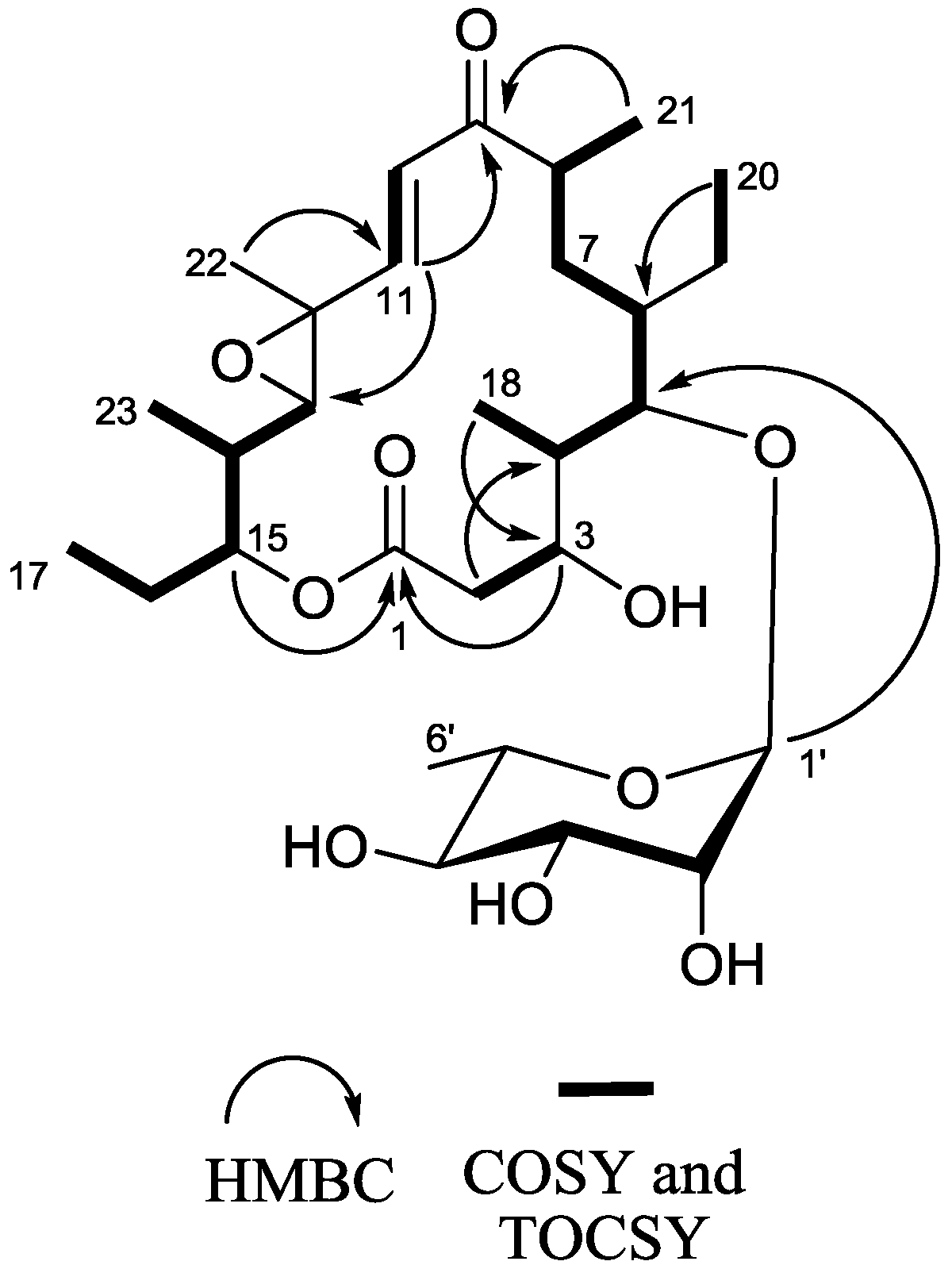
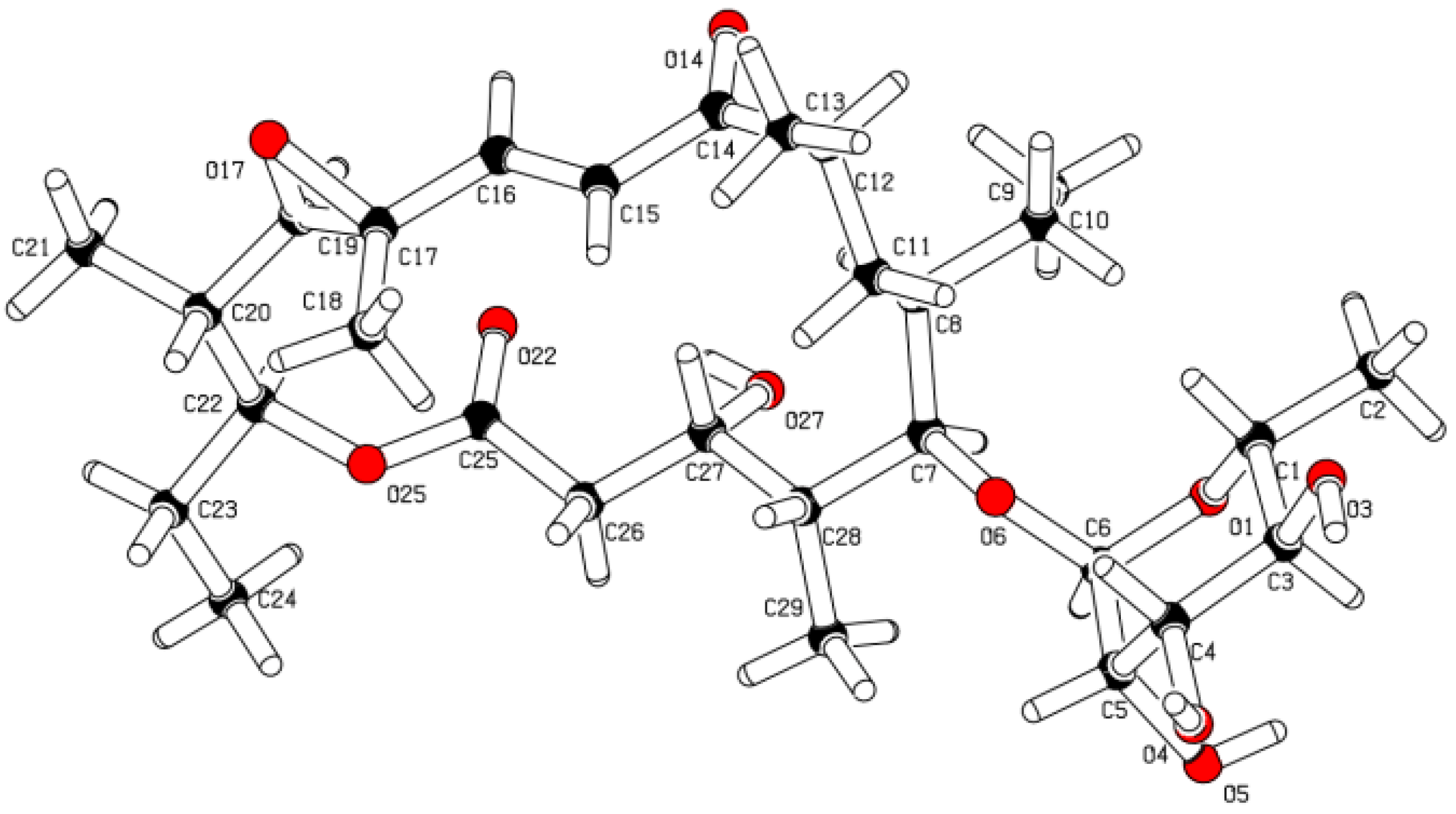
2.2. Chemopreventive Activity of 1 and 2
| Sample | QR1 IR | QR1 CD (μM) | Glutathione CD (μM) | Glutathione reductase * | Glutathione peroxidase * |
|---|---|---|---|---|---|
| Control | 1.0 | na | na | 9.6 | 63.7 |
| 1 | 4.3 | 10.1 | 27.7 | 12.2 | 138 |
| 2 | 1.4 | nd | nd | nd | nd |
| 4′-BF | 8.4 | 0.1 | 5.67 | 18.1 | 154 |
3. Experimental Section
3.1. General Experimental Procedures
3.2. Bacterial Isolation and Identification
3.3. Fermentation and Extraction
3.4. Isolation and Characterization of Juvenimicin C (1) and Isolation of 5-O-α-l-Rhamnosyltylactone (2)
3.5. Quinone Reductase 1 (QR1) Assay
3.6. Determination of GSH Levels in Cell Culture
3.7. Determination of Glutathione Reductase Activity
3.8. Determination of Glutathione Peroxidase Activity
4. Conclusions
Acknowledgements
References
- Talalay, P.; Fahey, J.W.; Holtzclaw, W.D.; Prestera, T.; Zhang, Y. Chemoprotection against cancer by phase 2 enzyme induction. Toxicol. Lett. 1995, 82–83, 173–179. [Google Scholar] [CrossRef]
- Dinkova-Kostova, A.T.; Talalay, P. Persuasive evidence that quinone reductase type 1 (DT diaphorase) protects cells against the toxicity of electrophiles and reactive forms of oxygen. Free Radic. Biol. Med. 2000, 29, 231–240. [Google Scholar] [CrossRef]
- Shen, L.; Park, E.-J.; Kondratyuk, T.P.; Guendisch, D.; Marler, L.; Pezzuto, J.M.; Wright, A.D.; Sun, D. Design, synthesis, and biological evaluation of callophycin A and analogues as potential chemopreventive and anticancer agents. Bioorg. Med. Chem. 2011, 19, 6182–6195. [Google Scholar] [CrossRef]
- Li, Y.; Zhao, M.; Parkin, K.L. β-Carboline derivatives and diphenols from soy sauce are in vitro quinone reductase (QR) inducers. J. Agric. Food Chem. 2011, 59, 2332–2340. [Google Scholar] [CrossRef]
- Mayhoub, A.S.; Marler, L.; Kondratyuk, T.P.; Park, E.-J.; Pezzuto, J.M.; Cushman, M. Optimization of thiazole analogues of resveratrol for induction of NAD(P)H: quinone reductase 1 (QR1). Bioorg. Med. Chem. 2012, 20, 7030–7039. [Google Scholar] [CrossRef]
- Song, L.L.; Kosmeder, J.W., II; Lee, S.K.; Gerhauser, C.; Lantvit, D.; Moon, R.C.; Moriarty, R.M.; Pezzuto, J.M. Cancer chemopreventive activity mediated by 4′-bromoflavone, a potent inducer of phase II detoxification enzymes. Cancer Res. 1999, 59, 578–585. [Google Scholar]
- Cheng, L.; Shen, L.-M.; Zhang, M.; Li, N.; Li, X.; Ma, Z.-J.; Qu, H.-B. Eleven new triterpenes from Eurycorymbus cavaleriei. Helv. Chim. Acta 2010, 93, 2263–2275. [Google Scholar] [CrossRef]
- Fahey, J.W.; Stephenson, K.K. Pinostrobin from honey and Thai ginger (Boesenbergia pandurata): A potent flavonoid inducer of mammalian phase 2 chemoprotective and antioxidant enzymes. J. Agric. Food Chem. 2002, 50, 7472–7476. [Google Scholar] [CrossRef]
- Wollenweber, E.; Stevens, J.F.; Klimo, K.; Knauft, J.; Frank, N.; Gerhauser, C. Cancer chemopreventive in vitro activities of isoflavones isolated from Iris germanica. Planta Med. 2003, 69, 15–20. [Google Scholar] [CrossRef]
- Carcache-Blanco, E.J.; Kang, Y.-H.; Park, E.J.; Su, B.-N.; Kardono, L.B.S.; Riswan, S.; Fong, H.H.S.; Pezzuto, J.M.; Kinghorn, A.D. Constituents of the stem bark of Pongamia pinnata with the potential to induce quinone reductase. J. Nat. Prod. 2003, 66, 1197–1202. [Google Scholar] [CrossRef]
- Yin, H.-Q.; Lee, B.-W.; Kim, Y.-C.; Sohn, D.-H.; Lee, B.-H. Induction of the anticarcinogenic marker enzyme, quinone reductase, by Dalbergiae Lignum. Arch. Pharmacal Res. 2004, 27, 919–922. [Google Scholar] [CrossRef]
- Yuan, Y.; Ji, L.; Luo, L.; Lu, J.; Ma, X.; Ma, Z.; Chen, Z. Quinone reductase (QR) inducers from Andrographis paniculata and identification of molecular target of andrographolide. Fitoterapia 2012, 83, 1506–1513. [Google Scholar] [CrossRef]
- Li, F.; Wang, Y.; Parkin, K.L.; Nitteranon, V.; Liang, J.; Yang, W.; Li, Y.; Zhang, G.; Hu, Q. Isolation of quinone reductase (QR) inducing agents from ginger rhizome and their in vitro anti-inflammatory activity. Food Res. Int. 2011, 44, 1597–1603. [Google Scholar] [CrossRef]
- Schell, U.; Haydock, S.F.; Kaja, A.L.; Carletti, I.; Lill, R.E.; Read, E.; Sheehan, L.S.; Low, L.; Fernandez, M.J.; Grolle, F.; et al. Engineered biosynthesis of hybrid macrolide polyketides containing d-angolosamine and d-mycaminose moieties. Org. Biomol. Chem. 2008, 6, 3315–3327. [Google Scholar] [CrossRef]
- Marler, L.; Conda-Sheridan, M.; Cinelli, M.A.; Morrell, A.E.; Cushman, M.; Chen, L.; Huang, K.; van Breemen, R.; Pezzuto, J.M. Cancer chemopreventive potential of aromathecins and phenazines, novel natural product derivatives. Anticancer Res. 2010, 30, 4873–4882. [Google Scholar]
- Zhang, L.; Xi, L.; Ruan, J.; Huang, Y. Micromonospora yangpuensis sp. nov., isolated from a sponge. Int. J. Syst. Evol. Microbiol. 2012, 62, 272–278. [Google Scholar] [CrossRef]
- Kim, O.S.; Cho, Y.J.; Lee, K.; Yoon, S.H.; Kim, M.; Na, H.; Park, S.C.; Jeon, Y.S.; Lee, J.H.; Yi, H.; et al. Introducing EzTaxon-e: A prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 2012, 62, 716–721. [Google Scholar] [CrossRef]
- Magarvey, N.A.; Keller, J.M.; Bernan, V.; Dworkin, M.; Sherman, D.H. Isolation and characterization of novel marine-derived actinomycete taxa rich in bioactive metabolites. Appl. Environ. Microbiol. 2004, 70, 7520–7529. [Google Scholar] [CrossRef]
- Zhang, Y.; Talalay, P.; Cho, C.G.; Posner, G.H. A major inducer of anticarcinogenic protective enzymes from broccoli: Isolation and elucidation of structure. Proc. Natl. Acad. Sci. USA 1992, 89, 2399–2403. [Google Scholar] [CrossRef]
- Prochaska, H.J.; Santamaria, A.B. Direct measurement of NAD(P)H:quinone reductase from cells cultured in microtiter wells: A screening assay for anticarcinogenic enzyme inducers. Anal. Biochem. 1988, 169, 328–336. [Google Scholar] [CrossRef]
- Gerhäuser, C.; You, M.; Liu, J.; Moriarty, R.M.; Hawthorne, M.; Mehta, R.G.; Moon, R.C.; Pezzuto, J.M. Cancer chemopreventive potential of sulforamate, a novel analogue of sulforaphane that induces phase 2 drug-metabolizing enzymes. Cancer Res. 1997, 57, 272–278. [Google Scholar]
- Carlson, G.P.; Turner, M.; Mantick, N.A. Effects of styrene and styrene oxide on glutathione-related antioxidant enzymes. Toxicology 2006, 227, 217–226. [Google Scholar] [CrossRef]
- Sample Availability: Available from the authors.
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Carlson, S.; Marler, L.; Nam, S.-J.; Santarsiero, B.D.; Pezzuto, J.M.; Murphy, B.T. Potential Chemopreventive Activity of a New Macrolide Antibiotic from a Marine-Derived Micromonospora sp. Mar. Drugs 2013, 11, 1152-1161. https://doi.org/10.3390/md11041152
Carlson S, Marler L, Nam S-J, Santarsiero BD, Pezzuto JM, Murphy BT. Potential Chemopreventive Activity of a New Macrolide Antibiotic from a Marine-Derived Micromonospora sp. Marine Drugs. 2013; 11(4):1152-1161. https://doi.org/10.3390/md11041152
Chicago/Turabian StyleCarlson, Skylar, Laura Marler, Sang-Jip Nam, Bernard D. Santarsiero, John M. Pezzuto, and Brian T. Murphy. 2013. "Potential Chemopreventive Activity of a New Macrolide Antibiotic from a Marine-Derived Micromonospora sp." Marine Drugs 11, no. 4: 1152-1161. https://doi.org/10.3390/md11041152